Abstract
CHIR-AB1 is a newly identified avian immunoglobulin receptor that includes both activating and inhibitory motifs and was therefore classified as a potentially bifunctional receptor. Recently, CHIR-AB1 was shown to bind the Fc region of chicken IgY and induce calcium mobilization via association with the common γ-chain, a subunit that transmits signals upon ligation of many different immunoreceptors. Here we describe the 1.8Å resolution crystal structure of the CHIR-AB1 ectodomain. The receptor ectodomain consists of a single C2-type immunoglobulin (Ig) domain resembling the Ig-like domains found in mammalian Fc receptors such as FcγRs and FcαRI. Unlike these receptors and other monomeric IgSF members, CHIR-AB1 crystallized as a two-fold symmetric homodimer that bears no resemblance to variable or constant region dimers in an antibody. Analytical ultracentrifugation demonstrated that CHIR-AB1 exists as a mixture of monomers and dimers in solution, and equilibrium gel filtration revealed a 2:1 receptor-ligand binding stoichiometry. Measurement of the 1:1 CHIR-AB1/IgY interaction affinity indicates a relatively low affinity complex, but a 2:1 CHIR-AB1/IgY interaction allows an increase in apparent affinity due to avidity effects when the receptor is tethered to a surface. Taken together, these results add to the structural understanding of Fc receptors and their functional mechanisms.
Keywords: Fc receptor, crystal structure, dimer, chicken, bifunctional receptor
Introduction
Antibodies are critical components of the adaptive immune system that allow specific recognition of a remarkable repertoire of pathogens. In addition to direct neutralization of target antigens, antibodies also participate in the regulation of both adaptive and innate immune mechanisms via interactions with Fc receptors (FcRs)1. Association with FcRs induces inflammatory responses mediated by macrophages, mast cells, neutrophils and natural killer (NK) cells1. Importantly, FcRs not only trigger the immune response, but also control and limit its magnitude, thus providing a crucial mechanism to balance between immunological tolerance and activation2; 3.
In light of the multiples roles of FcRs and their involvement in various pathological disorders, a detailed understanding of the structure and function of FcRs has become a subject of increasing interest. In mammals, immunoglobulin superfamily (IgSF) member FcRs have been identified that are specific for different immunoglobulin classes (including IgG, IgA, IgE and IgM)1; 4. Current crystallographic data (available for FcγRIIa, FcγRIIb, FcεRI and FcαRI)5; 6; 7; 8; 9; 10; 11; 12 show a similar overall structure composed of two extracellular immunoglobulin (Ig)-like domains known as D1 and D2 (corresponding to the membrane distal and membrane proximal domains, respectively). Each of the two domains adopts a typical Ig-like fold that includes two antiparallel β sheets connected by a conserved intradomain disulfide bond.
The origin of IgSF FcRs is not clear, but previous observations suggested that these receptors are evolutionary related to MHC class I-binding proteins13. Indeed, one FcR, FcαRI, is encoded within the leukocyte receptor cluster (LRC), a conserved genomic region that expresses a large number of IgSF receptors that are thought to have diverged from a common ancestor. In humans, these genes include the MHC class I-binding receptors KIRs and LIRs (killer and leukocyte Ig-like receptors, respectively) and the NK activating receptor NKp4614. A common feature shared by many LRC-encoded genes is the expression of both inhibitory and activating counterparts that deliver opposing signals upon binding to the same ligand15. Activating receptors are characterized by a relatively short cytoplasmic tail and a charged amino acid in the transmembrane domain that facilitates association with adaptor molecules such as the common γ-chain, a signaling protein that triggers activation15. The inhibitory receptors express a relatively longer cytoplasmic tail and carry at least one immunoreceptor tyrosine inhibitory motif (ITIM), which interacts with cytosolic phosphatases to attenuate activation signals15.
Recently, a new family of chicken Ig receptors (CHIRs) with homology to human LIRs and KIRs was identified13; 16; 17. The CHIRs are encoded on chicken chromosome 31 in a region that corresponds to the mammalian LRC17; 18 and includes a large family of highly polymorphic genes predicted to encode both activating (CHIR-A), inhibitory (CHIR-B), or bifunctional (CHIR-AB) receptors16; 17. Interestingly, one of these receptors, CHIR-AB1, was shown to function as a classical FcR expressed on chicken B cells, macrophages, monocytes and NK cells19. Unlike mammalian FcRs that include two or three extracellular Ig-like domains and carry either activating or inhibitory motifs1, CHIR-AB1 is composed of a single Ig-like domain and is classified as a bifunctional receptor due to the expression of both a charged amino acid in its transmembrane domain and an ITIM motif in its cytoplasmic tail. A specific interaction between CHIR-AB1 and the Fc portion of IgY, the avian counterpart of mammalian IgG, was shown to enhance calcium release in a chicken B cell line expressing CHIR-AB1 and the common γ chain19. Notably, the activation required aggregation of IgY, thus suggesting that immune complexes are required to trigger an activating response19.
Here we describe the 1.8Å crystal structure of the extracellular domain of CHIR-AB1. Although the overall structure of CHIR-AB1 closely resembles Ig-like domains found in other immunoreceptors, CHIR-AB1 differs from mammalian FcRs and LRC members in its ability to form homodimers. We provide biochemical evidence for the formation of CHIR-AB1 dimers in solution and characterize the stoichiometry and binding affinity to IgY. Based on these observations, we suggest a general mechanism by which bifunctional receptors mediate their dual function.
Results
Overview of the structure of CHIR-AB1
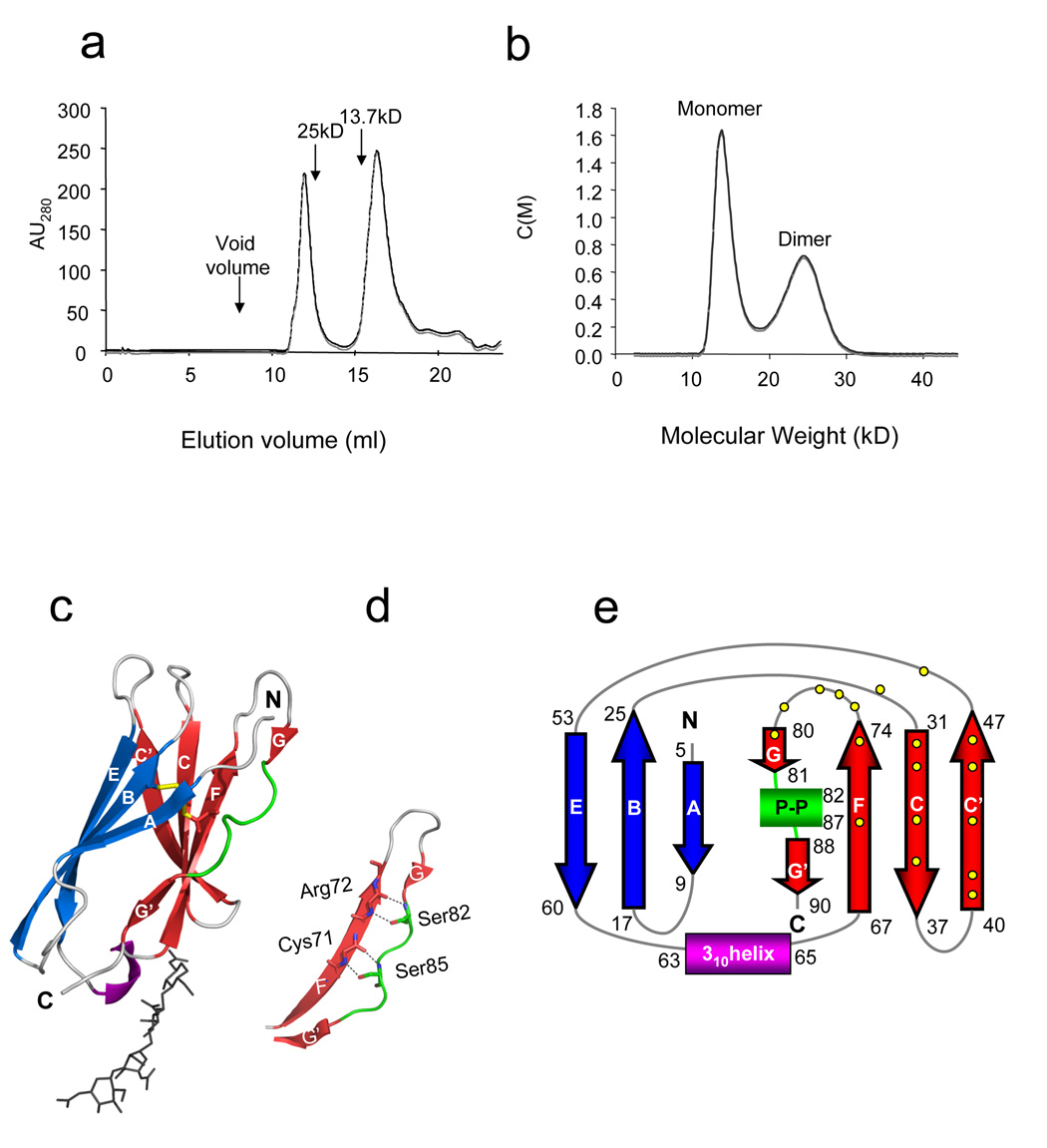
The extracellular domain of CHIR-AB1 (residues 1–93 of the mature protein) was expressed as a secreted protein in insect cells. Purified CHIR-AB1 migrated as two peaks on a gel filtration column suggesting more than one oligomeric state (Figure 1a). Re-injection of the dimer peak over a gel filtration column resulted in partial dissociation of the dimers and the appearance of monomers, thus suggesting that dimer formation is concentration dependent. To further characterize the oligomeric state of CHIR-AB1, we performed sedimentation velocity analytical ultracentrifugation. The sedimentation profile was analyzed by a continuous c(M) Lamm equation model resulting in two peaks with calculated average molecular weights corresponding to 14.3kD and 25.0kD (Figure 1b). As the molecular weight of the monomeric CHIR-AB1 is 13.4kD (determined by mass spectrometry, data not shown), these results demonstrated that CHIR-AB1 formed both monomers and dimers in solution. The monomeric fraction of CHIR-AB1 was isolated by gel filtration chromatography and crystallized in space group P3121 with one molecule per asymmetric unit. The structure was solved by molecular replacement using the D1 domain of KIR2DL120 as a search model and refined to 1.8 Å (Table 1).
Figure 1. Structure of CHIR-AB1.
(a). Elution profile of purified CHIR-AB1 on a S200 Superdex gel filtration column. The elution volumes of molecular weight standards are indicated by arrows. (b) Velocity sedimentation analytical ultracentrifugation analysis of purified CHIR-AB1 showing the existence of monomeric (14.3kD) and dimeric (25.0kD) forms. Data are displayed at the 95% confidence level. (c) Ribbon diagram of the CHIR-AB1 structure. β-strands A, B, and E are shown in blue, strands CC’FGG’ in red, a 310 helix in purple and a polyproline II helix in green. The disulfide bond is yellow and carbohydrate residues are shown in stick representation. (d) Close-up view of the region of the polyproline II helix showing hydrogen bonds between the side chains of Ser 82 and Ser 85 and main chain atoms of residues Arg72 and Cys71. Side chains are shown as sticks using atom-based color code (oxygen, red; nitrogen, blue). (e) Topology diagram of CHIR-AB1. β-strands, 310 helix and the polyproline II helix (P-P) are colored as above. Contact resides involved in dimerization (Figure 2 and Table 2) are represented by yellow dots.
Table 1. Crystallographic Data and Refinement Statistics.
| Unit cell | Space group | P3121 |
|---|---|---|
| Cell dimensions: | a, b, c (Å) | 63.9, 63.9, 55.4 |
| Data collection | ||
| Resolution (Å) | 1.8 | |
| aRmerge(%) | 4.4 (42.6) | |
| I / σI | 41.9 (4.3) | |
| Completeness (%) | 95.9 (93.9) | |
| Redundancy | 3.8 (3.8) | |
| Refinement statistics | ||
| Resolution (Å) | 27.7-1.8 | |
| No. of unique reflections | 12020 | |
| No. of reflections used | 10932 | |
| cRcryst / dRfree | 0.20/0.24 | |
| # atoms (B factor) | Protein | 737 (28.1 Å2) |
| Carbohydrate | 38 (43.4 Å2) | |
| Water | 116 (41.4 Å2) | |
| Model geometry | ||
| R.m.s. deviation from ideality: | ||
| Bond length (Å) | 0.014 | |
| Angles (°) | 1.667 | |
| Ramachandran plot | ||
| Most favored (%) | 91.4 | |
| Additionally allowed (%) | 8.6 | |
| Generously allowed (%) | 0.0 | |
| Disallowed (%) | 0.0 |
Rmerge = 100 × Σ|I − 〈I〉| /ΣI, where I is the integrated intensity of a given reflection. Values in parentheses are for the highest resolution shell (1.89-1.82 Å).
Rcryst = 100 × Σ Fobs − Fcalc ′ /Σ Fobs, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively, for reflections in the working set.
Fobs − Fcalc ′ /Σ Fobs, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively, for reflections in the working set.
Rfree was calculated for the test set containing randomly chosen reflections (5% of the data) that were not included in the refinement.
The CHIR-AB1 ectodomain is a C2-type IgSF domain21 with eight β-strands assembled into two antiparallel β-sheets. The first β-sheet is formed by strands A, B and E, and the second sheet is formed by strands C, C’, F, G and G’ (Figure 1c,e). The sheets are connected by a disulfide bridge involving Cys24 and Cys71 on strands B and F, respectively. The final β-strand in the structure is divided into two short segments, G and G’, which are separated by six residues in a left-handed polyproline II helical conformation. The side chains of two serines in this motive form hydrogen bonds with main chain atoms of strand F (Figure 1d). This type of interaction was suggested to contribute to the stability of the core Ig fold and is conserved among many Ig-like domains including the CHIR-related receptors such as KIRs20, LIRs22, NKp4623 and Fc receptors5; 6; 7; 8; 9; 10; 11; 12. Other features of the CHIR-AB1 structure include ordered carbohydrate in the CC’ loop attached to Asn38, one of two potential N-linked glycosylation sites in the CHIR-AB1 ectodomain, and a 310 helix in the EF loop (Leu63-Ala65).
CHIR-AB1 forms homodimers
Although the CHIR-AB1 protein used for crystallization was purified from the monomeric peak, crystal packing created a symmetric CHIR-AB1 dimer in which residues at the CC’FGG’ face interact across a crystallographic two-fold axis (Figure 2). Most of the dimerization surface involves the N-terminal tip of the CHIR-AB1 Ig-like domain. At the C-terminal tip of the Ig-like domain, the subunits of the dimer are separated. The C-termini of the model of each subunit connect to a linking region in an orientation that would allow anchoring to a common membrane (Figure 2c), compatible with dimerization in cis at the surface of a cell.
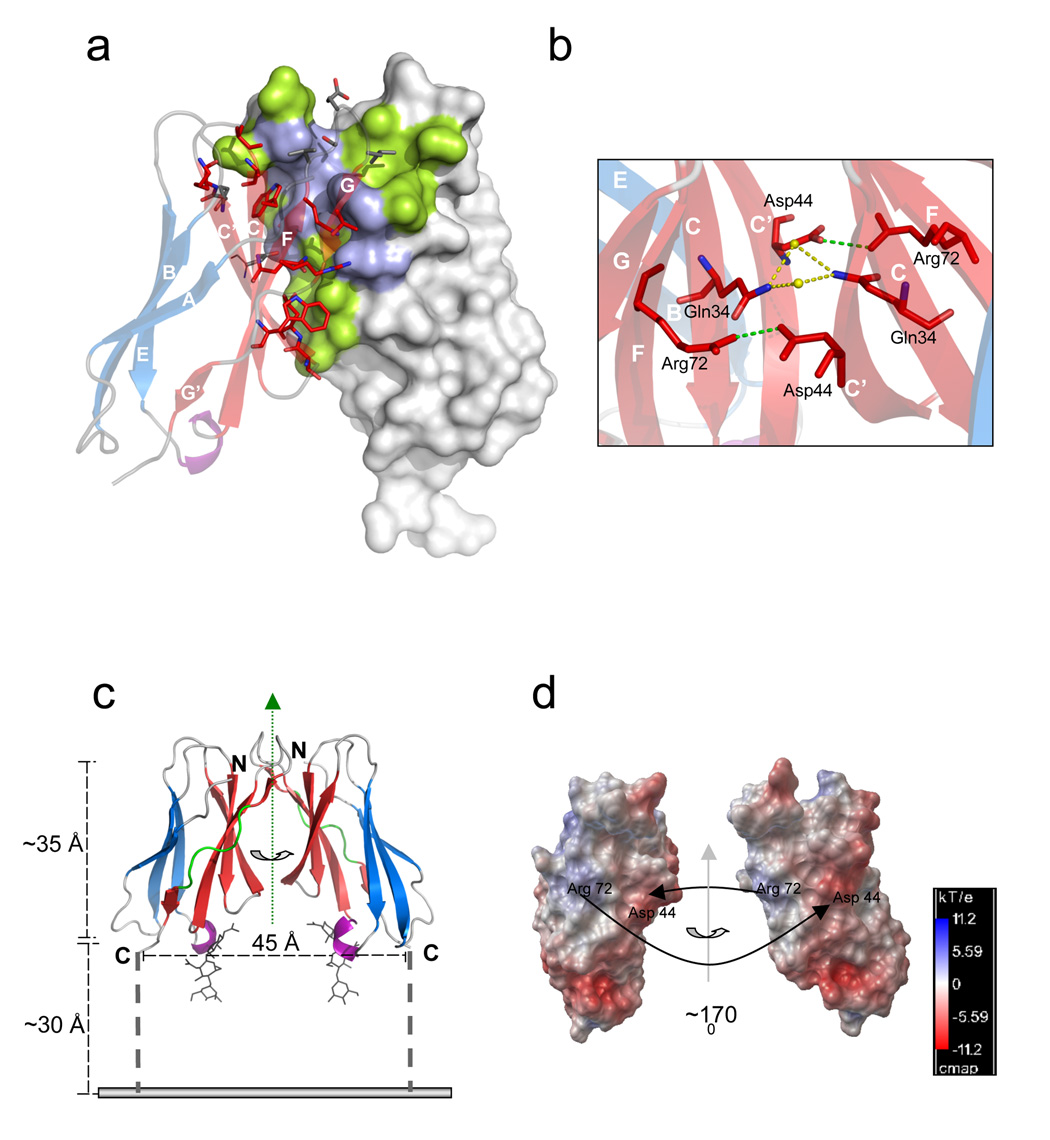
Figure 2. The dimer interface.
(a) Dimerization interface region with one subunit as a surface representation (hydrophilic contact residues in light blue and hydrophobic contact residues in green) and the other subunit as transparent ribbons diagram colored as in Figure 1b (β-strands A, B, and E are shown in blue, strands CC’FGG’ in red, a 310 helix in purple and loops are colored grey). Strands are labeled in white and contact residues are shown as sticks. (b) Cross section of the dimer interface illustrating the two symmetrical inter-subunit salt bridges (green lines) between Asp44 and Arg72. Water molecules (yellow spheres) that mediate a hydrogen bond network (yellow dotted line) between the Gln34 residues of each subunit are also shown. Strands are labeled in white and colored as above (c) Proposed orientation of a CHIR-AB1 dimer on a membrane (grey line). Monomers are colored as in Figure 1b. The two-fold symmetry axis relating the monomers is indicated by a vertical green dotted arrow in the plane of the page. The distance between the C-terminus of each ectodomain (45Å) and the approximate length of the ectodomain (~35Å) are indicated (black dashed lines). The 8-residue stem region connecting each ectodomain subunit to the transmembrane region is shown as a broken black bar with a maximum theoretical length of ~30Å (calculated assuming 3.8Å per residue). (d) Molecular surfaces of the two subunits in a CHIR-AB1 dimer with colors highlighting the electrostatic potential calculated with APBS tools 56. Electrostatic potential is plotted from −11.2 kT/e (electronegative; red) to +11.2 kT/e (electropositive; blue) with white indicating electroneutrality. The right monomer is related to the left monomer by a rotation about the indicated axis of ~170°. Black arrows point toward contact partner residues.
The dimer interface consists of a central hydrophobic patch surrounded by hydrophilic residues (Figure 2a,b) and includes 16 ordered water molecules involved in hydrogen bonds connecting the two subunits. To compare the CHIR-AB1 dimer to other protein-protein interactions, we calculated the shape complementarity index (SC)24, an index that varies from 0 (not complementary) to 1 (a perfect fit). The CHIR-AB1 dimer has a high SC value of 0.74, indicating a complementary fit, as compared to 0.64–0.68 for typical antibody/antigen interfaces24. However, the inclusion of water molecules at the dimer interface and the relatively small buried surface area (740 Å2 per subunit, as compared with 780 – 850 Å2 per subunit in typical protein-protein interactions25) is consistent with a monomer-dimer equilibrium, as observed in solution (Figure 1a,b), rather than obligate dimerization.
An analysis of electrostatic potentials reveals that the dimer interface includes a positive patch dominated by Arg72 and a negative patch dominated by Asp44 (Figure 2d). The negative and positive patches are paired symmetrically in the dimer, allowing complementary interactions between oppositely-charged counterions on each monomer – thus Arg72 from each subunit forms a salt bridge with Asp44 from the partner subunit (Table 2 and Figure 2a,b). Below the dimer interface (as oriented in Figure 2c), a cluster of negatively-charged residues creates electrostatic repulsion that pushes the subunits apart, and perhaps prevents a close interaction with negatively-charged lipid head groups at the membrane-proximal portion of the dimer. Steric restrictions caused by N-linked carbohydrate attached to Asn38 may also prevent a close approach of CHIR-AB1 to the membrane and facilitate orientation in the “upright” position depicted in Figure 2c.
Table 2. Amino acid contacts at the dimer interface.
Distance and geometry criteria for assigning hydrogen bonds (HB): a distance of <3.5Å and a hydrogen bond angle >90°. The maximum allowed distance for a van der Waals (VDW) interaction was 4Å and the salt bridge distance cutoff was ≤ 3.5Å.
| Monomer A residue | Partner residue on monomer B | Distance (Å) | Type of interaction | Buried surface area (Å2 per monomer) |
|---|---|---|---|---|
| Ala30 | Leu79 | 3.5 | VDW | 47.1 |
| Ala31 | Glu77 | 4 | VDW | 10.1 |
| Trp32 | Val75 | 3.1 | HB | 116.6 |
| Trp80 | 3.3 | HB | ||
| Gln34 | Gln34 | 3.6 | VDW | 20.9 |
| Trp36 | Leu41 | 3.8 | VDW | 14.4 |
| Thr40 | Thr40 | 3.6 | VDW | 33.0 |
| Leu41 | Trp36 | 3.8 | VDW | 62.8 |
| Asp44 | Gln34 | 3.2 | HB | 39.0 |
| Arg72 | 2.9 | Salt bridge | ||
| Glu46 | Leu79 | 3.8 | VDW | 24.8 |
| Lys47 | Leu79 | 3.7 | VDW | 2.8 |
| Asp48 | Leu79 | 3.8 | VDW | 10.8 |
| Arg72 | Asp44 | 2.9 | Salt bridge | 24.1 |
| Gln74 | Asp44 | 3.2 | HB | 55.6 |
| Val75 | Trp32 | 3.1 | HB | 15.7 |
| Ser76 | Ser76 | 3.3 | H-bond | 72.8 |
| Glu77 | Ala31 | 4 | VDW | 52.0 |
| Leu79 | Ala30 | 3.5 | VDW | 120.9 |
| Glu46 | 3.8 | |||
| Lys47 | 3.7 | |||
| Asp48 | 3.8 | |||
| Trp80 | Trp32 | 3.3 | HB | 4.9 |
Comparison of CHIR-AB1 with related IgSF immunoreceptors
Although CHIR-AB1 binds and responds to the Fc region of chicken IgY, thus establishing its function as a classical Fc receptor19, it shares only low sequence identity with other known IgSF FcRs (< 28%)13. Its closest mammalian relative is not a FcR, but rather the MHC class I-binding receptor KIR2DL1 (33% identity). Superimposition of CHIR-AB1 with the D1 domain of KIR2DL120 results in a root mean square (r.m.s) deviation of 2.5 Å (for 92 carbon-α atoms) (Figure S1a). While the overall folding topologies are similar, KIR2DL1 is monomeric20 and differs considerably from CHIR-AB1 in the length and conformations of its loops. In particularly, significant differences are found in the CC’ and FG loops, which participate in the CHIR-AB1 dimer interface. KIR2DL1 D1 also lacks counterparts of the CHIR-AB1 Asp44 and Arg72 residues and the corresponding positive and negative patches that facilitate CHIR-AB1 dimerization (Figure S1d). Another structural difference between CHIR-AB1 and KIR2DL1 is that the A strand in CHIR-AB1 does not switch to join the CC’FG face. Although the CHIR-AB1 structure includes a cis-proline (Pro10) in strand A that is conserved with KIRs, the break it introduces into strand A is not followed by a switch to the opposite β-sheet to create an A’ strand, as occurs in KIRs20; 26; 27; 28. The absence of an A’ strand in CHIR-AB1 is notable since this feature is shared by most C2-style Ig-like domains and is found among many related proteins besides KIRs, including LIRs22; 29 and mammalian FcRs5; 6; 7; 8; 9; 10; 11; 12.
A search of available structures in the Protein Data Bank30 using the DALI server31 identified the activating receptor of human NK cells, NKp46, as the closest structural homologue of CHIR-AB1. The NKp46 ectodomain contains two closely-related Ig-like domains23; 32 that share a low level of sequence identity with CHIR-AB1 (28.6% and 29.2% for D1 and D2, respectively). Structural alignments show that CHIR-AB1 most closely resembles NKp46-D1, resulting in a r.m.s deviation of 1.4 Å (89 carbon-α atoms, Figure S1c). Differences are observed mostly in the C’E and FG loops and in the absence of the A’ strand in CHIR-AB1.
When compared only to the structures of other IgSF FcRs, the FcR with the highest structural similarity to CHIR-AB1 is the D1 domain of FcαRI (r.m.s deviation of 2.9 Å for 88 carbon-α atoms; Supplemental Figure 1b). Given that the Fc binding site on FcαRI differs from the counterpart region on CHIR-AB1 (Supplemental Figure 1b), it is unlikely that the two proteins share a common mode of binding to Fc regions.
Stoichiometry of the CHIR-AB1/FcY complex
Classical IgSF FcRs have previously been shown to interact with their Fc ligands with a stoichiometry of either 1:1 (FcγRs and FcεRI) or 2:1 (FcαRI)4. To determine the stoichiometry of the CHIR-AB1/FcY interaction, we used equilibrium gel filtration chromatography. In this form of chromatography, gel filtration is conducted under equilibrium conditions by including one of the binding partners in the running buffer33, thereby preventing dissociation of weak complexes that could result in an altered stoichiometry.
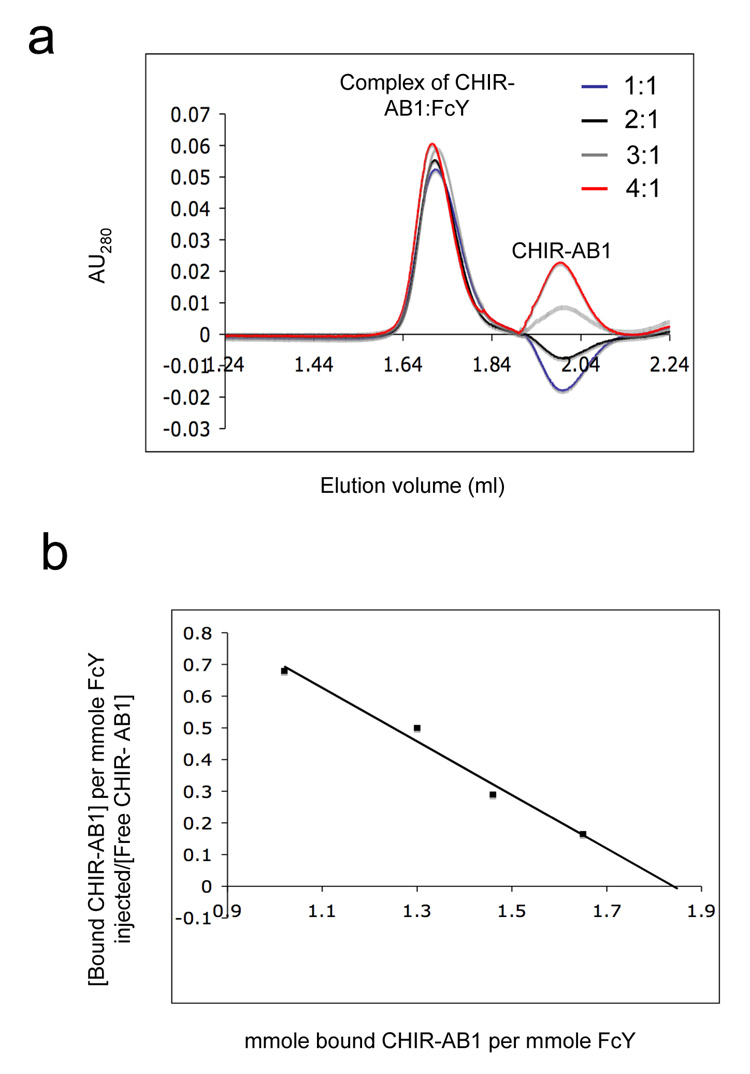
In our experiments, a gel filtration column was equilibrated with and run in a buffer containing a fixed concentration of purified monomeric CHIR-AB1. Samples containing different ratios of CHIR-AB1 to FcY were injected onto the column in the equilibration buffer. Figure 3a shows the elution pattern of CHIR-AB1/FcY complexes obtained at the indicated injected ratios over a column equilibrated with 2.5µM CHIR-AB1. A trough corresponding to the position where CHIR-AB1 migrates was observed when sample contained CHIR-AB1 mixed with FcY at 1:1 molar ratio, indicating a depletion of the receptor in the running buffer. The depth of the trough was reduced at a 2:1 ratio of CHIR-AB1 to FcY, and was replaced by a positive peak at CHIR-AB1:FcY ratios of 3:1 and 4:1 (Figure 3a). Similar sets of injections were performed with 1.5µM, 5µM and 10µM CHIR-AB1 in the equilibration buffer (data not shown). By determining the ratio of injected proteins at which no peak or trough appears at a series of concentrations of CHIR-AB1 in the equilibration buffer, the data could be converted into a Scatchard plot, allowing determination of both the stoichiometry and solution phase affinity34 (Figure 3b). The Scatchard plot shows an x-intercept of 1.8, which is most consistent with a 2:1 CHIR-AB1:FcY stoichiometry, such that two CHIR-AB1 molecules bind per homodimeric FcY. The derived solution phase affinity corresponds to an equilibrium dissociation constant (KD) of ~840nM.
Figure 3. CHIR-AB1-FcY binding stoichiometry.
(a) Equilibrium gel filtration analysis of the CHIR-AB1-FcY complex. The column was equilibrated with and run in a buffer containing 2.5 µM CHIR-AB1. Complexes were prepared by incubating 2.5 µM FcY with varying amounts of CHIR-AB1 at receptor:FcY molar ratios of 1:1, 2:1, 3:1 and 4:1. Similar experiments were conducted using columns equilibrated with 1.5µM, 5µM and 10 µM of CHIR-AB1 (data not shown). (b) Scatchard plot including data from equilibration buffers containing 1.5µM, 2.5µM, 5µM and 10µM CHIR-AB1. The best fit line to the data yields a KD of 840 nM and an x-intercept of 1.83.
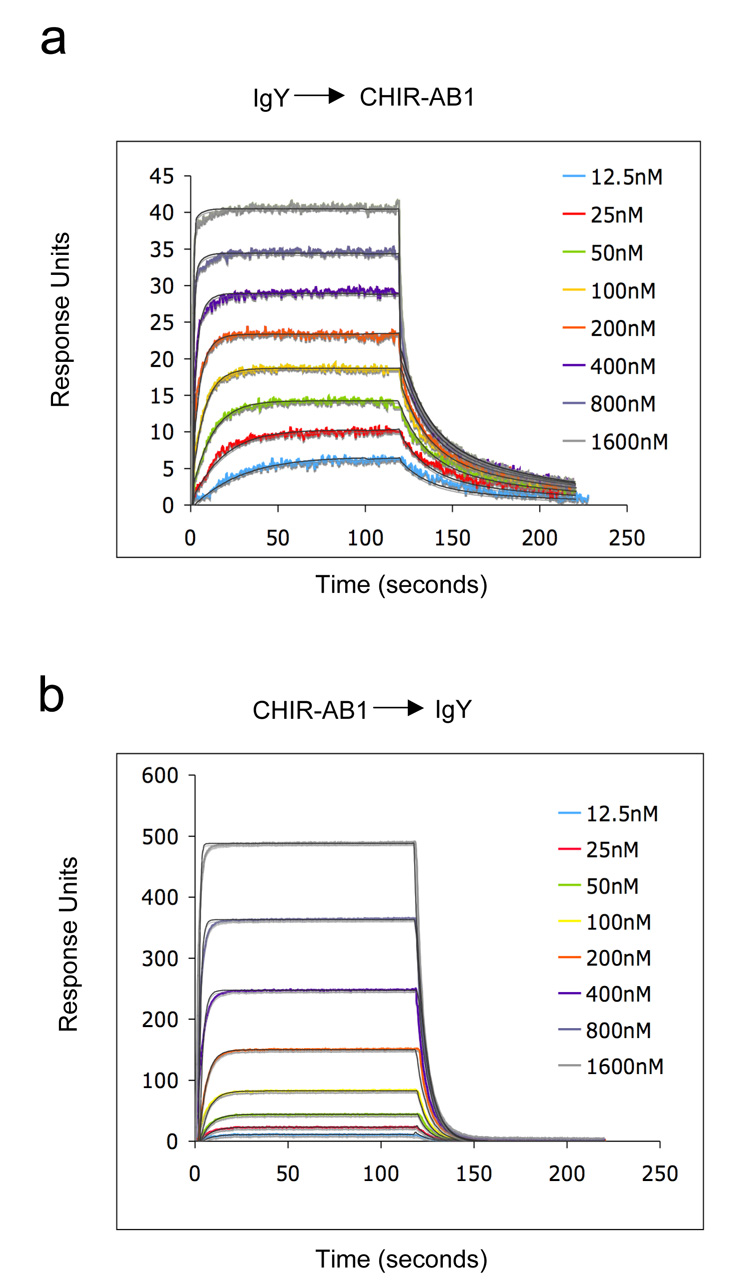
A solution phase affinity does not take into account potential avidity effects that can occur in a 2:1 receptor:ligand interaction. An increased apparent affinity due to avidity can result when a ligand cross-links receptors attached to a membrane or other surface. Indeed, when we used a 1:1 binding model to analyze binding data for IgY injected over CHIR-AB1 immobilized on a biosensor surface, we derived a macroscopic, or apparent, KD of 17 ± 9 nM (Figure 4a). This value is similar to a recently-reported KD derived from a SPR analysis of IgY binding to immobilized CHIR-AB1, which also assumed 1:1 binding19. We also analyzed the binding data in Figure 4a using a stepwise, or sequential, binding model (bivalent analyte binding model), which yielded microscopic KD values of 403 ± 30 nM and 3.9 ± 1.5 µM for the binding of the first and second CHIR-AB1 molecules to IgY. These values are consistent with KD values derived under conditions in which cross-linking could not increase the apparent affinity through avidity effects: ~840nM calculated from the equilibrium gel filtration experiments in solution (Figure 3b) and ~800nM derived from SPR experiments in which CHIR-AB1 was injected over immobilized IgY (Figure 4b).
Figure 4. Biosensor analyses of CHIR-AB1/IgY interactions.
Representative sensorgrams describing the binding response of increasing concentrations of IgY injected over immobilized CHIR-AB1 (58RUs) (a) or CHIR-AB1 injected over immobilized IgY (2000RUs) (b). Colored lines show the observed response overlaid with the calculated response (black line) based on a 2:1 binding model. Fitting of the same data with a 1:1 binding model resulted with KD values of 17 ± 9nM (a) and 800nM (b). Similar results were obtained when CHIR-AB1 was coupled to the chip at different densities (140RU, 320RU and 560RU; data not shown).
Localization of the FcY-binding site on CHIR-AB1
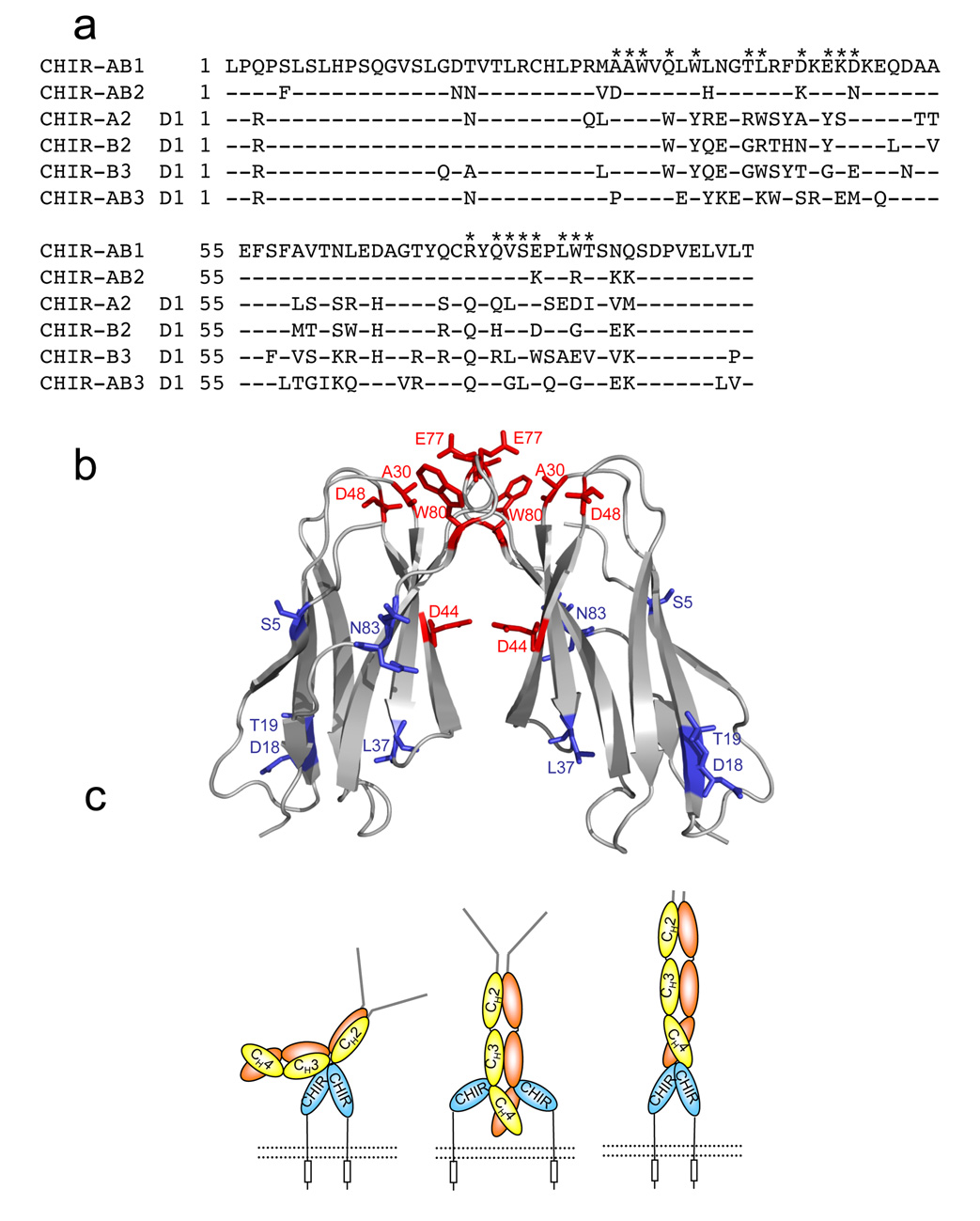
CHIR-AB1 belongs to a large family of highly polymorphic genes that show a high degree of sequence similarity17. Surprisingly, CHIR-AB1 is the only CHIR family member found to interact with IgY; other CHIR receptors, including CHIR-A2, CHIR-B2, CHIR-B3 and CHIR-AB3, exhibited no binding to IgY19. To gain insight into which residues confer the ability to bind IgY onto a CHIR protein, we mapped the amino acids differences between CHIR-AB1 and CHIR-AB2, which are related by 87% amino acid identity (Figure 5a), onto the CHIR-AB1 structure. To evaluate the potential significance of amino acid differences between CHIR-AB1 and CHIR-AB2, we compared these sequences to sequences of other non-IgY-binding CHIRs (CHIR-A2-D1, CHIR-B2-D1, and CHIR-B3-D1).
Figure 5. Models for the CHIR-AB1/FcY interaction.
(a) Sequence alignment of the extracellular domains of CHIR-AB1 and CHIR AB2 with the D1 domains of CHIR-A2, CHIR-B2, and CHIR-AB3 (GenBank accession numbers AJ745094, AJ745095, AJ745093, AJ639837 and AJ879909, respectively). Residues at the dimer interface of CHIR-AB1 are indicated by asterisks above the sequence. (b) Ribbon diagram of a CHIR-AB1 dimer with residues that differ from CHIR-AB2 highlighted as red (dimer interface residues) or blue (all others) sticks. The highlighted amino acids are labeled (c) Potential models for binding between CHIR-AB1 and IgY. The CHIR-AB1 ectodomain is blue, the ITIM motif in the cytoplasmic tail is represented by a rectangle, and the cell membrane is shown as a dotted black line. IgY is shown with a yellow and orange Fc region and grey Fabs. (Left) A dimer of CHIR-AB1 is bound asymmetrically to the lower hinge region between the CH2 and CH3 domains of FcY, analogous to the binding of FcγRs and FcεRI to Fcs. (Middle) CHIR-AB1 monomers bind to the CH3-CH4 interdomain interface to create a symmetrical 2:1 complex, analogous to the binding of FcαRI to Fc. (Right) The two-fold symmetry axis of a CHIR-AB1 dimer aligns with the two-fold symmetry axis of FcY to form a symmetric 2:1 complex in which each CHIR-AB1 monomer binds to the bottom of a FcY CH4 domain.
Comparison of the differences between CHIR-AB1 and -AB2 shows a cluster of unconserved amino acids located at the membrane distal portion of the receptor (Figure 5b), suggesting that the FcY binding site involves this surface. Interestingly, some of the non-conserved amino acids overlap with residues at the dimerization interface (highlighted in red in Figure 5a,b). For example, Asp44, which is critical for dimerization of CHIR-AB1, is not conserved in any of the non-IgY binding CHIR members. Substitution of Asp44 for an uncharged residue is unlikely to allow dimerization in the other CHIRs, since residue 44 pairs with a positively-charged residue, Arg72, in the CHIR-AB1 dimer. Notably, in the case of CHIR-AB2, residue 44 is a lysine, whose positive charge is likely to hinder dimerization due to electrostatic repulsion with Arg72. Whether dimerization is required for FcY binding remains to be demonstrated, but these results suggest that, in addition to being unable to bind FcY or IgY, CHIR-AB2 does not dimerize.
Discussion
Here we report the biochemical and structural characterization of CHIR-AB1, a non-mammalian classical FcR identified in the chicken genome. Based on chromosomal localization and sequence similarities, CHIR-AB1 was recognized as a member of the chicken LRC family17, suggesting a close evolutionary relationship with mammalian LRC members. In agreement, the crystal structure of CHIR-AB1 reveals structural similarity to the canonical Ig-like domains found in the membrane-distal D1 domains of mammalian LRC genes, and shows closer similarity to FcαRI, an LRC member, than to other FcRs. The relation between CHIR-AB1 and FcαRI is also highlighted by the 2:1 binding stoichiometry of both receptor:Fc interactions as compared with the 1:1 stoichiometry for the interactions of other IgSF FcRs with Fcs4.
Interestingly, despite low sequence identity (less than 29%), the closest structural homologue of CHIR-AB1 is the D1 domain of NKp46, an LRC member that is exclusively expressed on NK cells and plays a critical role in the recognition and killing of NK targets including tumors and virally infected cells35. Although NKp46 can bind to a variety of target cells, the nature and identity of its specific ligands is largely unknown – the only identified NKp46 ligand is influenza virus hemagglutinin, which binds and activates NKp46 in a sialic acid-dependent manner36; 37. The binding of NKp46 to hemagglutinin and putative tumor ligands depends entirely on the D2 domain of the receptor, thus the function of the NKp46 D1 domain is not clear38. In light of its striking similarity to CHIR-AB1, knowledge of the mode by which CHIR-AB1 binds its FcY ligand may shed light on the unknown function and binding properties of NKp46-D1.
Our findings reveal that CHIR-AB1 is unique among classical FcRs not only in its single Ig-like domain, but also in its ability to form symmetric homodimers. The dimerization of CHIR-AB1 was independent of FcY and was observed in crystals and in solution, as confirmed by size exclusion chromatography and analytical ultracentrifugation assays. Importantly, CHIR-AB1 is not an obligate homodimer; rather it exists in solution as a mixture of monomers and dimers. Such dimers may also occur at the surface of the cell where clustering could facilitate dimerization. Indeed, the presence of water molecules at the dimer interface is consistent with dimerization being a dynamic reversible process, which is also suggested by the observation that purified monomeric CHIR-AB1 crystallized as a dimer and purified dimers dissociated into monomers upon storage and/or dilution (data not shown).
The location of the CHIR-AB1 binding site on IgY is currently unknown. The Fc region of IgY, in common with Fc regions of mammalian IgM and IgE, contains three constant regions: CH2, CH3, and CH4, where the CH3 and CH4 domains are the counterparts of the CH2 and CH3 domains in an IgG Fc. In both this and a previous study19, the FcY fragment used for binding studies contained only the CH3 and CH4 domains, thus we know that the CH2 domains of FcY are not critical for binding to CHIR-AB1. Based on the 2:1 CHIR-AB1:FcY binding stoichiometry and the observation of CHIR-AB1 dimers, we can consider several options for the binding site within the CH3-CH4 domains of FcY (Figure 5c). One possibility is that CHIR-AB1 interacts with the N-terminal region of two FcY CH3 domains in an asymmetrical manner, analogous to the FcεRI-IgE and FcγR-IgG interactions4. In these examples, binding of the monomeric FcR to the lower hinge region and the CH2 domain of the IgG Fc or the CH3 domain of the IgE Fc induces structural asymmetry and steric interference that prevents binding of a second receptor to the symmetry-related location on the dimeric Fc, resulting in a 1:1 complex. Since CHIR-AB1:FcY forms a 2:1 complex, this mode of interaction can only be envisioned if FcY binds dimeric CHIR-AB1, thus implying that receptor dimerization is required for FcY binding. A second possibility, analogous to the interaction between FcαRI and IgA39, is that CHIR-AB1 binds to the interface between the FcY CH3 and CH4 domains, a region analogous to the “hotspot” for receptor binding to IgG Fc40. In this case, a 2:1 CHIR-AB1/FcY stoichiometry could only be achieved if monomers of CHIR-AB1 bind to each chain of the FcY homodimer. A third possibility, as yet unprecedented among FcRs, is that the two-fold symmetry axes of the CHIR-AB1 dimer and FcY align, such that each subunit of a CHIR-AB1 dimer binds an equivalent site on a symmetrical FcY dimer. This mode of binding would probably only be possible at the base of the FcY CH4 domains, as the analogous symmetric site on the CH3 domains would likely be blocked by the CH2 domains. Although this mode of interaction has not been seen in other FcR/Ig interactions, CHIR-AB1 is the only classical FcR that forms symmetrical dimers, thus it may have different binding properties than other known FcRs.
Cross-linking of surface receptors is recognized as a general mechanism by which ligand binding induces signal transduction and receptor activation. Since immune complexes of IgY are required for activation of cell surface CHIR-AB119, spontaneous dimerization of CHIR-AB1 is not likely to deliver activation signals. This assumption is supported by the structure of a CHIR-AB1 dimer, in which the C-termini of the two subunits are separated by ~45Å, suggesting that the connecting transmembrane domains of two interacting monomers would be too far apart to trigger a response. Binding of IgY-immune complexes may therefore be required to initiate clustering that would bring several receptors into proximity to trigger a signaling event.
Alternatively, it is possible that constitutive activation is inhibited by the presence of the ITIM motif in the CHIR-AB1 cytoplasmic tail. Although an inhibitory function for CHIR-AB1 has not been observed, the presence of an ITIM motif in its cytoplasmic tail is consistent with an inhibitory function that may be relevant under specific conditions or in particular cell types. The concept of activating receptors counterbalanced by inhibitory partners that interact with the same ligands is a repeating theme in the vertebrate immune system and is common among LRC members. In addition, bifunctional receptors have been identified in primates, including KIR2DL4 and NKp4441. Although both receptors were originally identified based on their ability to enhance NK-mediated cytotoxicity in activated NK cells42; 43; 44; 45 further studies demonstrated that KIR2DL426 and NKp4446 can also function as inhibitory receptors. Interestingly, the possibility that NKp44 can dimerize was recently suggested based on the observation of NKp44 dimers in the crystal lattice47. Further studies are required to evaluate the functional relevance of the ITIM motif in the cytoplasmic tail of CHIR-AB1 and the physiological significance of CHIR-AB1 dimerization.
Material and methods
Protein Expression and Purification
The cDNA encoding the extracellular domain of CHIR-AB1 (residues Gln1-Thr 95 of the mature protein) (GenBank accession no. AJ745094) was subcloned into the pAcGP67A baculovirus transfer vector (BD Biosynthesis), which includes a gp67 secretion signal, in frame with a C-terminal 6xHis tag sequence. Recombinant baculoviruses were generated by cotransfection of the transfer vector with linear DNA (Baculogold, BD Biosynthesis). Supernatants were harvested from Hi5 insect cells, and then concentrated and buffer exchanged against 40mM Tris, 300mM NaCl, pH 8. The expressed protein was purified using Ni-NTA affinity chromatography (Ni-NTA Superflow, Qiagen) followed by size exclusion column chromatography on a Superdex 75 16/60 column (Amersham Biosciences).
The cDNA encoding the CH3-CH4 domains of chicken IgY (residues 279–504 of the mature protein, referred to as ‘FcY’) was subcloned into the pAcGP67A baculovirus vector (BD Biosynthesis) in frame with an N-terminal 6xHis tag. FcY was expressed and purified as described for CHIR-AB1, except that the final purification step was done using a Superdex 200 16/60 column (Amersham Biosciences).
IgY was purchased from Jackson ImmunoResearch Laboratories and purified by gel filtration chromatography using a Superdex 200 16/60 column (Amersham Biosciences).
Protein concentrations were determined spectrophotometrically by measuring absorption at 280nm using extinction coefficients of 19,480 M−1cm−1 for CHIR-AB1, 11,200 M−1cm−1 for FcY, and 10,860 M−1cm−1 for IgY (calculated using the ProtParam tool on the ExPasy proteomic server).
Crystallization, Data Collection and Processing
Crystals of CHIR-AB1 were grown at 22°C in 1:1 hanging drops containing CHIR-AB1 (10–24mg/ml) and 2M ammonium sulfate. Single crystals were transferred to a cryoprotectant containing 2.2M ammonium sulfate and 25% glycerol prior to data collection. The crystals belong to space group P3121 (a = 63.69Å, b = 63.69Å, c = 55.45Å; one molecule per asymmetric unit). Data were collected to 1.8Å resolution at −170°C from a single crystal using an R-AXIS VII mounted on a Rigaku RU-200 rotating anode generator. Data were processed and scaled using HKL 2000.
Structure solution, Refinement and Analysis
To generate suitable protein models for molecular replacement calculations, a FFAS03 search against sequences with known structures was preformed48. For the ten highest scoring templates, an all-atom homology model and a 'mixed' model (non-identical sidechains replaced by serine) were built by the SCWRL server49. These models were input in various combinations into the programs PHASER50, MOLREP, and AMORE in space groups P321, P3121 and P3221. As crystallographic R-factors for most solutions were between 50% and 60%, a clear identification of a solution by data statistics alone was not possible, but visual inspection of the resulting electron density maps showed features not present in the original search model in a solution obtained by PHASER using a mixed model of PDB entry 1VDG in space group P3121. This potential solution (Z-score 7.1, LLG 67, initial Rcryst/Rfree 56%) was subjected to initial refinement using REFMAC51, which reduced the R values to 45% (Rcryst) and 53% (Rfree). The model was subsequently rebuilt using ARP/Warp52 and manual corrections. The complete model was refined at 1.8Å to a final Rcryst/Rfree of 22.1% and 24.6% with good geometry (Table 1). The final model consists of residues 1–93 of the mature CHIR-AB1 ectodomain, 116 water molecules and 38 atoms of carbohydrate [ordered carbohydrate was observed attached to the asparagine of one of the potential N-linked glycosylation sites (Asn38), but not to the other (Asn83)]. The sidechain of residue Glu63 was disordered and modeled as alanine. A disulphide bonds was observed between CHIR-AB1 residues Cys24 and Cys71.
Shape complementarity indices (SC) were calculated as described24 using the Sc program in the CCP4 suite53. Superimpositions were preformed using PyMol and the Combinatorial Extension (CE) Method58. Buried surface areas were calculated using the Protein interfaces, surfaces and assemblies service (PISA) at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html)57. Figures were generated using PyMOL40 (The PyMOL Molecular Graphics System, http://www.pymol.org).
Analytical Ultracentrifugation
Sedimentation velocity analytical ultracentrifugation experiments were performed at 20°C in a Beckman XL-I Ultima analytical ultracentrifuge using absorbance optics. CHIR-AB1 was freshly purified on a Superdex 75 16/60 column and loaded (420 µl, 9.4µM) into two-channel velocity cells with sapphire windows. Samples were spun at 55,000 rpm and 20°C, and absorbance scans were taken using the continuous scan mode as one replica with a point spacing of 0.003 mm. Data were analyzed using a continuous c(M) distribution model (direct fitting of the Lamm equation) with the program SedFit (www.analyticalultracentrifugation.com). Partial specific volume, buffer density, and viscosity values were calculated to be 0.712 ml/g, 1.00535 g/ml and 0.001002 poise, respectively, using the program Sednterp version 1.08.61. The partial specific volume of CHIR-AB1 was adjusted to reflect two N-linked glycosylation sites, predicted from the amino acid sequence and confirmed by mass spectrometry (data not shown).
Equilibrium Gel Filtration
The equilibrium gel filtration method33 including Scatchard analysis34 was used to analyze the association of CHIR-AB1 with FcY, as described previously54; 55. Chromatography was performed at a flow rate of 0.1 mL/min using a SMART micropurification system (Pharmacia). A Superdex 200 PC 3.2/30 gel filtration column was equilibrated with and run in 20 mM Tris pH 8.0, 150 mM NaCl containing 1.5µM, 2.5 µM, 5 µM or 10 µM monomeric CHIR-AB1 (equilibration buffer). For each concentration of CHIR-AB1 in the equilibration buffer, a series of injections were performed in which a fixed concentration of FcY (equal to the concentration of CHIR-AB1 in the equilibration buffer) was mixed with different concentrations of CHIR-AB1. Relative trough and peak areas were determined by integrating the CHIR-AB1 elution region using Excel. In each set of injections, the amount of bound CHIR-AB1 was calculated as previously described54; 55 and used for Scatchard analysis to determine the stoichiometry of CHIR-AB1:FcY binding and the solution KD.
Biosensor-based affinity measurements
The interaction between CHIR-AB1 and IgY was evaluated using a Biacore 2000 instrument (Pharmacia Biosensor, Uppsala, Sweden). To minimize signals resulting from aggregated proteins, samples were purified by gel filtration chromatography immediately prior to analysis. Purified CHIR-AB1 or IgY were covalently coupled to a CM5 sensor chip (Biacore) using the standard primary amine chemistry, as described in the Biacore manual. Proteins were coupled to a total of 58 resonance units (RUs) for CHIR-AB1 and 2000 RUs for IgY. One flow cell out of four on the biosensor chip was mock coupled with buffer only for background subtraction.
Measurements were performed at 20°C in 20 mM Hepes pH 7, 150 mM NaCl, and 0.005% surfactant P20. Increasing concentrations of analyte (injected protein) were injected at a flow rate of 50 µl/min, and association and dissociation phases were monitored for 7 min. Sensor signals returned to baseline after each injection without regeneration. After subtracting reference cell signals, the resulting binding data were globally fit to either a bivalent analyte or a 1:1 binding model (Biaevaluation software, Biacore). KD values were obtained by simultaneous fitting of the association and dissociation phases of all curves in the working set. The bivalent analyte model fits binding data to sequential reactions, deriving microscopic KD values for the binding reactions that produce singly- and doubly-liganded IgY proteins. The equilibrium binding constant KD2 was converted from RU units to molar units using the conversion factor M=RU/(100×molecular mass), based on the molar response in RUs and the volume of the flow-cell (Biacore AB). Corrections for statistical factors to KD1 and KD2 values derived from bivalent ligand models were applied as described54. A 1:1 binding model produces a macroscopic, or apparent, KD that does not take the avidity effects of a bivalent analyte (in this case, IgY) into account, thus it results in a higher apparent affinity for the binding of IgY to immobilized CHIR-AB1 than derived using a sequential binding model.
Supplementary Material
Acknowledgments
This work was supported by a Jane Coffin Childs Memorial Fund Fellowship for Medical Research (T.I.A), the National Institutes of Health (2 R37 AI041239-06A1 to P.J.B.), the Beckman Institute at Caltech (R.O.), funds from the Gordon and Betty Moore Foundation in support of the Molecular Observatory at Caltech, and a Deutsche Forschungsgemeinschaft grant GO489/3-6 (T.W.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Atomic coordinates have been deposited in the Protein Data Bank with accession code 2vsd.
References
- 1.Nimmerjahn F, Ravetch JV. Fc-receptors as regulators of immunity. Adv Immunol. 2007;96:179–204. doi: 10.1016/S0065-2776(07)96005-8. [DOI] [PubMed] [Google Scholar]
- 2.Nimmerjahn F, Ravetch JV. The antiinflammatory activity of IgG: the intravenous IgG paradox. J Exp Med. 2007;204:11–15. doi: 10.1084/jem.20061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 4.Woof JM, Burton DR. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat Rev Immunol. 2004;4:89–99. doi: 10.1038/nri1266. [DOI] [PubMed] [Google Scholar]
- 5.Garman SC, Kinet JP, Jardetzky TS. Crystal structure of the human high-affinity IgE receptor. Cell. 1998;95:951–961. doi: 10.1016/s0092-8674(00)81719-5. [DOI] [PubMed] [Google Scholar]
- 6.Garman SC, Wurzburg BA, Tarchevskaya SS, Kinet JP, Jardetzky TS. Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc epsilonRI alpha. Nature. 2000;406:259–266. doi: 10.1038/35018500. [DOI] [PubMed] [Google Scholar]
- 7.Maxwell KF, Powell MS, Hulett MD, Barton PA, McKenzie IF, Garrett TP, Hogarth PM. Crystal structure of the human leukocyte Fc receptor, Fc gammaRIIa. Nat Struct Biol. 1999;6:437–442. doi: 10.1038/8241. [DOI] [PubMed] [Google Scholar]
- 8.Radaev S, Motyka S, Fridman WH, Sautes-Fridman C, Sun PD. The structure of a human type III Fcgamma receptor in complex with Fc. J Biol Chem. 2001;276:16469–16477. doi: 10.1074/jbc.M100350200. [DOI] [PubMed] [Google Scholar]
- 9.Sondermann P, Huber R, Jacob U. Crystal structure of the soluble form of the human fcgamma-receptor IIb: a new member of the immunoglobulin superfamily at 1.7 A resolution. Embo J. 1999;18:1095–1103. doi: 10.1093/emboj/18.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- 11.Sondermann P, Kaiser J, Jacob U. Molecular basis for immune complex recognition: a comparison of Fc-receptor structures. J Mol Biol. 2001;309:737–749. doi: 10.1006/jmbi.2001.4670. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Boesen CC, Radaev S, Brooks AG, Fridman WH, Sautes-Fridman C, Sun PD. Crystal structure of the extracellular domain of a human Fc gamma RIII. Immunity. 2000;13:387–395. doi: 10.1016/s1074-7613(00)00038-8. [DOI] [PubMed] [Google Scholar]
- 13.Dennis G, Jr, Kubagawa H, Cooper MD. Paired Ig-like receptor homologs in birds and mammals share a common ancestor with mammalian Fc receptors. Proc Natl Acad Sci U S A. 2000;97:13245–13250. doi: 10.1073/pnas.230442897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin AM, Kulski JK, Witt C, Pontarotti P, Christiansen FT. Leukocyte Ig-like receptor complex (LRC) in mice and men. Trends Immunol. 2002;23:81–88. doi: 10.1016/s1471-4906(01)02155-x. [DOI] [PubMed] [Google Scholar]
- 15.Lanier LL. Natural killer cell receptor signaling. Curr Opin Immunol. 2003;15:308–314. doi: 10.1016/s0952-7915(03)00039-6. [DOI] [PubMed] [Google Scholar]
- 16.Viertlboeck BC, Crooijmans RP, Groenen MA, Gobel TW. Chicken Ig-like receptor B2, a member of a multigene family, is mainly expressed on B lymphocytes, recruits both Src homology 2 domain containing protein tyrosine phosphatase (SHP)-1 and SHP-2, and inhibits proliferation. J Immunol. 2004;173:7385–7393. doi: 10.4049/jimmunol.173.12.7385. [DOI] [PubMed] [Google Scholar]
- 17.Viertlboeck BC, Habermann FA, Schmitt R, Groenen MA, Du Pasquier L, Gobel TW. The chicken leukocyte receptor complex: a highly diverse multigene family encoding at least six structurally distinct receptor types. J Immunol. 2005;175:385–393. doi: 10.4049/jimmunol.175.1.385. [DOI] [PubMed] [Google Scholar]
- 18.Nikolaidis N, Makalowska I, Chalkia D, Makalowski W, Klein J, Nei M. Origin and evolution of the chicken leukocyte receptor complex. Proc Natl Acad Sci U S A. 2005;102:4057–4062. doi: 10.1073/pnas.0501040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viertlboeck BC, Schweinsberg S, Hanczaruk MA, Schmitt R, Du Pasquier L, Herberg FW, Gobel TW. The chicken leukocyte receptor complex encodes a primordial, activating, high-affinity IgY Fc receptor. Proc Natl Acad Sci U S A. 2007;104:11718–11723. doi: 10.1073/pnas.0702011104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan QR, Mosyak L, Winter CC, Wagtmann N, Long EO, Wiley DC. Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors. Nature. 1997;389:96–100. doi: 10.1038/38028. [DOI] [PubMed] [Google Scholar]
- 21.Harpaz Y, Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 22.Chapman TL, Heikeman AP, Bjorkman PJ. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity. 1999;11:603–613. doi: 10.1016/s1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- 23.Foster CE, Colonna M, Sun PD. Crystal structure of the human natural killer (NK) cell activating receptor NKp46 reveals structural relationship to other leukocyte receptor complex immunoreceptors. J Biol Chem. 2003;278:46081–46086. doi: 10.1074/jbc.M308491200. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 25.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faure M, Long EO. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J Immunol. 2002;168:6208–6214. doi: 10.4049/jimmunol.168.12.6208. [DOI] [PubMed] [Google Scholar]
- 27.Maenaka K, Juji T, Stuart DI, Jones EY. Crystal structure of the human p58 killer cell inhibitory receptor (KIR2DL3) specific for HLA-Cw3- related MHC class I. Structure. 1999;7:391–398. doi: 10.1016/s0969-2126(99)80052-5. [DOI] [PubMed] [Google Scholar]
- 28.Snyder GA, Brooks AG, Sun PD. Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. Proc Natl Acad Sci U S A. 1999;96:3864–3869. doi: 10.1073/pnas.96.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 2003;4:913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank. A computer-based archival file for macromolecular structures. Eur J Biochem. 1977;80:319–324. doi: 10.1111/j.1432-1033.1977.tb11885.x. [DOI] [PubMed] [Google Scholar]
- 31.Holm L, Sander C. Structural alignment of globins, phycocyanins and colicin A. FEBS Lett. 1993;315:301–306. doi: 10.1016/0014-5793(93)81183-z. [DOI] [PubMed] [Google Scholar]
- 32.Ponassi M, Cantoni C, Biassoni R, Conte R, Spallarossa A, Pesce A, Moretta A, Moretta L, Bolognesi M, Bordo D. Structure of the human NK cell triggering receptor NKp46 ectodomain. Biochem Biophys Res Commun. 2003;309:317–323. doi: 10.1016/j.bbrc.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Hummel JP, Dreyer WJ. Measurement of protein-binding phenomena by gel filtration. Biochim Biophys Acta. 1962;63:530–532. doi: 10.1016/0006-3002(62)90124-5. [DOI] [PubMed] [Google Scholar]
- 34.Gegner JA, Dahlquist FW. Signal transduction in bacteria: CheW forms a reversible complex with the protein kinase CheA. Proc Natl Acad Sci U S A. 1991;88:750–754. doi: 10.1073/pnas.88.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 36.Gazit R, Gruda R, Elboim M, Arnon TI, Katz G, Achdout H, Hanna J, Qimron U, Landau G, Greenbaum E, Zakay-Rones Z, Porgador A, Mandelboim O. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol. 2006;7:517–523. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- 37.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 38.Arnon TI, Achdout H, Lieberman N, Gazit R, Gonen-Gross T, Katz G, Bar-Ilan A, Bloushtain N, Lev M, Joseph A, Kedar E, Porgador A, Mandelboim O. The mechanisms controlling the recognition of tumor-and virus-infected cells by NKp46. Blood. 2004;103:664–672. doi: 10.1182/blood-2003-05-1716. [DOI] [PubMed] [Google Scholar]
- 39.Herr AB, Ballister ER, Bjorkman PJ. Insights into IgA-mediated immune responses from the crystal structures of human FcalphaRI and its complex with IgA1-Fc. Nature. 2003;423:614–620. doi: 10.1038/nature01685. [DOI] [PubMed] [Google Scholar]
- 40.DeLano WL. The PyMOL molecular graphics system on the world wide web. 2002 ( http://www.pymol.org).
- 41.Biassoni R, Cantoni C, Pende D, Sivori S, Parolini S, Vitale M, Bottino C, Moretta A. Human natural killer cell receptors and co-receptors. Immunol Rev. 2001;181:203–214. doi: 10.1034/j.1600-065x.2001.1810117.x. [DOI] [PubMed] [Google Scholar]
- 42.Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production. J Immunol. 2003;171:3415–3425. doi: 10.4049/jimmunol.171.7.3415. [DOI] [PubMed] [Google Scholar]
- 43.Rajagopalan S, Fu J, Long EO. Cutting edge: induction of IFN-gamma production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD158d) in resting NK cells. J Immunol. 2001;167:1877–1881. doi: 10.4049/jimmunol.167.4.1877. [DOI] [PubMed] [Google Scholar]
- 44.Campbell KS, Yusa S, Kikuchi-Maki A, Catina TL. NKp44 triggers NK cell activation through DAP12 association that is not influenced by a putative cytoplasmic inhibitory sequence. J Immunol. 2004;172:899–906. doi: 10.4049/jimmunol.172.2.899. [DOI] [PubMed] [Google Scholar]
- 45.Cantoni C, Bottino C, Vitale M, Pessino A, Augugliaro R, Malaspina A, Parolini S, Moretta L, Moretta A, Biassoni R. NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J Exp Med. 1999;189:787–796. doi: 10.1084/jem.189.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuchs A, Cella M, Kondo T, Colonna M. Paradoxic inhibition of human natural interferon-producing cells by the activating receptor NKp44. Blood. 2005;106:2076–2082. doi: 10.1182/blood-2004-12-4802. [DOI] [PubMed] [Google Scholar]
- 47.Cantoni C, Ponassi M, Biassoni R, Conte R, Spallarossa A, Moretta A, Moretta L, Bolognesi M, Bordo D. The three-dimensional structure of the human NK cell receptor NKp44, a triggering partner in natural cytotoxicity. Structure. 2003;11:725–734. doi: 10.1016/s0969-2126(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 48.Jaroszewski L, Rychlewski L, Godzik A. Improving the quality of twilight-zone alignments. Protein Sci. 2000;9:1487–1496. doi: 10.1110/ps.9.8.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 51.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 52.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 53.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 54.West AP, Jr, Giannetti AM, Herr AB, Bennett MJ, Nangiana JS, Pierce JR, Weiner LP, Snow PM, Bjorkman PJ. Mutational analysis of the transferrin receptor reveals overlapping HFE and transferring binding sites. J Mol Biol. 2001;313:385–397. doi: 10.1006/jmbi.2001.5048. [DOI] [PubMed] [Google Scholar]
- 55.West AP, Jr, Herr AB, Bjorkman PJ. The chicken yolk sac IgY receptor, a functional equivalent of the mammalian MHC-related Fc receptor, is a phospholipase A2 receptor homolog. Immunity. 2004;20:601–610. doi: 10.1016/s1074-7613(04)00113-x. [DOI] [PubMed] [Google Scholar]
- 56.Holst M, Kozack RE, Saied F, Subramaniam S. Protein electrostatics: rapid multigrid-based Newton algorithm for solution of the full nonlinear Poisson-Boltzmann equation. J Biomol Struct Dyn. 1994;11:1437–1445. doi: 10.1080/07391102.1994.10508078. [DOI] [PubMed] [Google Scholar]
- 57.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 58.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.