Abstract
Polyethylenimine (PEI) is a cationic polymer that has shown significant potential for delivering genes in vitro and in vivo. Mixing cationic PEI with negatively charged plasmid DNA (pDNA) results in the spontaneous electrostatic formation of stable nanoparticle complexes. The structure of PEI can be branched or linear. In this study, we show that branched PEI has a stronger electrostatic interaction with pDNA than linear PEI, which accounts for greater compaction, higher zeta potentials and smaller nanoparticle sizes at equivalent pDNA concentrations. For both linear and branched PEI, increasing the concentration of pDNA mixed in the same volume and at the same nitrogen to phosphate (N:P) ratio results in larger average particle sizes. Increasing the N:P ratio increases luciferase activity generated by branched PEI-pDNA nanoparticles and linear PEI-pDNA nanoparticles in HEK293, COS7 and HeLa cell lines. Increasing the N:P ratio at which branched PEI-pDNA nanoparticles are prepared also increases luciferase expression in HepG2 cells but does not increase luciferase expression generated by linear PEI-pDNA nanoparticles. In all of the cell lines, branched PEI-pDNA nanoparticles prepared at N:P ratios of 10 and above generated significantly higher luciferase activity than linear PEI-pDNA nanoparticles. Luciferase activity was highest in the HEK293 cells and luciferase expression in each of the cell lines followed the order of HEK293<COS7<HepG2<HeLa. Intraperitoneal (IP) injection of PEI-pDNA nanoparticles is attractive because it is simple, reproducible and often leads to a depot effect of nanoparticle complexes residing in the peritoneum. The IP route of administration avoids PEI-pDNA nanoparticle accumulation in the lung and the nanoparticles do not pass through the blood-brain barrier. In this study, using bioluminescent imaging (BLI), we show that changing the PEI structure and dose of the PEI-pDNA nanoparticles has a significant impact on the strength and duration of transgene expression after IP injection in vivo but increasing the N:P ratio does not. Increasing the dose and N:P ratio for all the PEI-pDNA nanoparticle formulations injected IP did not reduce mice survival and all mice remained in good health as determined by the Body Condition Scoring (BCS) technique.
Keywords: Non-Viral Gene Delivery, Polyethylenimine (PEI), Branched, Linear, Intraperitoneal (IP), Plasmid DNA
1. Introduction
Gene therapy aims to treat diseases by delivering genes or DNA to target cells or tissues to produce therapeutics proteins or suppress defective genes.[1–3] Successful application of gene therapy is dependent on optimization of the delivery carrier.[2–6] Gene therapy can be achieved using viral or non-viral vectors.[7, 8] Viral vectors generate significantly higher transgene expression than non-viral vectors.[7, 8] Viral vectors, however, also have several limitations. These include the requirement of cell mitosis for most retroviruses, immunogenicity of adenoviruses, safety concerns with HIV-like viruses and packaging constraints of adeno-associated viruses (AAV).[4] Promising results with some viral vectors such as AAV have progressed into human clinical trials, but the long-term safety concerns of viral vectors still exist and therapeutic levels of expression remain transient in the clinic.[9, 10] In contrast, non-viral vectors display substantially reduced immunogenicity. Advantages to non-viral vectors include ease of scale-up, storage stability, and improved quality control.[4]
One of the most efficient non-viral vectors in vitro and in vivo is the cationic polymer, polyethylenimine (PEI).[11–25] PEI is a water-soluble polymer in which the repeat unit of PEI is two carbon atoms followed by a nitrogen atom. Under physiological conditions, approximately 20% of the nitrogens are protonated.[18, 26, 27] The positive charge of the PEI results in effective binding to the negatively charged plasmid DNA (pDNA), and this condensation protects the pDNA from digestion in serum and as the nanoparticle complex enters cells.[28, 29] Once in the endosomal compartment, PEI can act as a buffer or “proton sponge” to induce osmotic swelling and cause release from the endosome. This is necessary to avoid degradation of the plasmid DNA when the endosome fuses with the lysosome.[29]
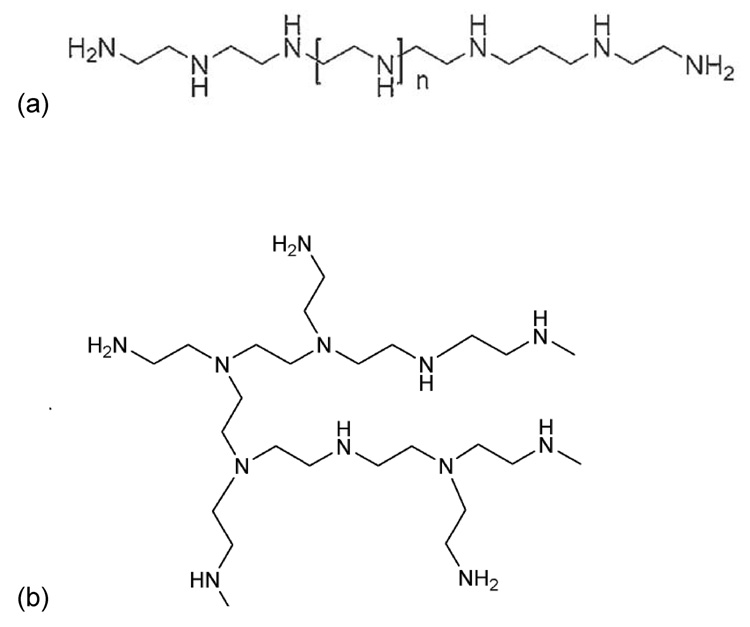
PEI comes in two structural forms: linear and branched (figure 1).[13, 30, 31] Several studies have shown that linear PEI generates stronger transgene expression in vivo when injected intravenously or administered via the lung in comparison to branched PEI at equivalent molecular weights.[24, 31–35] In contrast, other studies have shown that branched PEI generates stronger transgene expression than linear PEI in vitro in selected cell lines.[18] Some studies have shown that linear PEI is more efficient than branched PEI in vitro and in vivo.[31, 34] No study, to date, has compared linear and branched PEI after intraperitoneal (IP) injection in vivo or compared these results to transgene expression in vitro.
Figure 1.
Chemical structure of (a) linear and (b) branched polyethylenimine
Linear PEI has shown strong potential for gene and siRNA delivery when injected intraperitoneally (IP).[36] Intraperitoneal injection of PEI is attractive because organs in the peritoneal cavity are normally covered with peritoneum and subperitoneal connective tissue whereas tumor nodules on the peritoneal surfaces are not.[37] The peritoneum lining and underlying fibrous components significantly reduce the entry of nanoparticles through the surface of non-targeted organs. Intraperitoneal injections have been shown to be simple, reproducible and often lead to a depot effect of nanoparticle complex residing in the peritoneum for several hours.[37, 38] In addition, a number of studies have shown that polymers of similar chemistry but different structure induce changes in transgene activity that can readily be correlated between IP and intravenous (IV) injections.[6] The IP route of administration is considered promising for treatment of a variety of diseases including cancer. Ovarian, pancreatic, and gastric cancers are particularly attractive targets for IP injections.[6, 37, 39–46]
In this study, for the first time, we comprehensively compare the transgene expression generated by IP injection of branched or linear PEI-pDNA nanoparticles using bioluminescent imaging (BLI). BLI is a modern transgene expression imaging technique from which the results strongly correlate to transgene expression results measured using traditional tissue and organ harvesting methodologies.[47] We also compare the transgene expression generated by linear and branched PEI-pDNA nanoparticles in a series of cell lines that are commonly used as transfection hosts for evaluating non-viral vector efficiency. Finally, we compare the transgene expression generated by branched or linear PEI-pDNA nanoparticles in vitro and after IP injection in vivo. The results from this study highlight the differences in transgene expression generated by PEI-pDNA nanoparticles in vitro and in vivo and provide a robust time-dependent profile of transgene expression generated by linear or branched PEI-pDNA nanoparticles after IP injection in a murine model.
2. Materials and Methods
2.1 Materials
Branched polyethylenimine (PEI, MW 25kDa) was purchased from Sigma-Aldrich (St. Louis, MO). Linear polyethylenimine (PEI, MW 25kDa) was obtained from Polysciences Inc. (Warrington, PA). Luciferin was obtained from Xenogen (Hopkinton, MA).
2.2 Cell culture
Human Embryonic Kidney cells (HEK293), Monkey African Green Kidney cells (COS7), Human Liver Hepatocellular Carcinoma cells (HepG2) and Human Cervix Adenocarcinoma cells (HeLa) were purchased from American Type Culture Collection (ATCC, Rockville, MD). The cells were maintained in DMEM supplemented with 10% Fetal Bovine Serum (FBS), streptomycin at 100µg/ml, penicillin at 100U/ml, and 4mM L-glutamine at 37°C in a humidified 5% CO2-containing atmosphere.
2.3 Amplification and purification of pDNA
The firefly luciferase gene was used as a reporter gene to monitor gene expression. The pDNA (VR1255) is a 6.4-kb cDNA encoding firefly luciferase driven by the cytomegalovirus (CMV) promoter/enhancer. The pDNA was transformed in Escherichia coli DH5α and amplified in Terrific Broth media at 37°C overnight on a plate shaker set at 300rpm. The pDNA was then purified by an endotoxin-free QIAGEN Giga plasmid purification kit (QIAGEN, Valencia, CA) according to the manufacturer’s protocol. Purified pDNA was dissolved in Tris-EDTA solution, and its purity and concentration were determined by UV absorbance at 260 and 280nm.
2.4 Preparation of PEI-pDNA nanoparticles
Branched or linear PEI-pDNA nanoparticles were formulated in 5% glucose solution at various ratios of PEI nitrogen to pDNA phosphate (N/P ratio). Different amounts of pDNA were diluted and branched or linear PEI was then transferred into the pDNA solution. The solution was vortex mixed for 20 seconds and left at room temperature for 30 minutes.
2.5 Particle size and zetapotential analysis
Nanoparticle size measurements were conducted using the Zetasizer Nano ZS (Malvern, Southborough, MA). Briefly, the nanoparticles were suspended in deionized water at a concentration of 1 mg/ml. The size measurements were performed at 25°C at a 173° scattering angle. The mean hydrodynamic diameter was determined by cumulative analysis. The zeta potential determinations were based on electrophoretic mobility of the nanoparticles in the aqueous medium, which were performed using folded capillary cells in automatic mode.
2.6 Evaluation of luciferase expression in COS7, HEK293, HeLa and HepG2 cells
Cells were seeded into a 24-well plate at a density of 8×104/well of COS7, HeLa, HepG2 and HEK293 cells 24 hours before transfection. One hundred µl of PEI-pDNA nanoparticle solution containing 1 µg of pDNA was added to the cells in transfection medium (serum-free) and incubated for 4 hours at 37 °C, followed by further incubation in serum containing medium for 44 hours. After 44 hours incubation, cells were treated with 200 µl of lysis buffer (Promega, Madison, WI). The lysate was subjected to two cycles of freezing and thawing, then transferred into tubes and centrifuged at 13,200 rpm for 5 minutes. Twenty µl of supernatant was added to 100 µl of luciferase assay reagent (Promega, Madison, WI) and samples were measured on a luminometer for 10 seconds (Lumat LB 9507, EG&G Berthold, Bad Wildbad, Germany). The relative light units (RLU) were normalized against protein concentration in the cell extracts, measured by a micro BCA protein assay kit (Pierce). Luciferase activity is expressed as relative light units (RLU/mg protein in the cell lysate). The data was reported as mean ± standard deviation for triplicate samples. Every transfection experiment was repeated at least twice.
2.7 Evaluation of luciferase expression in vivo
All animal experiments were conducted in accordance with the principles and procedures described in the University of Iowa’s Guidelines for Care and Use of Experimental Animals. Female Balb/C 6–8 week-old mice were obtained from Charles-River Laboratories and maintained in a pathogenfree environment. Mice were injected IP with pDNA alone or PEI-pDNA nanoparticles prepared with branched PEI or linear PEI at varing N:P ratios and doses. Each PEI-pDNA nanoparticle solution was prepared in 1 ml 5% glucose solution in triplicate. Twenty-four hours later, the mice were injected IP with 200 µl of luciferin (15 mg/ml) and were then anesthetized by isoflurane administered by a gas manifold at a flow rate of 2%. The mice were placed onto a warmed stage in a sealed chamber of an IVIS Imaging 100 Series instrument (Xenogen, Hopkinton, MA). Images of luciferase activity were continuously acquired at a medium binning level until maximal activity was detected. Auto-functions were utilized to determine the minimum (5% of maximum) for the scale at each time-point. The images were analyzed using Live image 2.5 software to provide a quantitative amount of luciferase activity. Region of interest (ROI) from displayed images were drawn around the abdominal and quantified as photons/second (ph/s) and background bioluminescence noted for each experiment.
2.8 Statistical analysis
Group data are reported as mean+/−SD. Differences between groups were analyzed by one way analysis of variance with a Tukey post-test analysis. Levels of significance were accepted at the P<0.05 level. Statistical analyses were performed using Prism 3.02 software (Graphpad Software, Inc., San Diego, CA.)
3. Results
3.1 Zeta potential and particle size analysis of PEI-pDNA nanoparticles
Mixing cationic PEI with negatively charged pDNA results in the spontaneous electrostatic formation of stable nanoparticle complexes. At a constant N:P ratio of 5, increasing the concentration of pDNA mixed with branched PEI from 1 to 5 µg increases the average particle size from 77.9 nm +/− 11.5 to 109.1 nm +/− 2.2 (table 1). A further 5-fold increase in concentration to 25 µg increases the particle size to 155.2 nm +/− 2.66. Increasing the concentration of pDNA mixed with branched PEI to 100 µg increased the average nanoparticle size to 238.5 nm +/− 5.26. For linear PEI, at a constant N:P ratio of 5, increasing the concentration of pDNA mixed with linear PEI from 1 to 5 µg resulted in a non-significant increase in particle size from 106.3 nm +/− 2.49 to 112.3 nm +/− 8.11 (table 1). A further 5-fold increase in concentration to 25 µg increased the particle size to 138.3 nm +/− 3.90. Increasing the concentration of pDNA mixed with linear PEI to 100 µg increased the average nanoparticle size to 265.7 nm +/− 6.40. Branched PEI has a stronger electrostatic interaction with pDNA than linear PEI, which accounts for the greater compaction, higher zeta potentials and smaller nanoparticle sizes at equivalent pDNA concentrations. For both linear and branched PEI, increasing the concentration of pDNA mixed in similar volumes but at the same N:P ratio presumably results in greater aggregation of the nanoparticles and larger average particle sizes (table 1). For branched PEI, at a fixed pDNA concentration of 25 µg, increasing the N:P ratio from 1 to 2 increases the zeta potential from −33.1 +/− 2.05 to −13.36 +/− 1.88 and the average particle size from 96.2 nm +/− 2.09 to 112.5 nm +/− 1.45. Upto an N:P ratio of 10, the zeta potential and particle size continue to increase to 36.01 +/− 1.33 and 131.1 nm +/− 1.74 respectively. At an N:P ratio of 20, nanoparticles formed with branched PEI and pDNA have a lower particle size of 97.5 nm +/− 1.03 and a zeta potential of 36.21 +/− 0.75. For linear PEI, at a fixed pDNA concentration of 25 µg, increasing the N:P ratio from 1 to 3 does not significantly change the zeta potential or particle size. At an N:P ratio of 10, the particle size drops to 141.5 nm +/− 3.26. At an N:P ratio of 20, nanoparticles formed with linear PEI and pDNA have a lower particle size of 179.0 nm +/− 3.45 and a positive zeta potential of 13.05 +/− 0.56.
Table 1.
Characterization of the size of PEI-pDNA nanoparticle complexes.
| Group | Amount of DNA (μg/mouse) | PEI Structure | N:P ratio | Size (nm) |
|---|---|---|---|---|
| 1 | 1 | Branched | 5 | 77.9 ± 11.5 |
| 2 | 5 | Branched | 5 | 109.1 ± 2.20 |
| 3 | 10 | Branched | 5 | 136.4 ± 2.84 |
| 4 | 25 | Branched | 5 | 155.2 ± 2.66 |
| 5 | 50 | Branched | 5 | 165.3 ± 4.30 |
| 6 | 100 | Branched | 5 | 238.5 ± 5.26 |
| 7 | 1 | Linear | 5 | 106.3 ± 2.49 |
| 8 | 5 | Linear | 5 | 112.3 ± 8.11 |
| 9 | 10 | Linear | 5 | 122.8 ± 0.85 |
| 10 | 25 | Linear | 5 | 138.3 ± 3.90 |
| 11 | 50 | Linear | 5 | 161.9 ± 3.88 |
| 12 | 100 | Linear | 5 | 265.7 ± 6.40 |
| 13 | 25 | Branched | 1 | 96.2 ± 2.09 |
| 14 | 25 | Branched | 2 | 112.5 ± 1.45 |
| 15 | 25 | Branched | 3 | 128.1 ± 5.35 |
| 16 | 25 | Branched | 10 | 131.1 ± 1.74 |
| 17 | 25 | Branched | 20 | 97.5 ± 1.03 |
| 18 | 25 | Linear | 1 | 433.9 ± 6.76 |
| 19 | 25 | Linear | 2 | 581.7 ± 9.10 |
| 20 | 25 | Linear | 3 | 415.1 ± 3.26 |
| 21 | 25 | Linear | 10 | 141.5 ± 3.26 |
| 22 | 25 | Linear | 20 | 179.0 ± 3.45 |
N/P ratio is defined as the ratio of primary amino group in PEI to phosphate group (in DNA).
3.2 PEI-pDNA nanoparticle mediated transgene expression of luciferase in HeLa Cells: Effect of N:P ratio
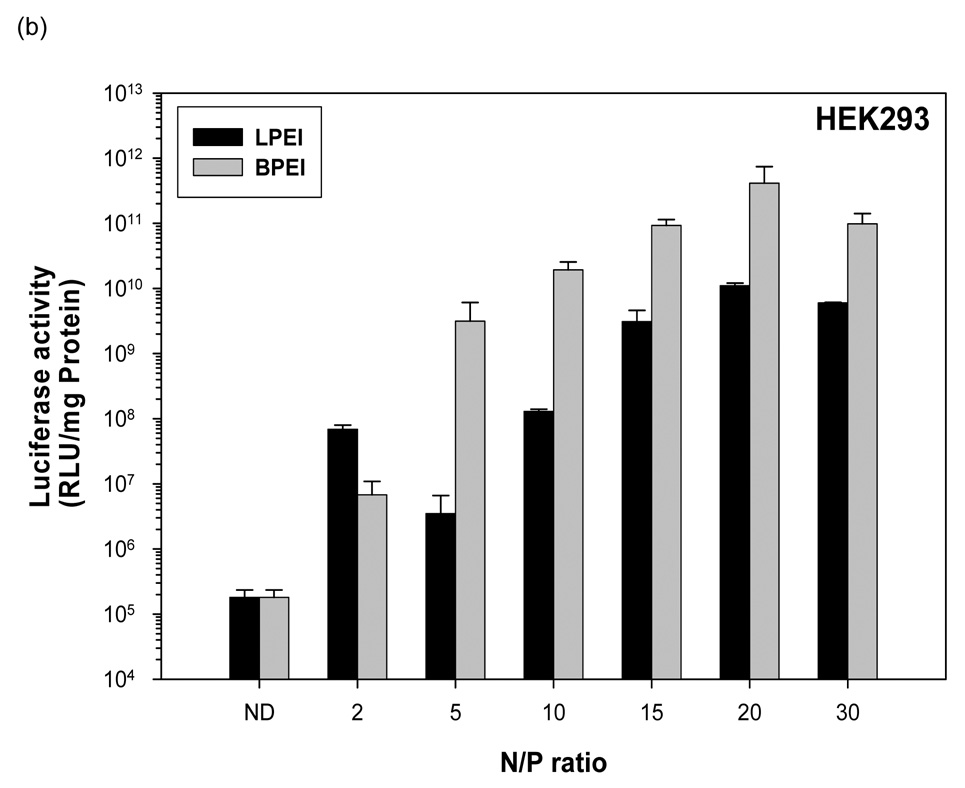
Naked pDNA generated 5.07 × 103 RLU/mg protein mean luciferase activity in HeLa cells (figure 2a). Linear PEI-pDNA nanoparticles prepared at an N:P ratio of 2 generated 1.3 × 104 RLU/mg protein mean luciferase activity in HeLa cells. Branched PEI-pDNA nanoparticles prepared at an N:P ratio of 2 generated 1.1 × 104 RLU/mg protein mean luciferase activity in HeLa cells which was not significantly different from the transgene expression generated by linear PEI-pDNA nanoparticles prepared at an N:P ratio of 2 (P>0.05). When the N:P ratio was increased to 5, linear PEI-pDNA nanoparticles generated 9.7 × 103 RLU/mg protein mean luciferase activity in HeLa cells which is not significantly different from luciferase activity generated by linear PEI-pDNA nanoparticles prepared at an N:P ratio of 2 (P>0.05). In contrast branched PEI-pDNA nanoparticles prepared at an N:P ratio of 5 generated 3 ×107 RLU/mg protein mean luciferase activity in HeLa cells which is significantly higher than luciferase activity generated by linear PEI-pDNA nanoparticles prepared at an N:P ratio of 5 and branched PEI-pDNA nanoparticles prepared at an N:P ratio of 2. As the N:P ratio increased to 20, the PEI-pDNA nanoparticles generated increasing mean luciferase activity upto 1.7 × 108 RLU/mg protein in the HeLa cells. From N:P ratios of 5 and upwards, the branched PEI-pDNA nanoparticles generated significantly higher luciferase activity than linear PEI-pDNA nanoparticles. No significant increases in luciferase activity was observed for branched PEI-pDNA nanoparticles prepared at N:P ratios above 20 and no significant increases in luciferase activity was observed for linear PEI-pDNA nanoparticles prepared at N:P ratios above 15 (P>0.05). The maximal luciferase activity generated by branched PEI-pDNA nanoparticles prepared at an N:P ratio of 20 in HeLa cells was approximately 2400-fold lower than that achieved in HEK293 cells, 60-fold lower than COS7 cells and similar to the luciferase activity generated in HepG2 cells.
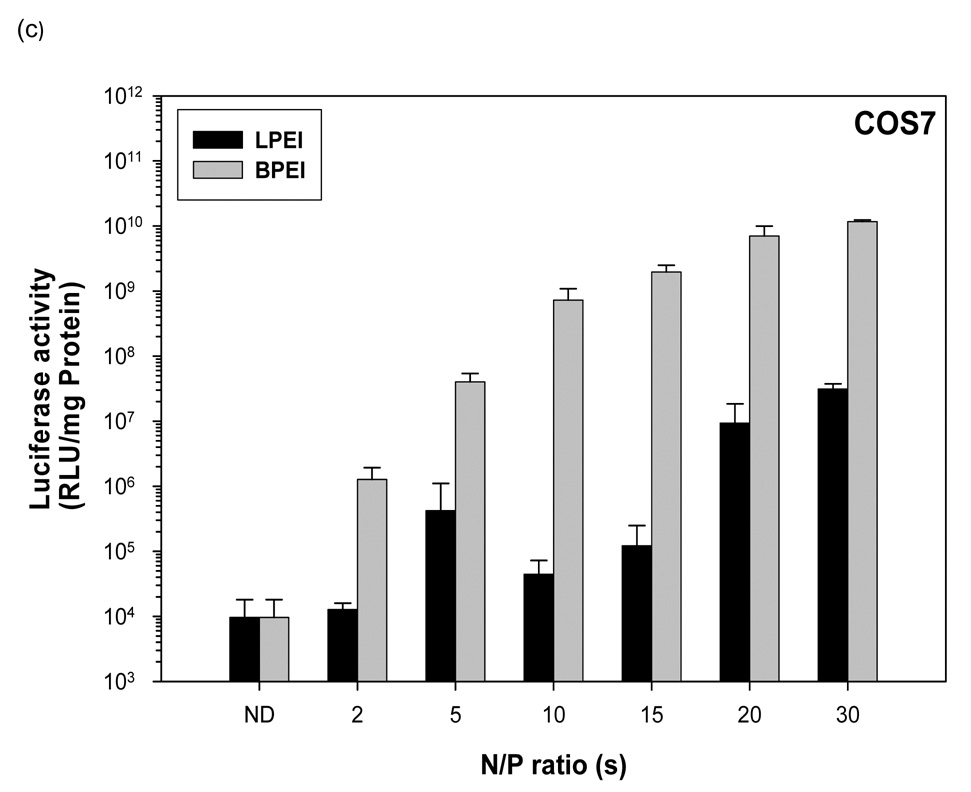
Figure 2.
Transfection mediated by linear polyethylenimine (25 KDa) (LPEI)-pDNA (1 µg) nanoparticle complexes and branched polyethylenimine (25 KDa) (BPEI)-pDNA nanoparticle complexes at various N/P ratios in (a) HeLa, (b) HEK, (c) COS7 and (d) HepG2. Luciferase activity is shown as means ± STD (n=3, error bars represent standard deviation).
3.3 PEI-pDNA nanoparticle mediated transgene expression of luciferase in HEK293 Cells: Effect of N:P ratio
Naked pDNA generated 1.8 × 105 RLU/mg protein mean luciferase activity in HEK293 cells (figure 2b). Linear PEI-pDNA nanoparticles prepared at an N:P ratio of 2 generated 6.9 × 107 RLU/mg protein mean luciferase activity in HEK293 cells. Branched PEI-pDNA nanoparticles prepared at an N:P ratio of 2 generated 6.8 × 106 RLU/mg protein mean luciferase activity in HEK293 cells which was 10-fold lower than the mean luciferase activity generated by linear PEI-pDNA nanoparticles prepared at the same N:P ratio. When the N:P ratio was increased to 5, linear PEI-pDNA nanoparticles generated 3.5 × 106 RLU/mg protein mean luciferase activity in HEK293 cells which was 20-fold lower than linear PEI-pDNA nanoparticles prepared at an N:P ratio of 2. In contrast branched PEI-pDNA nanoparticles prepared at an N:P ratio of 5 generated 3.1 × 109 RLU/mg protein mean luciferase activity in HEK293 cells which is significantly higher than linear PEI-pDNA nanoparticles prepared at an N:P ratio of 5 and branched PEI-pDNA nanoparticles prepared at an N:P ratio of 2, which is similar to results observed in the HeLa cells. As the N:P ratio increased to 20, the branched and linear PEI-pDNA nanoparticles generated increasing mean luciferase activity upto 4.1 × 1011 RLU/mg protein in the HEK293 cells. Consistent with results from the HeLa cells, branched PEI-pDNA nanoparticles prepared at N:P ratios of 5 and upwards, generated significantly higher luciferase activity than linear PEI-pDNA nanoparticles (P<0.05). In HEK293 cells, no significant increases in luciferase activity was observed for branched PEI-pDNA nanoparticles prepared at N:P ratios above 20 and no significant increases in luciferase activity was observed for linear PEI-pDNA nanoparticles prepared at N:P ratios above 15 (P>0.05).
3.4 PEI-pDNA nanoparticle mediated transgene expression of luciferase in COS7 Cells: Effect of N:P ratio
Naked pDNA generated 9.6 × 103 RLU/mg protein mean luciferase activity in COS7 cells (figure 2c). Linear PEI-pDNA nanoparticles prepared at an N:P ratio of 2 generated 1.2 × 104 RLU/mg protein mean luciferase activity in COS7 cells. Branched PEI-pDNA nanoparticles prepared at an N:P ratio of 2 generated 1.2 × 106 RLU/mg protein mean luciferase activity in COS7 cells which was 100-fold higher than the transgene expression generated by linear PEI-pDNA nanoparticles prepared at the same N:P ratio. When the N:P ratio was increased to 5, linear PEI-pDNA nanoparticles generated 4.2 × 105 RLU/mg protein mean luciferase activity in COS7 cells which was 35-fold higher than linear PEI-pDNA nanoparticles prepared at an N:P ratio of 2. Similarly, branched PEI-pDNA nanoparticles prepared at an N:P ratio of 5 generated 4 × 107 RLU/mg protein mean luciferase activity in COS7 cells which is significantly higher than luciferase activity generated by linear PEI-pDNA nanoparticles prepared at an N:P ratio of 5 and branched PEI-pDNA nanoparticles prepared at an N:P ratio of 2. As the N:P ratio increased to 20, the branched PEI-pDNA nanoparticles generated increasing mean luciferase activity upto 7 × 109 RLU/mg protein in the COS7 cells. Consistent with results from the HeLa cells and the HEK293 cells, branched PEI-pDNA nanoparticles prepared at N:P ratios of 5 and upwards generated significantly higher luciferase activity than linear PEI-pDNA nanoparticles (P<0.001). Consistent with results from HeLa cells and HEK293 cells, the COS7 cells showed no significant further increases in luciferase activity when treated with branched PEI-pDNA nanoparticles prepared at N:P ratios above 20 (P>0.05).
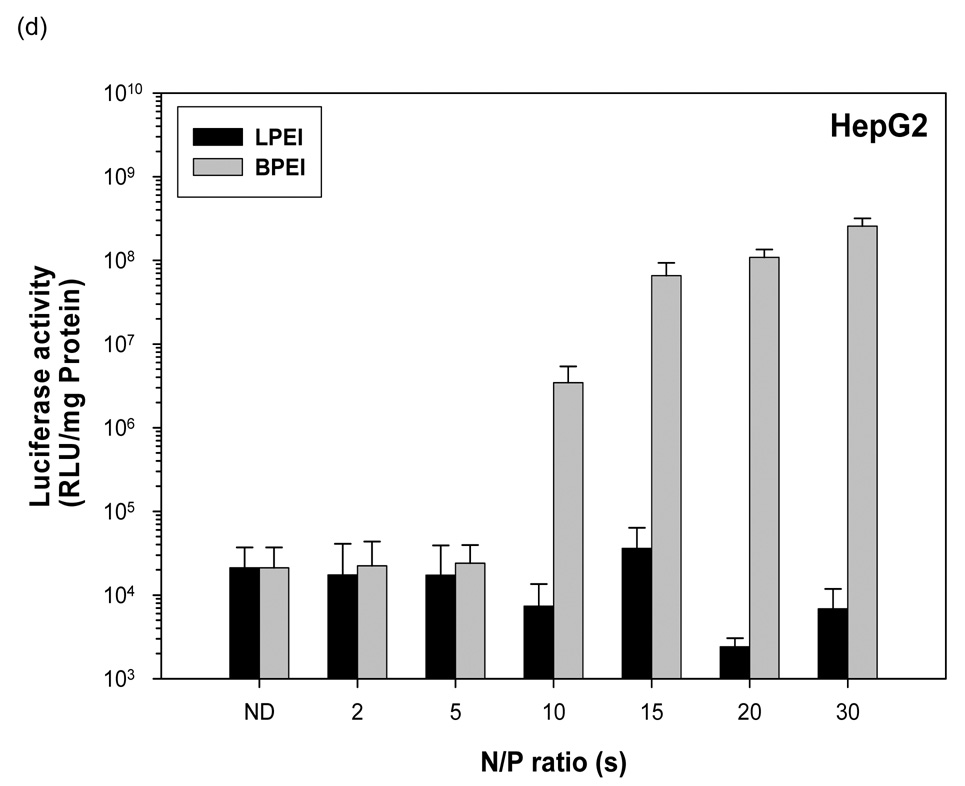
3.5 PEI-pDNA nanoparticle mediated transgene expression of luciferase in HepG2 Cells: Effect of N:P ratio
Naked pDNA generated 2 × 104 RLU/mg protein mean luciferase activity in HepG2 cells (figure 2d). In contrast to HEK293, HeLa and HepG2 cells, linear PEI-pDNA nanoparticles and branched PEI-pDNA nanoparticles prepared at N:P ratios upto 5 generated no significant difference in luciferase activity in HepG2 cells, when compared to naked pDNA (P>0.05). Linear PEI-pDNA nanoparticles resulted in no significant increase in luciferase expression at all N:P ratios (P>0.05). This result is in contrast to the improved luciferase activity observed in HEK293, HeLa and COS7 cells as the N:P ratio of the linear PEI-pDNA nanoparticles increased. Branched PEI-pDNA nanoparticles prepared at an N:P ratio of 10, however, increased mean luciferase expression by 170-fold to 3.4 × 106 RLU/mg protein. As the N:P ratio of the branched PEI-pDNA nanoparticles increased to 20, the mean luciferase activity increased to 1.08 × 108 RLU/mg protein (P>0.001 compared to linear PEI-pDNA nanoparticles at the same N:P ratio).
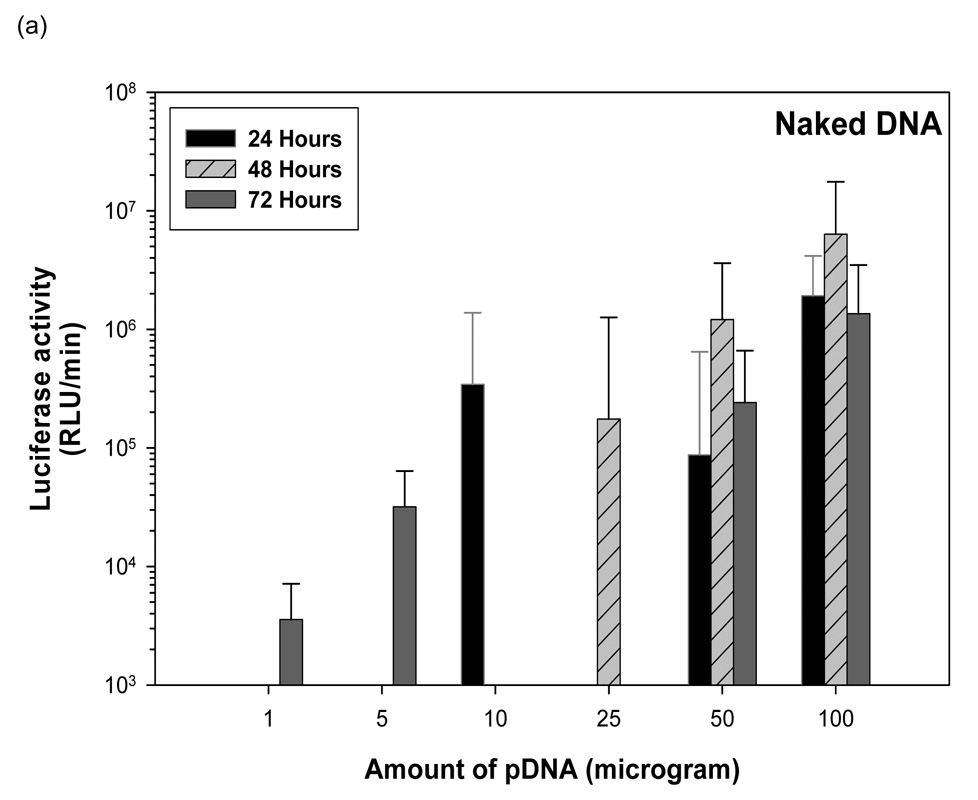
3.6 Naked pDNA mediated transgene expression of luciferase after IP injection in Balb/c mice: Effect of dose
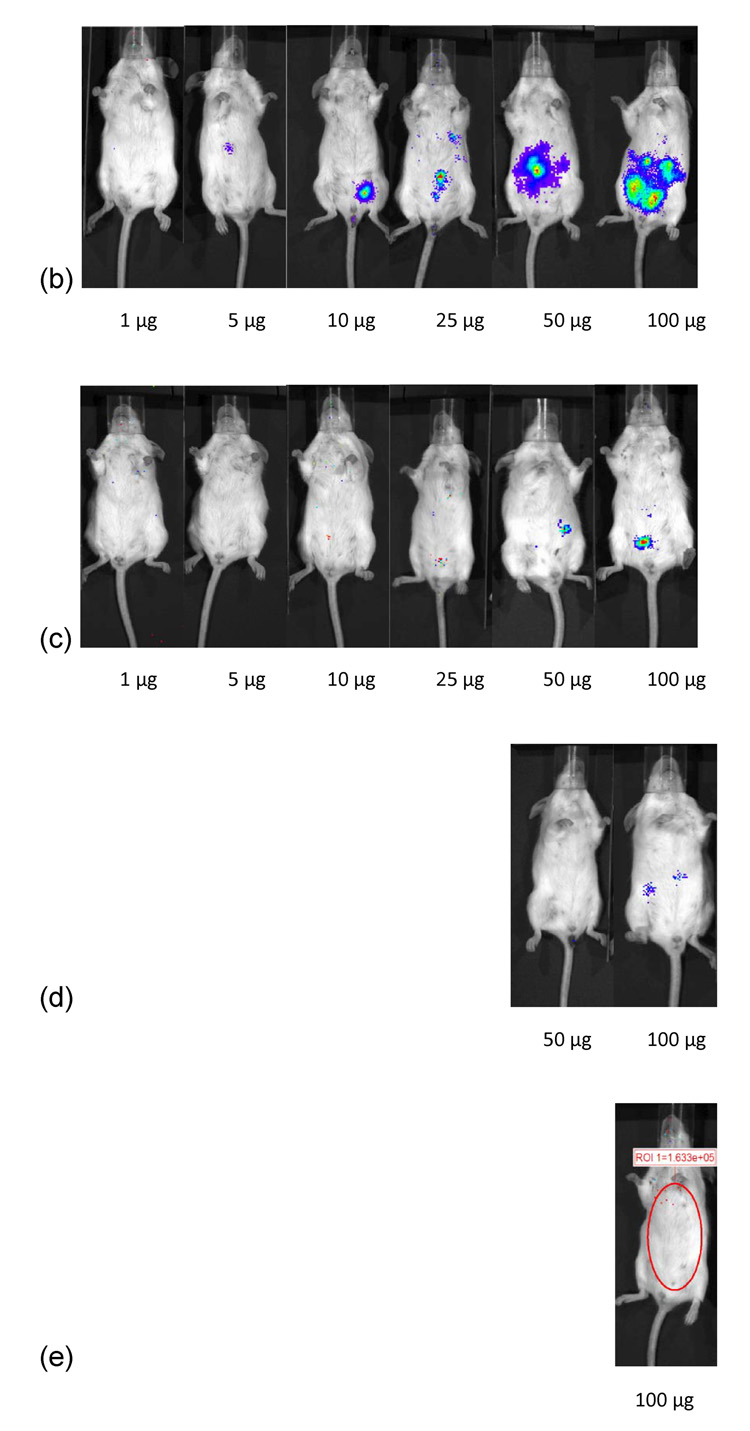
One µg of pDNA generated 3.57 × 103 RLU/minute at 72 hours (figure 3). Five µg of pDNA generated 3.57 × 103 RLU/minute at 72 hours. Ten µg of pDNA generated 3.44 × 105 RLU/minute at 24 hours. Twenty-five µg of pDNA generated an average 1.75 × 105 RLU/min at 48 hours. At fifty µg dosing of pDNA, 8.7 × 104 RLU/minute was observed at 24 hours. This increased to 1.2 × 106 RLU/minute by 48 hours folowed by a decrease to 2.4 × 105 RLU/minute by 72 hours. Increasing the dose from 50 µg to 100 µg pDNA increased luciferase expression 20-fold to 1.9 × 106 RLU/minute at 24 hours. This increased to 6.34 × 106 by 48 hours followed by a fall to 1.36 × 106 by 72 hours. No luciferase expression was detected beyond 72 hours for mice injected with naked pDNA.
Figure 3.
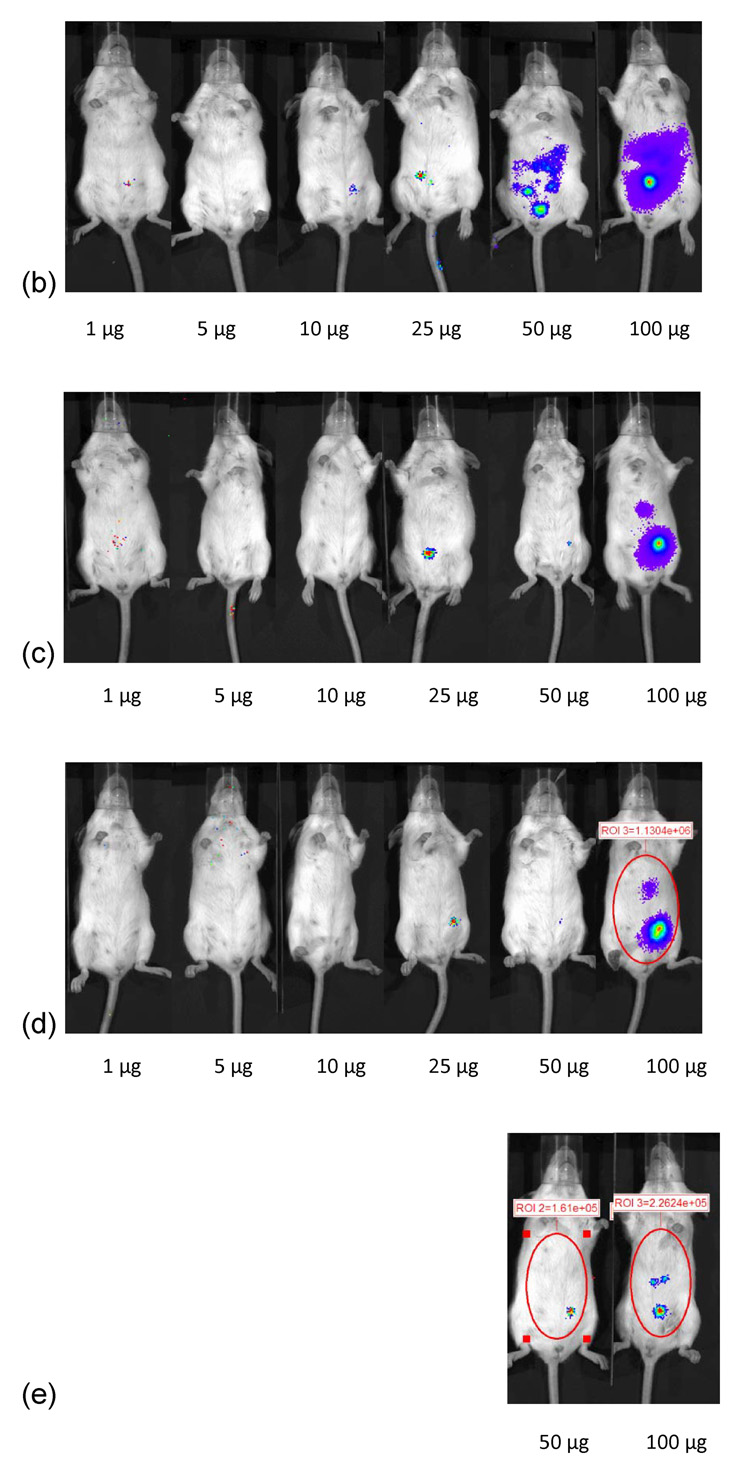
Dose dependent study of naked DNA. (a) Quantification of the signal produced in Balb/c mice after IP injection of each formulation. Values shown are mean ± Standard deviation (n=3). (b)–(d) Luciferase expression in Balb/C mice measured by IVIS. (b), (c), and (d) represent 24 hours, 48 hours and 72 hours after injection. The number of photons is depicted in a color image superimposed on a video image of the animal. Each mouse presented in the image is representative of at least three mice in each experiment.
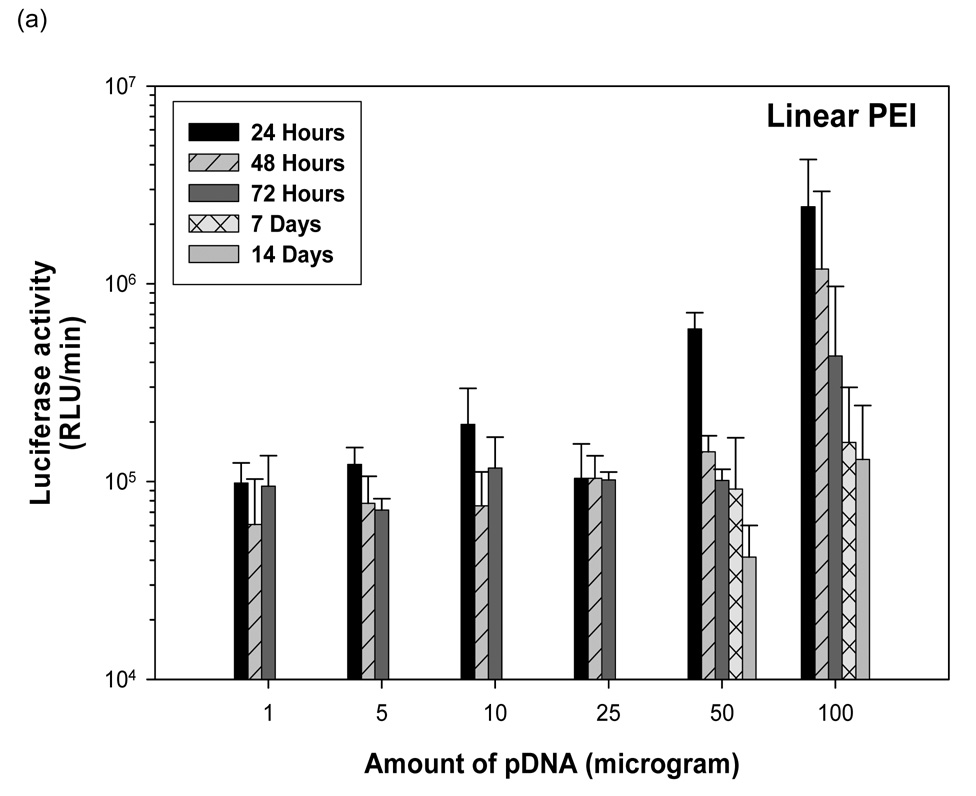
3.7 Linear PEI-pDNA nanoparticle mediated transgene expression of luciferase after IP injection in Balb/c mice: Effect of dose
One µg of pDNA complexed with PEI at an N:P ratio of 5 resulted in 9.8 × 104 RLU/minute mean luciferase activitiy at 24 hours (figure 4). This expression stayed relatively constant upto 72 hours (P>0.05). After 72 hours, no luciferase expression could be detected from mice injected with 1 µg linear PEI-pDNA nanoparticles. This luciferase expression was not significantly different for mice injected IP with upto 25 µg linear PEI-pDNA nanoparticles (P>0.05). However, mice dosed with 50 µg linear PEI-pDNA nanoparticles produced a 6-fold increase in mean luciferase activity to 5.9 × 105 RLU/minute at 24 hours. Doubling the 50 µg dose to 100 µg increased luciferase activity by a further 4-fold at 24 hours. Both the 50 µg and 100 µg doses of pDNA-linear PEI nanoparticles generated dose-dependent mean luciferase activity that was still clearly detectable by 14 days. In contrast with naked pDNA injected into mice IP, which produced maximal mean luciferase activity at 48 hours or 72 hours, IP injected linear PEI-pDNA nanoparticles resulted in the strongest expression levels after 24 hours.
Figure 4.
Dose dependent study of linear PEI-pDNA nanoparticle complexes. (a) Quantification of the signal produced in Balb/c mice after IP injection of each formulation. Values shown are mean ± standard deviation (n=3). (b)–(f) Luciferase expression in Balb/C mice measured by IVIS. (b), (c), (d), (e) and (f) represent 24 hours, 48 hours, 72 hours, 7 days and 14 days after injection. The number of photons is depicted in a color image superimposed on a video image of the animal. Each mouse presented in the figure is representative of at least three mice in each experiment.
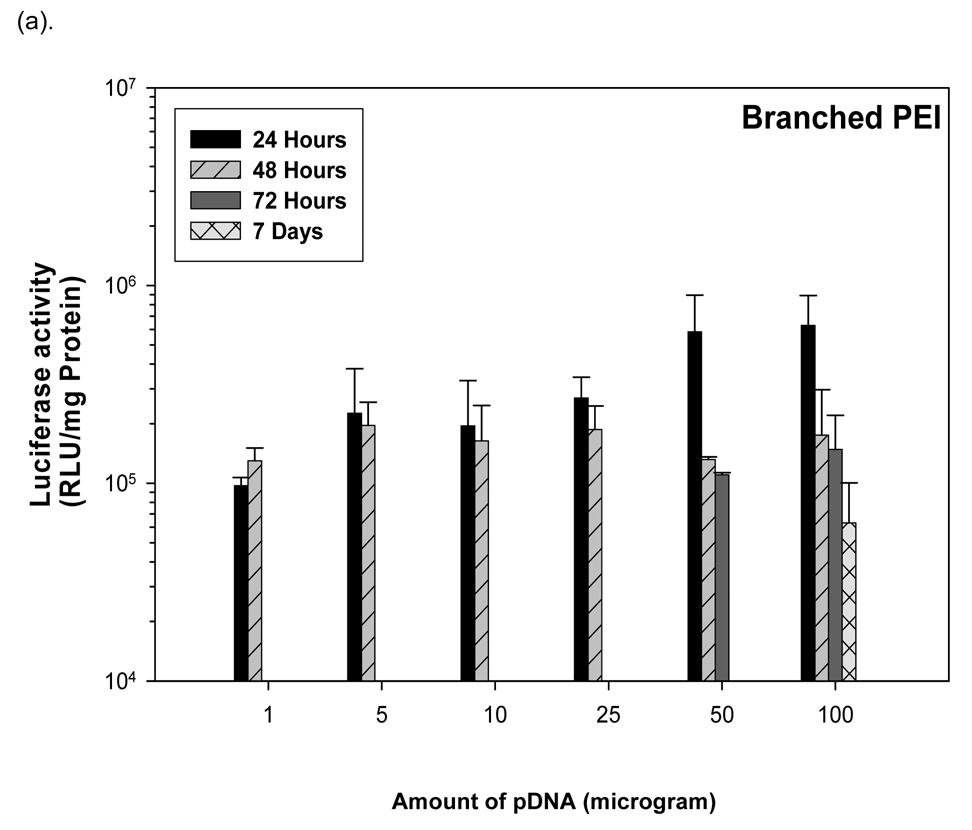
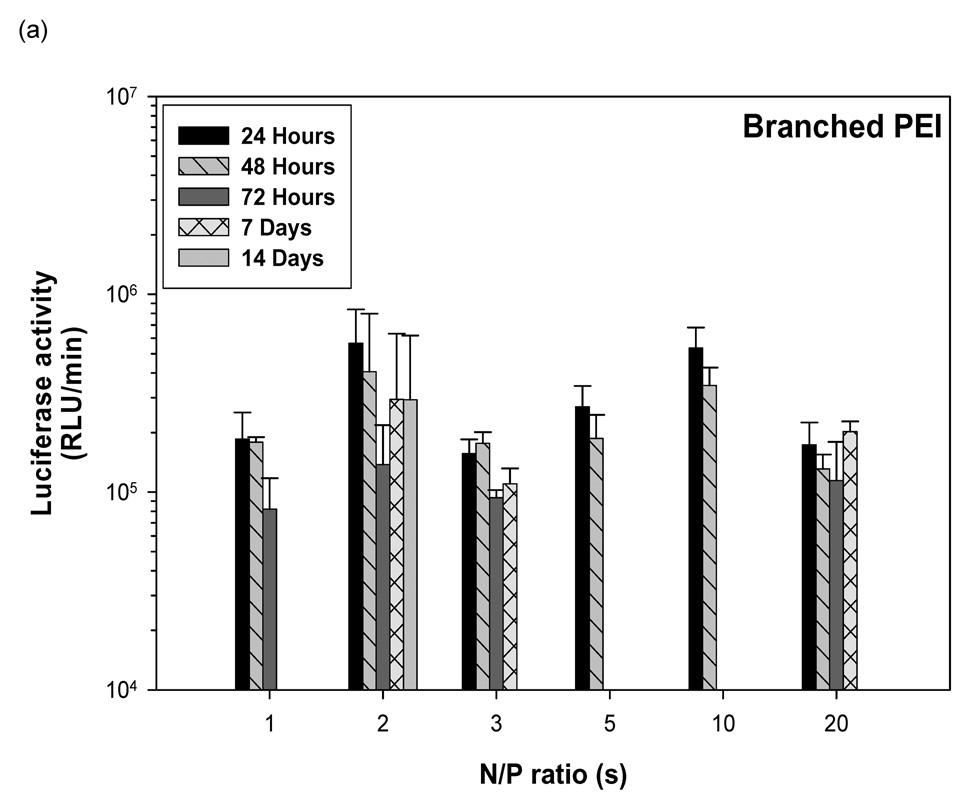
3.8 Branched PEI-pDNA nanoparticle mediated transgene expression of luciferase after IP injection in Balb/c mice: Effect of dose
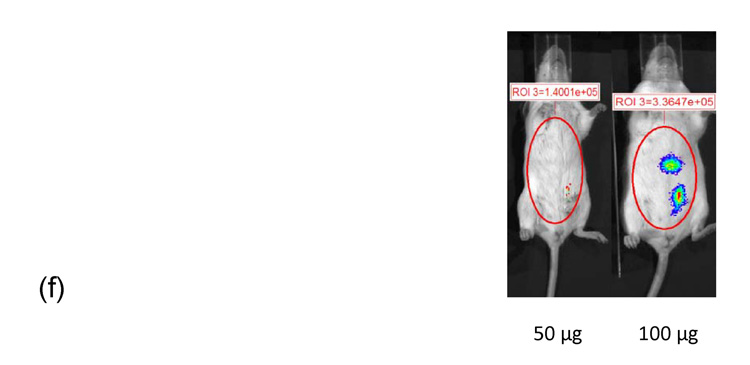
Similar to the results from IP injection of linear PEI-pDNA, 1 µg of pDNA complexed with branched PEI at an N:P ratio of 5 resulted in 9.7 × 104 RLU/minute mean luciferase activitiy at 24 hours (figure 5). In contrast to linear PEI, however, luciferase expression could only be detected upto 48 hours. A 5-fold increase in the branched PEI-pDNA nanoparticle dose resulted in a 2-fold increase in luciferase activity at 24 hours. After 48 hours, no luciferase expression could be detected from mice injected with upto 25 µg branched PEI-pDNA nanoparticles. However, mice dosed with 50 µg branched PEI-pDNA nanoparticles produced a 2-fold increase in mean luciferase activity to 5.8 × 104 at 24 hours and luciferase expression at this dose could still be detected at 72 hours but not 7 days. Doubling the 50 µg dose to 100 µg did not significantly increase the strength of luciferase activity any further at 24 hours but luciferase expression could still be detected at 7 days. For mice injected with branched PEI-pDNA nanoparticles, no luciferase expression could be detected by 14 days at any dose.
Figure 5.
Dose dependent study of branched PEI-pDNA nanoparticle complexes. (a) Quantification of the signal produced in Balb/c mice after IP injection of each formulation. Values shown are mean ± standard deviation (n=3). (b)–(e) Luciferase expression in Balb/C mice measured by IVIS. (b), (c), (d) and (e) represent 24 hours, 48 hours, 72 hours, and 7 days after injection. The number of photons is depicted in a color image superimposed on a video image of the animal. Each mouse presented in the figure is representative of at least three mice in each experiment.
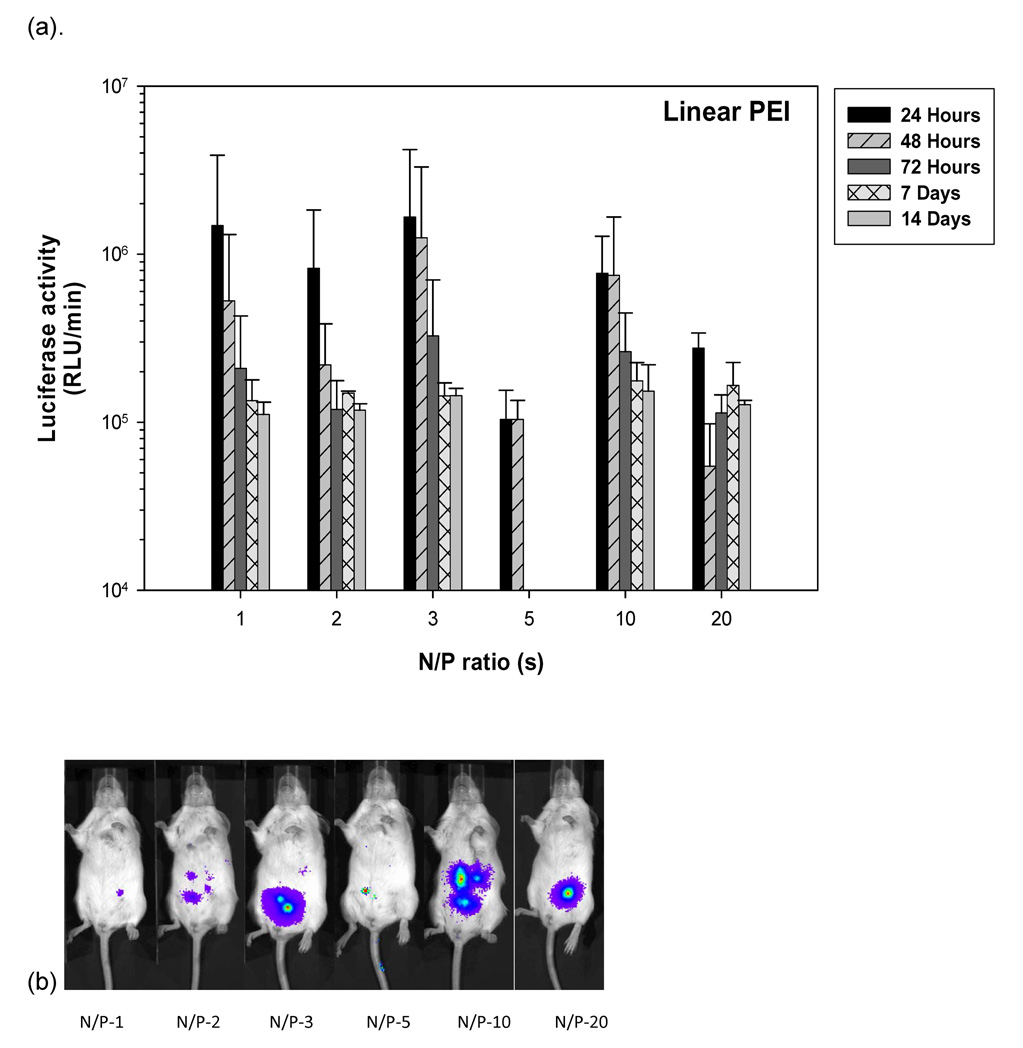
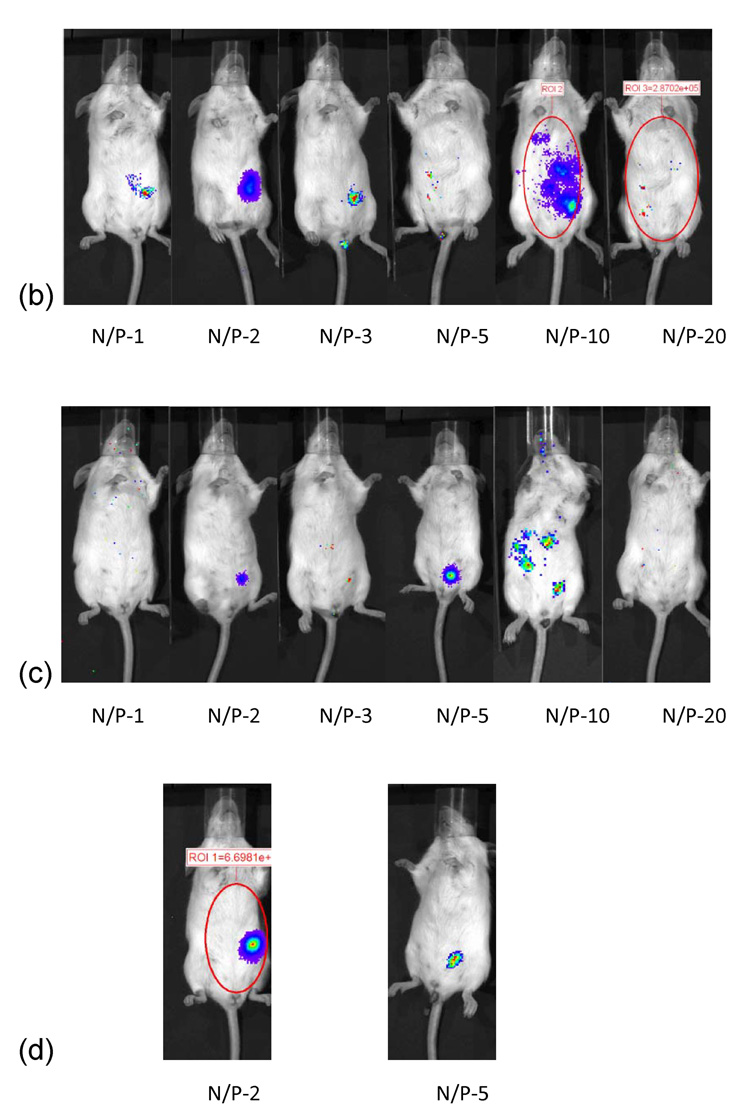
3.9 Linear PEI-pDNA nanoparticle mediated transgene expression of luciferase after IP injection in Balb/c mice: Effect of N:P ratio
Mice injected with 25 µg linear PEI-pDNA nanoparticles prepared at an N:P ratio of 1 generated 1.48 × 106 RLU/minute mean luciferase activity by 24 hours (figure 6). This dropped 3-fold to 5.3 × 105 RLU/minute mean luciferase activity by 48 hours and further 2.65 fold by 72 hours. Luciferase activity could still be detected upto 1.1 × 105 RLU/minute by 14 days. Similar profiles were observed for mice injected with linear PEI-pDNA nanoparticles prepared with N:P ratios upto 20. In contrast to luciferase activity observed in vitro, increasing the N:P ratio at which linear PEI-pDNA nanoparticles were prepared did not change luciferase expression significantly (P>0.05).
Figure 6.
Effect of linear PEI complexed with 25 micrograms of pDNA at different N/P ratios on gene expression in Balb/C. (a) Quantification of the signal produced in Balb/c mice after IP injection of each formulation. Values shown are mean ± standard deviation (n=3). (b)–(f) Luciferase expression in Balb/C mice measured by IVIS. (b), (c), (d), (e) and (f) represent 24 hours, 48 hours, 72 hours, 7 days and 14 days after injection. The number of photons is depicted in a color image superimposed on a video image of the animal. Each mouse presented in the figure is representative of at least three mice in each experiment.
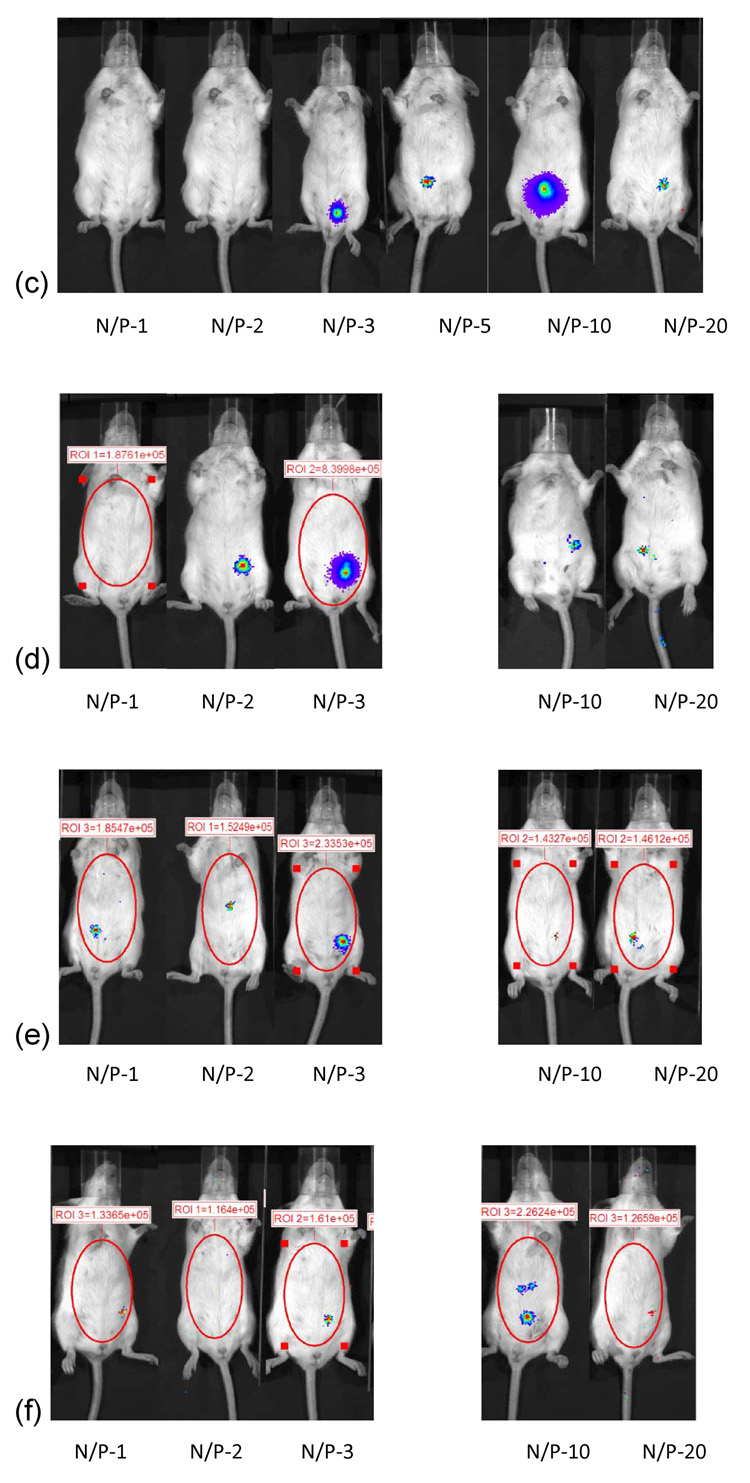
3.10 Branched PEI-pDNA nanoparticle mediated transgene expression of luciferase after IP injection in Balb/c mice: Effect of N:P ratio
Mice injected with branched PEI-pDNA (25 µg) nanoparticles prepared at an N:P ratio of 1 generated 1.85 × 105 RLU/minute mean luciferase activity by 24 hours (figure 7). This was 8-fold lower than luciferase expression generated by linear PEI-pDNA nanoparticles prepared at an N:P ratio of 1. This dropped 2-fold to 8.2 × 104 RLU/minute mean luciferase activity by 72 hours. No further luciferase expression could be detected beyond 72 hours. The maximal luciferase activity was generated from branched PEI-pDNA nanoparticles prepared at an N:P ratio of 2. This was 5.6 × 105 RLU/minute luciferase activity at 24 hours. Luciferase activity could still be detected upto 2.93 × 105 RLU/minute by 14 days. In contrast to linear PEI-pDNA nanoparticles, branched PEI-pDNA nanoparticles prepared at all other N:P ratios did not generate any luciferase expression by day 14.
Figure 7.
Effect of branched PEI complexed with 25 micrograms of pDNA at different N/P ratios on gene expression in Balb/C. (a) Quantification of the signal produced in Balb/c mice after IP injection of each formulation. Values shown are mean ± standard deviation (n=3). (b)–(f) Luciferase expression in Balb/C mice measured by IVIS. (b), (c), (d), (e) and (f) represent 24 hours, 48 hours, 72 hours, 7days and 14 days after injection. The number of photons is depicted in a color image superimposed on a video image of the animal. Each mouse presented in the figure is representative of at least three mice in each experiment.
4. Discussion
PEI has shown significant potential for delivery and protection of siRNA and pDNA.[14, 38] The structure of the PEI significantly impacts the transfection efficiency.[20] A number of studies have shown that, in selected cell lines, branched PEI generates stronger transgene expression that linear PEI.[18, 31] In contrast, when PEI is injected intravenously or via the lungs, linear PEI, generates stronger transgene expression than branched PEI.[31, 34] In vitro studies show that increasing the N:P ratio of the PEI-pDNA nanoparticles increases luciferase activity and toxicity.
In this study, we show that increasing the concentration of PEI-pDNA nanoparticle concentration results in aggregation and larger particle sizes but that this does not reduce the transfection efficiency in vivo. We prepared PEI-pDNA nanoparticles in salt-free conditions with 5% glucose because these preparation conditions have been shown to generate the strongest transgene expression in vivo.[34] Linear PEI-pDNA nanoparticles show lower zeta potentials than branched PEI-pDNA nanoparticles when prepared at the same N:P ratio. Branched PEI-pDNA nanoparticles prepared at the same N:P ratio as linear PEI-pDNA nanoparticles are smaller in size and more compact. The higher zetapotential and smaller size of the branched PEI-pDNA nanoparticles translate into higher transfection efficiencies in vitro but not in vivo. We selected four cell lines that are commonly used as transfection hosts but rarely compared side by side when evaluating non-viral gene delivery systems. Specifically, we show that increasing the N:P ratio increases luciferase activity for branched PEI-pDNA nanoparticles and linear PEI-pDNA nanoparticles in HEK293, COS7 and HeLa cell lines. Increasing the N:P ratio for branched PEI-pDNA nanoparticles also increases luciferase expression in HepG2 cells but does not increase luciferase expression generated by linear PEI-pDNA nanoparticles. In all of the cell lines, branched PEI-pDNA nanoparticles prepared at N:P ratios of 10 and above generated significantly higher luciferase activity than linear PEI-pDNA nanoparticles. These results are based on PEI-pDNA nanoparticles prepared in salt-free conditions. It should be noted that Wightman et al have shown that preparing linear and branched PEI-pDNA nanoparticles in salt containing buffer reversed these results with linear PEI-pDNA nanoparticles generating stronger luciferase activity than branched PEI-pDNA nanoparticles.[34] Choosakoonkirang et al, have previously reported that a minimum N:P ratio of 4 and above is necessary for transfection of COS7 and CHO-K1 cells.[18] Transfection results from PEI-pDNA nanoparticles prepared at N:P ratios less than 4 were not presented in their studies. Our results are consistent with the observations of Choosakoonkriang et al for linear PEI. However, we show that branched PEI-pDNA nanoparticles can significantly improve transfection efficiencies in COS7 cells even when prepared at an N:P ratio of 2. Although linear and branched PEI display similar rates of internalization, the higher charge associated with branched PEI in comparison to linear PEI is reported to enhance extracellular binding by 5-fold.[13] In addition, branched PEI is internalized by cholesterol dependent pathways whilst the internalization of linear PEI appears to be independent of clathrin and cholesterol pathways.[13] Luciferase activity was highest in the HEK293 cells and luciferase expression in each of the cell lines followed the order of HEK293>COS7>HepG2>HeLa. These results suggest that HEK293 cells are the most sensitive cell line for evaluating cationic polymers as non-viral vectors. It also suggests that HeLa cells may be a suitable starting candidate cell line for testing the development of new non-viral vectors targeted to less transfection permissive cell and tissue targets. These results show that the optimal N:P ratio for PEI-pDNA nanoparticles is cell line dependent and that separate optimization is necessary dependent on the target tissue or cell being transfected.
For IP injection of PEI-pDNA nanoparticles in vivo, we have shown that two dominant parameters in increasing luciferase activity are dose and structure of PEI but not increasing N:P ratio. To measure this, we used bioluminescent imaging (BLI), which is a method that in recent years, is increasingly being utilized to determine location and strength of luciferase activity in a variety of animal models.[47–51] Luciferase activity is observed following an IP dose of D-luciferin, which results in bioluminescence that is detected in anesthetized mice by a cooled charge-coupled device camera. BLI is now an established method for quantitatively detecting transgene expression in vivo.[52] BLI is rapid, sensitive and does not generate false positives.[47, 52] In addition, Rettig et al have shown that luciferase expression measured by tissue harvesting and luciferase expression measured using BLI are directly correlated. This therefore eliminates the need to harvest tissue and allows for the same reduced number of animals to be assayed over time.[47]
In all experiments, branched PEI-pDNA nanoparticles generated significantly lower luciferase activity than linear PEI-pDNA nanoparticles following IP injection. For the in vitro studies, increasing the N:P ratio at which the PEI-pDNA nanoparticles are prepared increased the transfection efficiency. Increasing the N:P ratio at which PEI-pDNA nanoparticles are prepared for IP injection in vivo, in contrast, does not significantly improve the strength or duration of luciferase activity. For example, for branched PEI-pDNA nanoparticles, the strongest and longest duration of luciferase activity was for nanoparticles prepared at an N:P ratio of 2. Branched PEI-pDNA nanoparticles prepared at all other N:P ratios did not generate any luciferase activity at day 14. Linear PEI-pDNA nanoparticles generated higher than 1 × 105 RLU/minute mean luciferase activity at all N:P ratios apart from 5 at day 14. However, increasing the N:P ratio at which the linear PEI-pDNA nanoparticles were prepared did not significantly change luciferase activity. These results for IP injections are in contrast to those reported for intraventricular injections and intratracheal intubation where increasing N:P ratio increased transfection efficiencies. This suggests that the N:P ratio may need to be optimized for each different route of administration.[20, 53]
Our results have shown that increasing zeta potential and decreasing particle size by strengthening the electrostatic interactions between PEI and pDNA can substantially improve transfection efficiencies in vitro. In contrast, for in vivo gene delivery, there is an optimal balance between having interactions between the PEI that are strong enough to pack the pDNA so it is protected from enzymatic degradation whilst simultaneously having interactions that are weak enough to allow for eventual release of the pDNA. Bertschinger et al have shown that linear PEI-pDNA particles are much more efficiently disassembled than branched PEI-pDNA nanoparticles in the presence of molecules typically found in vivo. These molecules include heparin, RNA, non-genomic DNA and proteins. This is important because compacted PEI-pDNA nanoparticles that enter the nucleus cannot function as a template for transcription and even low levels of PEI have been reported to disrupt efficient transcription of DNA.[54] Another factor that may reduce the in vivo efficacy of branched PEI, is that it has been reported to induce proinflammatory chemokine receptors. Linear PEI in comparison does not.[11] One parameter in these studies that requires further investigation is the effect of number of doses. Previous studies have shown that re-dosing mice after 7 days with linear PEI-pDNA nanoparticles results in significantly reduced luciferase activity in comparison to the first dose. This reduced expression has been attributed to a variety of factors including CpG sequences in the pDNA stimulating immune responses [55–57] and possibly the effect of anaesthesia on the mouse.[31] Kawakami et al have suggested that the buffering capacity of the PEI in the endosome abrogate the pDNA (CpG)-induced activation of NFkB.[30] Increasing the time after which the second dose of PEI-pDNA nanoparticles is given has been reported to be the dominant parameter in improving the luciferase activity generated by the second dose.[31]
In our studies, despite the reported toxicity that is associated with PEI, including deterioration in lung function when lungs are exposed to PEI,[30, 31] increasing dose or N:P ratio of the PEI-pDNA nanoparticles did not result in reduced mice survival in vivo. The toxicity that arises with PEI administration is associated with charged PEI-pDNA nanoparticles interacting with blood components such as erythrocytes, causing aggregation in the lung capillaries and subsequent lung embolism.[34, 58, 59] Injecting PEI-pDNA nanoparticles intravenously or intratracheally results in gene expression that is stronger in the lung than liver, heart, spleen or kidneys.[30, 34] Because the peritoneal cavity acts as barrier to PEI-pDNA nanoparticles entering the lung or bypassing the blood-brain barrier, PEI-pDNA nanoparticles injected via this route are expected to demonstrate significantly reduced toxicity.[6, 36–38] Consistent with these expectations, in all of the formulations we tested, mice tolerated the PEI-pDNA nanoparticles and remained in good health as determined by the Body Condition Scoring (BCS) technique.[60] The mice appeared well conditioned. The vertebrae and dorsal pelvis were not prominent but palpable with slight pressure indicating a state of good health in the mice. No adverse injection site reactions such as infection, redness or wounding were observed. These observations suggest that the intraperitoneal route of administering PEI-pDNA nanoparticles is safe for the doses and compositions tested.
5. Conclusion
Polyethylenimine is a cationic polymer that has shown promising potential as a non-viral carrier for gene delivery. The structure of PEI can be branched or linear. PEI binds to pDNA electrostatically to form nanoparticles. In this study, we show that branched PEI-pDNA nanoparticles generate stronger luciferase activity than linear PEI-pDNA nanoparticles in vitro. In contrast, we show that linear PEI-pDNA nanoparticles generate stronger and more sustained luciferase activity when injected intraperitoneally in vivo. Further differences between in vitro and in vivo results include the impact of N:P ratio on the preparation of PEI-pDNA nanoparticles. Increasing the N:P ratio at which PEI-pDNA nanoparticles are prepared increased luciferase activity in vitro. The optimal N:P ratio is dependent on the cell line. Of the cell lines tested, HEK293 cells are the most sensitive whilst HeLa cells were the least transfection permissive. For in vivo experiments, increasing the N:P ratio did not significantly increase the luciferase activity generated by linear or branched PEI-pDNA nanoparticles after IP injection. These studies highlight the need for non-viral delivery systems to be optimized in vitro and in vivo for specific cell and tissue targets prior to utilization in therapeutic applications. The IP route of administration has been demonstrated to be relatively non-toxic for all the PEI-pDNA nanoparticle formulations tested in this study, can be optimized by dose and polymer structure and should have significant potential for future non-viral gene delivery applications.
Acknowledgments
We gratefully acknowledge support aided by grant number IRG-77-004-28 from the American Cancer Society and grant number 1R21CA133456-01 from the National Institutes of Health. J. Intra acknowledges support from the Parenteral Drug Association for a pre-doctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Salem AK, Searson PC, Leong KW. Multifunctional nanorods for gene delivery. Nature Materials. 2003;2(10):668–671. doi: 10.1038/nmat974. [DOI] [PubMed] [Google Scholar]
- 2.Wiethoff CM, Middaugh CR. Barriers to nonviral gene delivery. Journal of Pharmaceutical Sciences. 2003;92(2):203–217. doi: 10.1002/jps.10286. [DOI] [PubMed] [Google Scholar]
- 3.Wong SY, Pelet JM, Putnam D. Polymer systems for gene delivery-past, present, and future. Progress in Polymer Science. 2007;32:799–837. [Google Scholar]
- 4.Abbas AO, Donovan MD, Salem AK. Formulating PLGA particles for plasmid DNA delivery. Journal of Pharmaceutical Sciences. 2007 doi: 10.1002/jps.21215. [DOI] [PubMed] [Google Scholar]
- 5.Kircheis R, Wightman L, Wagner E. Design and gene delivery activity of modified polyethylenimines. Advanced Drug Delivery Reviews. 2001;53(3):341–358. doi: 10.1016/s0169-409x(01)00202-2. [DOI] [PubMed] [Google Scholar]
- 6.Zugates GT, Peng WD, Zumbuehl A, Jhunjhunwala S, Huang YH, Langer R, Sawicki JA, Anderson DG. Rapid optimization of gene delivery by parallel end-modification of poly(beta-amino ester)s. Molecular Therapy. 2007;15(7):1306–1312. doi: 10.1038/mt.sj.6300132. [DOI] [PubMed] [Google Scholar]
- 7.Zhang XJ, Godbey WT. Viral vectors for gene delivery in tissue engineering. Advanced Drug Delivery Reviews. 2006;58(4):515–534. doi: 10.1016/j.addr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Kamiya H, Tsuchiya H, Yamazaki J, Harashima H. Intracellular trafficking and transgene expression of viral and non-viral gene vectors. Advanced Drug Delivery Reviews. 2001;52(3):153–164. doi: 10.1016/s0169-409x(01)00216-2. [DOI] [PubMed] [Google Scholar]
- 9.Manno CS, Arruda VR, Pierce GF, Glader B, Ragni M, Rasko J, Ozelo MC, Hoots K, Blatt P, Konkle B, Dake M, Kaye R, Razavi M, Zajko A, Zehnder J, Nakai H, Chew A, Leonard D, Wright JF, Lessard RR, Sommer JM, Tigges M, Sabatino D, Luk A, Jiang HY, Mingozzi F, Couto L, Ertl HC, High KA, Kay MA. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nature Medicine. 2006;12(3):342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 10.Mingozzi F, Maus MV, Hui DJ, Sabatino DE, Murphy SL, Rasko JEJ, Ragni MV, Manno CS, Sommer J, Jiang HY, Pierce GF, Ertl HCJ, High KA. CD8+ T-cell responses to adeno-associated virus capsid in humans. Nature Medicine. 2007;13(4):419–422. doi: 10.1038/nm1549. [DOI] [PubMed] [Google Scholar]
- 11.Jeong GJ, Byun HM, Kim JM, Yoon H, Choi HG, Kim WK, Kim SJ, Oh YK. Biodistribution and tissue expression kinetics of plasmid DNA complexed with polyethylenimines of different molecular weight and structure. Journal of Controlled Release. 2007;118(1):118–125. doi: 10.1016/j.jconrel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Neu M, Germershaus O, Behe M, Kissel T. Bioreversibly crosslinked polyplexes of PEI and high molecular weight PEG show extended circulation times in vivo. Journal of Controlled Release. 2007;124:69–80. doi: 10.1016/j.jconrel.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Seib FP, Jones AT, Duncan R. Comparison of the endocytic properties of linear and branched PEIs, and cationic PAMAM dendrimers in B16f10 melanoma cells. Journal of Controlled Release. 2007;117(3):291–300. doi: 10.1016/j.jconrel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 14.Zhang SB, Zhao B, Jiang HM, Wang B, Ma BC. Cationic lipids and polymers mediated vectors for delivery of siRNA. Journal of Controlled Release. 2007;123:1–10. doi: 10.1016/j.jconrel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Zhou XF, Liu B, Yu XH, Zha X, Zhang XZ, Chen Y, Wang XY, Jin YH, Wu YG, Chen Y, Shan YM, Chen Y, Liu JQ, Kong W, Shen JC. Controlled release of PEI/DNA complexes from mannose-bearing chitosan microspheres as a potent delivery system to enhance immune response to HBV DNA vaccine. Journal of Controlled Release. 2007;121:200–207. doi: 10.1016/j.jconrel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Intra J, Glasgow JM, Mai HQ, Salem AK. Pulsatile release of biomolecules from polydimethylsiloxane (PDMS) chips with hydrolytically degradable seals. Journal of Controlled Release. 2008 doi: 10.1016/j.jconrel.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Nimesh S, Goyal A, Pawar V, Jayaraman S, Kumar P, Chandra R, Singh Y, Gupta KC. Polyethylenimine nanoparticles as efficient transfecting agents for mammalian cells. Journal of Controlled Release. 2006;110(2):457–468. doi: 10.1016/j.jconrel.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Choosakoonkriang S, Lobo BA, Koe GS, Koe JG, Middaugh CR. Biophysical characterization of PEI/DNA complexes. Journal of Pharmaceutical Sciences. 2003;92(8):1710–1722. doi: 10.1002/jps.10437. [DOI] [PubMed] [Google Scholar]
- 19.Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, Demeneix BA. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: Polyethylenimine. Human Gene Therapy. 1996;7(16):1947–1954. doi: 10.1089/hum.1996.7.16-1947. [DOI] [PubMed] [Google Scholar]
- 20.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in-Vivo - Polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boussif O, Zanta MA, Behr JP. Optimized galenics improve in vitro gene transfer with cationic molecules up to 1000-fold. Gene Therapy. 1996;3(12):1074–1080. [PubMed] [Google Scholar]
- 22.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. Journal of Controlled Release. 1999;60(2–3):149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 23.Goula D, Benoist C, Mantero S, Merlo G, Levi G, Demeneix BA. Polyethylenimine-based intravenous delivery of transgenes to mouse lung. Gene Therapy. 1998;5(9):1291–1295. doi: 10.1038/sj.gt.3300717. [DOI] [PubMed] [Google Scholar]
- 24.Goula D, Remy JS, Erbacher P, Wasowicz M, Levi G, Abdallah B, Demeneix BA. Size, diffusibility and transfection performance of linear PEI/DNA complexes in the mouse central nervous system. Gene Therapy. 1998;5(5):712–717. doi: 10.1038/sj.gt.3300635. [DOI] [PubMed] [Google Scholar]
- 25.Kircheis R, Kichler A, Wallner G, Kursa M, Ogris M, Felzmann T, Buchberger M, Wagner E. Coupling of cell-binding ligands to polyethylenimine for targeted gene delivery. Gene Therapy. 1997;4(5):409–418. doi: 10.1038/sj.gt.3300418. [DOI] [PubMed] [Google Scholar]
- 26.Suh J, Paik HJ, Hwang BK. Ionization of Poly(Ethylenimine) and Poly(Allylamine) at Various Phs. Bioorganic Chemistry. 1994;22(3):318–327. [Google Scholar]
- 27.Tang MX, Szoka FC. The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes. Gene Therapy. 1997;4(8):823–832. doi: 10.1038/sj.gt.3300454. [DOI] [PubMed] [Google Scholar]
- 28.Zhang XQ, Intra J, Salem AK. Conjugation of polyamidoamine dendrimers on biodegradable microparticles for nonviral gene delivery. Bioconjugate Chemistry. 2007;18:2068–2076. doi: 10.1021/bc070116l. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XQ, Intra J, Salem AK. Comparative study of poly (lactic-co-glycolic acid)-poly ethyleneimine-plasmid DNA microparticles prepared using double emulsion methods. Journal of Microencapsulation. 2008;25:1–12. doi: 10.1080/02652040701659347. [DOI] [PubMed] [Google Scholar]
- 30.Kawakami S, Ito Y, Charoensit P, Yamashita F, Hashida M. Evaluation of proinflammatory cytokine production induced by linear and branched polyethylenimine/plasmid DNA complexes in mice. Journal of Pharmacology and Experimental Therapeutics. 2006;317(3):1382–1390. doi: 10.1124/jpet.105.100669. [DOI] [PubMed] [Google Scholar]
- 31.Wiseman JW, Goddard CA, McLelland D, Colledge WH. A comparison of linear and branched polyethylenimine (PEI) with DCChol/DOPE liposomes for gene delivery to epithelial cells in vitro and in vivo. Gene Therapy. 2003;10(19):1654–1662. doi: 10.1038/sj.gt.3302050. [DOI] [PubMed] [Google Scholar]
- 32.Bragonzi A, Dina G, Villa A, Calori G, Biffi A, Bordignon C, Assael BM, Conese M. Biodistribution and transgene expression with nonviral cationic vector/DNA complexes in the lungs. Gene Therapy. 2000;7(20):1753–1760. doi: 10.1038/sj.gt.3301282. [DOI] [PubMed] [Google Scholar]
- 33.Goula D, Becker N, Lemkine GF, Normandie P, Rodrigues J, Mantero S, Levi G, Demeneix BA. Rapid crossing of the pulmonary endothelial barrier by polyethylenimine/DNA complexes. Gene Therapy. 2000;7(6):499–504. doi: 10.1038/sj.gt.3301113. [DOI] [PubMed] [Google Scholar]
- 34.Wightman L, Kircheis R, Rossler V, Carotta S, Ruzicka R, Kursa M, Wagner E. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. Journal of Gene Medicine. 2001;3(4):362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 35.Zou SM, Erbacher P, Remy JS, Behr JP. Systemic linear polyethylenimine (L-PEI)-mediated gene delivery in the mouse. Journal of Gene Medicine. 2000;2(2):128–134. doi: 10.1002/(SICI)1521-2254(200003/04)2:2<128::AID-JGM95>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 36.Aigner A, Fischer D, Merdan T, Brus C, Kissel T, Czubayko F. Delivery of unmodified bioactive ribozymes by an RNA-stabilizing polyethylenimine (LMW-PEI) efficiently down-regulates gene expression. Gene Therapy. 2002;9(24):1700–1707. doi: 10.1038/sj.gt.3301839. [DOI] [PubMed] [Google Scholar]
- 37.Aoki K, Furuhata S, Hatanaka K, Maeda M, Remy JS, Behr JP, Terada M, Yoshida T. Polyethylenimine-mediated gene transfer into pancreatic tumor dissemination in the murine peritoneal cavity. Gene Therapy. 2001;8(7):508–514. doi: 10.1038/sj.gt.3301435. [DOI] [PubMed] [Google Scholar]
- 38.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Therapy. 2005;12(5):461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 39.Furgeson DY, Yockman JW, Janat MM, Kim SW. Tumor efficacy and biodistribution of linear polyethylenimin-cholesterol/DNA complexes. Molecular Therapy. 2004;9(6):837–845. doi: 10.1016/j.ymthe.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Alvarez RD, Curiel DT. A phase I study of recombinant adenovirus vector-mediated intraperitoneal delivery of herpes simplex virus thymidine kinase (HSV-TK) gene and intravenous ganciclovir for previously treated ovarian and extraovarian cancer patients. Human Gene Therapy. 1997;8(5):597–613. doi: 10.1089/hum.1997.8.5-597. [DOI] [PubMed] [Google Scholar]
- 41.Jayne DG. The molecular biology of peritoneal carcinomatosis from gastrointestinal cancer. Annals Academy of Medicine Singapore. 2003;32(2):219–225. [PubMed] [Google Scholar]
- 42.Kasuya H, Nishiyama Y, Nomoto S, Hosono J, Takeda S, Nakao A. Intraperitoneal delivery of hrR3 and ganciclovir prolongs survival in mice with disseminated pancreatic cancer. Journal of Surgical Oncology. 1999;72(3):136–141. doi: 10.1002/(sici)1096-9098(199911)72:3<136::aid-jso5>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 43.Kasuya H, Nomoto S, Nakao A. The potential of gene therapy in the treatment of pancreatic cancer. Drugs of Today. 2002;38(7):457–464. doi: 10.1358/dot.2002.38.7.820114. [DOI] [PubMed] [Google Scholar]
- 44.Lee MJ, Cho SS, You JR, Lee Y, Kang BD, Choi JS, Park JW, Suh YL, Kim JA, Kim DK, Park JS. Intraperitoneal gene delivery mediated by a novel cationic liposome in a peritoneal disseminated ovarian cancer model. Gene Therapy. 2002;9(13):859–866. doi: 10.1038/sj.gt.3301704. [DOI] [PubMed] [Google Scholar]
- 45.Louis MH, Dutoit S, Denoux Y, Erbacher P, Deslandes E, Behr JP, Gauduchon P, Poulain L. Intraperitoneal linear polyethylenimine (L-PEI)-mediated gene delivery to ovarian carcinoma nodes in mice. Cancer Gene Therapy. 2006;13(4):367–374. doi: 10.1038/sj.cgt.7700893. [DOI] [PubMed] [Google Scholar]
- 46.Rocconi RP, Numnum TM, Stoff-Khalili M, Makhija S, Alvarez RD, Curiel DT. Targeted gene therapy for ovarian cancer. Current Gene Therapy. 2005;5(6):643–653. doi: 10.2174/156652305774964668. [DOI] [PubMed] [Google Scholar]
- 47.Rettig GR, McAnuff M, Liu DJ, Kim JS, Rice KG. Quantitative bioluminescence imaging of transgene expression in vivo. Analytical Biochemistry. 2006;355(1):90–94. doi: 10.1016/j.ab.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 48.De A, Gambhir SS. Noninvasive imaging of protein-protein interactions from live cells and living subjects using bioluminescence resonance energy transfer. Faseb Journal. 2005;19(12) doi: 10.1096/fj.05-4628fje. 2017-+ [DOI] [PubMed] [Google Scholar]
- 49.Doyle TC, Nawotka KA, Kawahara CB, Francis KP, Contag PR. Visualizing fungal infections in living mice using bioluminescent pathogenic Candida albicans strains transformed with the firefly luciferase gene. Microbial Pathogenesis. 2006;40(2):82–90. doi: 10.1016/j.micpath.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 50.Jawhara S, Mordon S. In vivo imaging of bioluminescent Escherichia coli in a cutaneous wound infection model for evaluation of an antibiotic therapy. Antimicrobial Agents and Chemotherapy. 2004;48(9):3436–3441. doi: 10.1128/AAC.48.9.3436-3441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenkins DE, Oei Y, Hornig YS, Yu SF, Dusich J, Purchio T, Contag PR. Bioluminescent imaging (BLI) to improve and refine traditional murine models of tumor growth and metastasis. Clinical & Experimental Metastasis. 2003;20(8):733–744. doi: 10.1023/b:clin.0000006815.49932.98. [DOI] [PubMed] [Google Scholar]
- 52.Sato A, Klaunberg B, Tolwani R. In vivo bioluminescence imaging. Comparative Medicine. 2004;54(6):631–634. [PubMed] [Google Scholar]
- 53.Rudolph C, Lausier J, Naundorf S, Muller RH, Rosenecker J. In vivo gene delivery to the lung using polyethylenimine and fractured polyamidoamine dendrimers. Journal of Gene Medicine. 2000;2(4):269–278. doi: 10.1002/1521-2254(200007/08)2:4<269::AID-JGM112>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 54.Bertschinger M, Backliwal G, Schertenleib A, Jordan M, Hacker DL, Wurm FM. Disassembly of polyethylenimine-DNA particles in vitro: Implications for polyethylenimine-mediated DNA delivery. Journal of Controlled Release. 2006;116(1):96–104. doi: 10.1016/j.jconrel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Salem AK, Hung CF, Kim TW, Wu TC, Searson PC, Leong KW. Multi-component nanorods for vaccination applications. Nanotechnology. 2005;16(4):484–487. [Google Scholar]
- 56.Zhang XQ, Dahle CE, Baman NK, Rich N, Weiner GJ, Salem AK. Potent antigen-specific immune responses stimulated by codelivery of CpG ODN and antigens in degradable microparticles. Journal of Immunotherapy. 2007;30(5):469–478. doi: 10.1097/CJI.0b013e31802fd8c6. [DOI] [PubMed] [Google Scholar]
- 57.Zhang XQ, Dahle CE, Weiner GJ, Salem AK. A comparative study of the antigen-specific immune response induced by co-delivery of CpG ODN and antigen using fusion molecules or biodegradable microparticles. Journal of Pharmaceutical Sciences. 2007;96:3283–3292. doi: 10.1002/jps.20978. [DOI] [PubMed] [Google Scholar]
- 58.Gharwan H, Wightman L, Kircheis R, Wagner E, Zatloukal K. Nonviral gene transfer into fetal mouse livers (a comparison between the cationic polymer PEI and naked DNA) Gene Therapy. 2003;10(9):810–817. doi: 10.1038/sj.gt.3301954. [DOI] [PubMed] [Google Scholar]
- 59.Ogris M, Walker G, Blessing T, Kircheis R, Wolschek M, Wagner E. Tumor-targeted gene therapy: strategies for the preparation of ligand-polyethylene glycol-polyethylenimine/DNA complexes. Journal of Controlled Release. 2003;91(1–2):173–181. doi: 10.1016/s0168-3659(03)00230-x. [DOI] [PubMed] [Google Scholar]
- 60.Ullman-Cullere MH, Foltz CJ. Body condition scoring: A rapid and accurate method for assessing health status in mice. Laboratory Animal Science. 1999;49(3):319–323. [PubMed] [Google Scholar]