Abstract
The evolution of haemagglutinin (HA), an important influenza virus antigen, has been the subject of intensive research for more than two decades. Many characteristics of HA's sequence evolution are captured by standard Markov chain substitution models. Such models assign equal fitness to all accessible amino acids at a site. We show, however, that such models strongly underestimate the number of homoplastic amino acid substitutions during the course of HA's evolution, i.e. substitutions that repeatedly give rise to the same amino acid at a site. We develop statistics to detect individual homoplastic events and find that they preferentially occur at positively selected epitopic sites. Our results suggest that the evolution of the influenza A HA, including evolution by positive selection, is strongly affected by the long-term site-specific preferences for individual amino acids.
Keywords: directional selection, dN/dS, haemagglutinin, homoplasy, influenza A
1. Introduction
Influenza viruses offer an extraordinary opportunity for improving our understanding of molecular evolution. Several hundred complete genomes of the influenza A virus have been sequenced, as well as several thousand variants of its primary surface antigen, haemagglutinin (HA). Over the past four decades, roughly 20% of sites, concentrated within the epitopic regions of the HA1 domain of HA, have undergone amino acid substitutions, representing the equivalent of millions of years of evolutionary change in a typical mammalian protein (Carroll 2003). These substitutions were driven primarily by selection to evade the antibody response in the host population (Fitch et al. 1991; Nelson & Holmes 2007).
Aside from recent studies of recombination (Lindstrom et al. 2004; Holmes et al. 2005), most research on the evolution of HA has focused on identifying the sites that experience positive selection for amino acid substitutions (Fitch et al. 1997; Bush et al. 1999a,b; Yang 2000; Plotkin & Dushoff 2003; Suzuki 2006; Wolf et al. 2006; Shih et al. 2007) among the majority of sites that evolve under negative selection. One of the key tools used for detecting genes or sites under positive selection is the concept of the dN/dS ratio, the ratio of the rates of non-synonymous (amino acid altering) and synonymous (amino acid preserving) substitutions along a phylogenetic tree. Assuming that the rate of synonymous substitutions is a good approximation of the neutral standard, a dN/dS ratio exceeding unity is an indication of positive Darwinian selection (Ina & Gojobori 1994; Yang & Bielawski 2000). This idea stems from a prediction of the neutral theory that the rate of amino acid substitutions in a gene must be smaller than or equal to the rate of synonymous substitutions (Kimura 1977, 1983). Later, this idea was carried over to subregions of genes and even individual sites. In several recent studies, a dN/dS analysis was applied to HA sequences, and sites with dN/dS ratios significantly greater than unity were identified (Ina & Gojobori 1994; Bush et al. 1999a; Yang 2000; Suzuki 2006; Wolf et al. 2006), suggesting that positive selection plays an important role in the evolution of influenza. Although this research sheds light on HA evolution, and although it may help to calibrate appropriate vaccines (Bush et al. 1999b; Plotkin et al. 2002), a simple catalogue of sites experiencing positive selection fails to address the full spectrum of possibilities for the action of natural selection on HA. In particular, an important question—which amino acids were fixed in HA due to natural selection as opposed to random drift—has remained largely untouched in the literature (but see Wolf et al. 2006; Shih et al. 2007). In this work, we attempt to take an initial step in this direction.
It is instructive to re-examine the substitutions models in which the dN/dS concept is developed. Such models, introduced by Goldman & Yang (1994) and by Muse & Gaut (1994), are based on the theory of finite-state, continuous-time Markov chains. This general approach has proved extremely fruitful, and various modifications of the original models have been suggested (Nielsen & Yang 1998; Yang & Nielsen 2000, 2002, 2008; Yang et al. 2000; Forsberg & Christiansen 2003; Guindon et al. 2004; Mayrose et al. 2007). We refer to this whole family of models as ‘codon-based Markov chain models’ or simply ‘Markov chain models’. The key feature of these models is that codons rather than nucleotides are treated as the evolving unit in the coding sequences. Each position in the evolving sequence can assume one of 61 states (stop codons are usually excluded), and transitions between states occur with certain rates. The synonymous substitution rates equal the corresponding mutation rates, and the non-synonymous substitution rates equal the mutation rates scaled by the dN/dS ratio (usually labelled as ω), so that sites with dN/dS ratios smaller (greater) than unity evolve, at the amino acid level, slower (faster) than expected from neutrality.
Sites under positive selection are typically identified by reconstructing the phylogenetic history among sampled sequences, and inferring the evolutionary parameters of a Markov chain model, either within the maximum-likelihood or Bayesian setting. A statistical test is then applied to determine whether the dN/dS ratios of any sites significantly exceed unity.
However, what exactly does it mean that a site evolves according to a Markov chain model with ω≠1?
Standard Markov chain substitution models assume independence of sites and assign an ω value to each site. In such models, substitution rates towards different amino acids are entirely determined by the mutation rates, and no preference is given to one substitution over another. This simplification ignores an important aspect of real protein evolution, the fact that different amino acids are preferred at different sites. More generally, one can picture a site as evolving on a fitness landscape whereby each amino acid at the site provides the organism with certain fitness, given a genetic background. This marginal fitness is what we call the ‘fitness of amino acid X at site n’. The fitness landscape of a site is not static: the fitness of an amino acid can change depending on the environment, genetic background, etc.
At a site, substitutions towards amino acids with higher fitness values will occur faster than lower fitness values. One straightforward consequence of this is the well-known fact that substitutions between biochemically similar amino acids occur more readily than dissimilar amino acids (Miyata & Yasunaga 1980). Although many widely used Markov chain models ignore biochemical properties, models have been developed that account for them (Goldman & Yang 1994; Yang et al. 1998; Sainudiin et al. 2005; Wong et al. 2006).
Another, more serious, consequence of the aforementioned simplification is the fact that Markov chain models with a single ω value per site describe a site evolving on a fitness landscape that changes with each substitution event. In particular, in the ‘negative selection mode’ (ω<1), the amino acid currently fixed in the population provides a local fitness maximum, whereas in the ‘continual positive selection mode’ (ω>1), it provides a local fitness minimum. If a site is characterized by ω<1 (ω>1), then any new arising amino acid at the site is assigned a negative (positive) selection coefficient (Nielsen & Yang 2003). Even though it is conceivable that fitness landscapes of some proteins are dynamic, one rarely expects this to be the case at the level of individual sites. Indeed, consider the evolution of the influenza A HA, a canonical example of evolution on a highly dynamic fitness landscape shaped by constantly changing immune status of the human population. Epitopic sites in HA are described by a Markov chain model with ω>1 (Bush et al. 1999b; Yang 2000). Suppose that amino acid A was recently substituted by amino acid B at such site, for example, because amino acid B provided a protein conformation that was less recognizable by human antibodies. According to the Markov chain model with ω>1, immediately after the substitution occurs, amino acid B becomes deleterious and any amino acid other than B is preferred at the site. This implies that, as soon as A is replaced by B, there is immediately pressure to change again, to any other amino acid (including A), even though the immune status of the population has hardly changed. Analogously, consider a site that evolves under negative selection and is described by a Markov chain model with ω<1. Suppose that a certain amino acid A is currently fixed in the population and provides a local fitness maximum. Any other amino acid at this site is therefore deleterious. Nevertheless, in a finite population, A can be substituted by some other amino acid B. In the negative selection regime, one would expect to observe a rapid reversion to the locally optimal A. However, in a Markov chain model with ω<1 such substitution is unlikely, and B becomes the local fitness maximum. This implies that the fitness landscape has changed, and the substitution A→B was, in fact, adaptive.
These examples illustrate that standard Markov chain models may describe unrealistic evolutionary trajectories for individual sites and/or lead to wrong interpretations of evolution under an elevated/decreased dN/dS ratio. In reality, the fitness landscape of a site is expected to be less dynamic than assumed by such Markov chain models. In order to treat this problem, considerable effort has been made in recent years to create substitution models that attempt to use more fully the information about the fitness landscape on which a protein evolves. This can be done at various levels of detail and complexity. In the most complex models, substitution rates are specified based on the three-dimensional protein structure (Fornasari et al. 2002; Rodrigue et al. 2005; Robinson et al. 2007). In another type of model, sites are treated independently but a distinct fitness value is ascribed to each amino acid at each site (Halpern & Bruno 1998). In yet another type of model, sites are grouped into classes and different fitness landscapes are modelled in different site classes (Thorne et al. 1996; Koshi & Goldstein 1998; Koshi et al. 1999; Dimmic et al. 2000; Lartillot & Philippe 2004; Sainudiin et al. 2005; Wong et al. 2006). These models have been used to more accurately estimate evolutionary distances between sequences (Halpern & Bruno 1998), reconstruct phylogenies (Thorne et al. 1996; Koshi et al. 1999; Lartillot & Philippe 2004) and infer the dN/dS ratios (Sainudiin et al. 2005; Wong et al. 2006; Robinson et al. 2007). Although some of these models, notably those by Halpern & Bruno (1998) and Koshi & Goldstein (1998), are capable of capturing evolution at a site on a static fitness landscape, no conclusions about the fitness landscapes on which real proteins evolve have been drawn from these models.
Very recently, Kosakovsky Pond et al. (in press) developed a maximum-likelihood (ML) approach for detecting directional selection for amino acid substitutions and applied it to the influenza A sequence data. Their method, although potentially very useful, has two important limitations. First, it uses an amino acid- rather than codon-based Markov chain model and, as such, may not properly account for the underlying mutational biases, which may potentially lead to spuriously significant results. Second, the method lacks power because, in order to detect directional selection towards a certain amino acid, it requires an increase in the overall frequency of that amino acid in the entire gene.
Here we take an alternative approach to studying directionality in the evolution of an amino acid site in a protein. We use an independent site codon-based Markov chain substitution model with site-specific dN/dS ratios as our null model and quantify the departures from it that stem from the violation of the assumption of a fitness landscape that changes with every substitution event. In reality, the changes in the fitness landscape of a site are unlikely to be precisely synchronized with the substitution events, so that a specific amino acid or a set of specific amino acids are selectively advantageous at a site for a prolonged period of time. If this set is small, convergent and/or parallel evolution may be observed (Yeager et al. 1997; Zhang & Kumar 1997). This fact can be used to develop an analogue of the dN/dS test to detect not only sites but also amino acids at those sites that are substituted more frequently than expected under neutrality. A similar approach has been suggested earlier by Chen et al. (2004) who have detected the majority of known as well as many previously unknown amino acid substitutions that are likely to have arisen in HIV as a response to drug therapy. Their approach was later formalized within the ML framework by Seoighe et al. (2007). In this paper, we provide a characterization of the fitness landscape of influenza A HA. Our approach differs from that by Chen et al. and Seoighe et al., in which we (i) take into account the phylogenetic relationships between sequences to infer the amino acid substitution opportunities and (ii) quantify the departures of the HA evolution from a Markov chain model with site-specific dN/dS values rather than from a purely neutral model.
Unlike the dN/dS measure that discriminates rapidly evolving sites from slowly evolving sites, our method is aimed at detecting individual amino acids that have evolved under directional selection at specific sites. Directed substitutions can occur at either rapidly or slowly evolving sites, and we do not a priori expect a consistent relationship between a site's dN/dS ratio and the presence of directionally selected amino acids.
2. Material and methods
(a) Data
We downloaded all HA nucleotide sequences from human influenza A virus subtype H3N2, which were available in the NCBI database of April 2006. We excluded isolates that were processed in chicken eggs (Bush et al. 2000). The remaining sequences were aligned using Clustal W v. 1.83 (Thompson et al. 1994) and coding regions for the HA1 part were extracted. The alignment length was 987 nucleotides. Occasional gaps were filled if more than 90% of sequences agreed on the symbol at the gap position, otherwise the sequence with a gap was excluded from further analysis. The resulting alignment contained 1249 sequences. The list of sequences is available upon request.
(b) Phylogeny reconstruction
We reconstructed parsimonious phylogenetic trees for the HA protein of subtype H3N2 using the parsimony ratchet algorithm (Nixon 1999) built on top of PAUP* (Swofford 2002). We choose the maximum parsimony (MP) algorithm in favour of ML because (i) ML algorithms, especially those based on realistic evolutionary models, are unreasonably time consuming for such a large number of sequences and (ii) theoretically, the MP algorithm should perform well on this dataset because the sequences are sufficiently similar that multiple substitutions in a single position on a single branch are rare. However, our results are robust with respect to the method of phylogeny reconstruction (see the electronic supplementary material).
The following parameters were used for the parsimony ratchet: fraction of sites whose weight was set to zero in the ‘jump step’, 0.1; number of trees kept in memory, from 1 to 2; and number of iterations, 30 (Nixon 1999). The minimum tree length obtained was 3349. We found 34 islands containing trees with this score. We randomly selected 20 trees from these islands and performed our analysis on each of them. The selected 20 trees were rooted by designating the isolate V01085/Aichi/1968 as an out-group. For each of these trees, we inferred internal nodes by the parsimony algorithm using PAUP with the Acctran option. It turned out that the root sequence was the same for all 20 trees and was equal to the sequence of the out-group V01085/Aichi/1968.
(c) Simulation of sequence evolution
We simulate the evolution of the HA along the phylogeny using a modified version of the codon-based Goldman–Yang (GY) model with site-specific dN/dS ratios (Goldman & Yang 1994; Yang 2000). Our algorithm takes the following objects as input: (i) a rooted phylogenetic tree with branch lengths that correspond to the number of nucleotide differences, (ii) the sequence at the root node, (iii) the 4×3 mutation matrix, (iv) the list of site-specific dN/dS ratios, ωk, k=1, 2, …, L, and (v) the codon-position correction coefficients, λi, i=1, 2, 3 (see below).
Starting from the root of the tree, the algorithm recursively generates sequences for all other nodes. A sequence at a node is generated by the following rule, given that the sequence of its ancestor is already known. Consider a node A connected to its ancestral node B by a branch of length n; in other words, A differs from B in n nucleotides. To generate the sequence in B, we change exactly n different nucleotides of sequence in A. As a result, the simulated tree differs from the original tree only in the sequences at the nodes, while the original topology and branch lengths are preserved.
We distribute n nucleotide changes along the sequence as follows. Suppose m (0≤m<n) changes have already been distributed along the sequence and fell on nucleotide positions from the set . To simulate the m+1th change, we first assign the following weights to each possible mutation of nucleotide xk at position k (k=1, 2, …, 3L) in the parental sequence (yk is any nucleotide other than xk):
| (2.1) |
where, ci is the codon at the position i in the ancestral sequence; p(k)=⌊(k−1)/3⌋+1 is the amino acid position corresponding to nucleotide position k; q(k)=(k−1) mod 3+1 is the position of nucleotide k in the codon cp(k); is a codon that equals cp(k) at all positions except for q(k) where it has nucleotide yk instead of xk; and A(c) is the amino acid encoded by codon c.
Then, the m+1th change becomes the change of nucleotide xk with nucleotide yk at position k with probability proportional to the weight of the corresponding change,
After the change falls at position km+1, we set and repeat the procedure until all n substitutions have been distributed.
This procedure is similar to the original GY model, with the differences as follows.
GY implements a continuous-time Markov process, drawing the number of changes that occurred on a branch from a distribution whose mean equals the branch length. As a result, the branch lengths of the simulated tree generally differ from those of the real tree. This difference could lead to biases in some of the kinds of comparisons between observed and simulated data presented below. Instead, we distribute the same exact number of changes along a branch in the simulation as observed in the data. Therefore, the simulated branch lengths exactly match those in the data.
We have included an additional parameter, the codon-position correction coefficient λ that weights amino acid substitutions according to the codon position in which a substitution occurs (see equation (2.1)). The model with this parameter captures the evolution of HA better than the model in which the probability of a non-synonymous substitution is independent of the position in a codon (see the electronic supplementary material).
We refer to the evolutionary model with codon-position correction coefficient as mGY+λ for ‘modified GY with codon-position corrections’, and to the model where as mGY.
The code implementing the mGY+λ model written in Objective Caml is available upon request.
(d) Statistics
(i) Excess-of-substitution statistics
The excess-of-substitutions (ES) statistic is designed to detect the individual amino acid substitutions that occurred with unusually high frequency. Unlike the homoplasy statistic (see the electronic supplementary material), it controls for variation in mutational opportunities of different substitutions. The ES statistic is elevated by either homoplastic or individual substitutions that occur at a rate higher than expected on the basis of their mutational opportunities.
First, we define the opportunity ρ(c→Y) for a sense codon c to change by a single nucleotide substitution to any codon that encodes amino acid Y other than A(c),
where M(c)is the set of one-mutational neighbours of codon c excluding the stop codons; ϕ(c,c′) is the position at which codons c and c′ differ; and is the rate of the nucleotide mutation that transforms codon c into codon c′; for example, if r(A→G) is the mutation rate between adenine and guanine, then . We put ρ(c→Y)=0 if A(c)=Y. Next, the opportunity for amino acid X to be substituted by amino acid Y at site k on the whole tree is given by
where, is the set of branches of the tree whose parental node sequence encodes amino acid X at position k; is the total number of non-synonymous substitutions that occurred at branch i. Then,
is the opportunity of gaining amino acid Y at site k.
Finally, let be the number of observed (or inferred) substitutions towards amino acid X at site k, and let be the total number of amino acid substitutions at site k. Then,
is the number of substitutions leading to amino acid X at site k expected under the assumptions of the mGY+λ model.
We define the ES statistic for site k and amino acid X as
A positive ES statistic implies that the number of amino acid substitutions resulting in amino acid X at site k is larger than expected from the mGY+λ model.
We define the grand excess-of-substitutions (GES) statistic as
Because for each site k, a value of GES significantly different from 0 implies an excess of substitutions into one or several amino acids and a deficit of substitutions into other amino acids, compared to what is expected from opportunities Ok.
To separately compute the GES statistic for epitopes (denoted by ) and non-epitopes (denoted by ), the outer summation is taken over the corresponding sites.
(ii) Finding selectively advantageous amino acids
We use the ES statistic described above to identify amino acids that were selectively advantageous or disadvantageous at individual sites. For that, we group together all potential amino acid substitutions at a site that have the same target amino acid and for which the opportunity on the phylogenetic tree is non-zero (such substitutions may or may not have actually occurred on the phylogenetic tree). For example, all potential amino acid substitutions that resulted in asparagine at site 23, no matter what the substituted amino acid was, would fall in the same group. We only include a group in further analysis if it was observed for each of the 20 trees; this is true for 2490 groups (more than 98% of all observed groups). Each such group of potential substitutions is associated with a corresponding value of the ES statistic. By comparison with the distribution of the ES statistic expected from the mGY+λ model, we identify the groups of potential substitutions resulting in the same amino acid whose ES statistic (averaged over 20 trees) is significantly elevated or depressed. Since we perform multiple hypothesis testing, we expect to find N·x false positives if we are looking at the x-quantile of the null distribution, where N is the number of tests. Under the stringent Bonferroni correction, we treat as significant only those target amino acids whose ES statistic falls into the x/N-quantile of the null distribution.
3. Results
Our analysis of HA sequence data is designed to quantify aspects of the HA fitness landscape that is not captured by a simple Markov chain model of substitutions. Our basic methodology is straightforward: we design statistics that quantify aspects of protein evolution, such as directionality, and we assess the significance of these statistics by comparing the observed data with a null distribution generated by simulating a Markov chain substitution model. The substitution model used to generate the null distribution is a modified version of the GY codon-based model, which we call the mGY+λ model. Under this model, the probabilities of synonymous substitutions at a codon site are determined by a neutral mutation matrix (constant across the gene), and the probabilities of non-synonymous substitutions are determined by the mutation matrix, the site-specific ω values and codon-position correction coefficients to ω (see §2). Thus, the probabilities of non-synonymous substitutions differ between different codon sites and the three positions in the codon, but no consideration is given to the identities of the target amino acids. For the sake of comparison, we also use the mGY model in which the probabilities of non-synonymous substitutions are equal between three codon positions.
In order to fit the parameters of our null model, we reconstruct 20 equally parsimonious phylogenies for 1249 sequences of the HA1 part of the HA protein (Wilson & Cox 1990) sampled from human patients between 1968 and 2005. From each of the reconstructed trees, we infer the sequences at internal nodes, the mutation rates, the site-specific dN/dS ratios and the codon-position correction coefficients (see the electronic supplementary material). The inferred parameter values can be found in table S1 in the electronic supplementary material. Then, using these parameters and the inferred root sequence, we produce null distributions of various statistics by simulating the mGY+λ substitution model 50 times along each tree, obtaining a total of 1000 replicate sequences of all leaf and internal nodes.
We compared the features of real HA protein evolution as inferred from the 20 parsimonious trees with the features of the null distribution generated by the mGY+λ model using three types of statistics. First, we assessed the degree of HA protein conservation by measuring the average Miyata distance for an amino acid substitution, as well as the Simpson diversity index for the amino acid distribution at a typical site in HA (see the electronic supplementary material). Second, we used the GES statistic to assess the overall overabundance of substitutions in HA that gave rise to the same amino acid at a site. Third, we used the ES statistic to find particular amino acids that arose at certain sites more frequently than expected under the mGY+λ model.
(a) Model consistency
The mGY+λ model captures both the synonymous evolution and the coarse aspects of non-synonymous evolution of the HA gene. It provides a good fit to the observed numbers of synonymous and non-synonymous substitutions on the tree, the numbers of synonymous and non-synonymous substitutions that fall at different codon positions, and the time trajectory of the nucleotide composition at fourfold degenerate sites and second position sites (see the electronic supplementary material). The mGY model, which does not control for differences in rates of non-synonymous substitutions between the three codon positions, fails to capture the distribution of substitutions between the positions; in particular, it overestimates the number of non-synonymous substitutions at the second codon position (see the electronic supplementary material).
(b) Protein conservation
We computed the average Miyata distance of an amino acid substitution for the mGY+λ simulations and the real HA protein. The average Miyata distance of an amino acid substitution in the HA protein as a whole is significantly smaller than in the null distribution, which ignores physicochemical constraints (p<0.05; see the electronic supplementary material). This result holds for non-epitopic regions of the protein (p<0.01), but is not statistically significant for the epitopes (p=0.16). Here and below, p-values are computed for the average (over 20 parsimonious trees) value of each statistic, and all tests are two tailed unless noted otherwise.
We also measured protein conservation using the average Simpson index of the amino acid frequency distribution at each site (see the electronic supplementary material). The inverse of the Simpson index quantifies the effective number of different amino acids that have been present at a typical site during the course of its evolution (Simpson 1949). In other words, if the average Simpson index is equal to 1, then most sites have seen many amino acids in the course of evolution; while if the Simpson index is close to 1, then most sites have been dominated by a single amino acid. The latter situation would indicate strong protein conservation. The mGY+λ model captures the degree of conservation of the HA protein as a whole relatively well (p=0.2; see the electronic supplementary material). However, separating sites into epitopes and non-epitopes reveals an important pattern: non-epitopic sites are strongly conserved (the Simpson index is significantly greater than expected, p<10−3) while the epitopic sites are not (the Simpson index does not significantly differ from expectation, p=0.14). The effective number of amino acids at a typical epitopic site is 1.20 while at a non-epitopic site it is 1.02.
(c) Directional selection
At each amino acid site, an average of 2.0 different amino acids arose by non-synonymous substitutions at least once somewhere on the phylogenetic tree, compared to 2.3 in simulations of the mGY+λ model (standard errors are less than 0.1). 334.5±0.3 (49%) of these amino acids arose on average more than once, compared with 364.1±0.4 (48%) in simulations. In other words, the actual pattern of substitutions during HA evolution exhibits characteristics of directional selection: substitutions tend to lead to a specific set of amino acids, producing a smaller number of amino acids observed at each site.
In order to estimate whether the substitution rates towards certain amino acids are significantly higher in the data than expected under the mGY+λ model, we use the ES and GES statistics. The ES statistic is defined as the difference between the observed number of substitutions towards a specific amino acid at a site and the corresponding number expected if the substitution probabilities were determined according to the mGY+λ model (see §2 for details). This statistic is elevated if substitutions leading to the amino acid under consideration at a given site occurred more frequently than expected. The GES statistic is the sum of squares of the ES values for each amino acid at each site; it is elevated if many amino acid substitutions in the whole gene occurred at unexpectedly high rates.
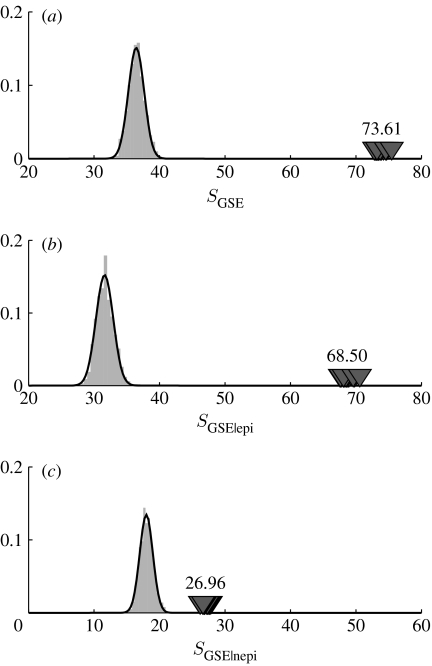
Using the GES statistic, we find that significantly more substitutions gave rise to the same amino acids at a site in the data than in simulations. The value of the GES statistic is significantly larger in the data than in simulations, for the HA protein as a whole and for both the epitopic and the non-epitopic parts of the HA protein (p<10−3; figure 1). We also explored other statistics that measure the same quantity, and the results are qualitatively the same (see the electronic supplementary material).
Figure 1.
GES statistic. Grey bars represent the histogram for the GES statistic distribution obtained from the simulation based on the mGY+λ model. The corresponding best-fit Gaussian curves are indicated in black. Dark grey triangles show the values of the GES statistic for the real HA protein as inferred from each of the 20 MP trees, with the mean value shown above the triangles. Data for all sites pulled together are shown in (a), for epitopic sites in (b) and for the non-epitopic sites in (c).
Using the ES statistic, we identified the individual amino acids that were significantly more and less likely to be the targets for non-synonymous substitutions at a given site than expected from the corresponding mutational opportunities, site-specific dN/dS ratios and codon-position corrections. The ES statistic is significantly elevated at the 0.1 per cent level (one-tailed test without the Bonferroni correction) for 28 amino acids at 25 sites (table 1); the ES statistic is significantly depressed at the 0.1 per cent level for 19 amino acids at 13 sites (table 1). The expected number of false positives in each case is 2.5. Therefore, most (although probably not all) of the target amino acids presented in table 1 have, in fact, been favoured or disfavoured by natural selection. In each of the cases presented in table 1 with the elevated ES statistic, the target amino acid arose more than once on the phylogenetic tree. Of the 25 reported sites (84%), 21 are located in the epitopic regions of the protein; 20 sites (80%) have dN/dS values exceeding 1, indicative of positive selection as the predominant evolutionary force (table 1).
Table 1.
Amino acids whose average ES statistic is significantly elevated or depressed (grey shade) at the 0.1% level, without the Bonferroni correction (one-tailed tests). (The expected number of false positives is 2.5. Notation k denotes the site; dN/dS denotes the average dN/dS ratio for the site; X denotes the amino acid at the site; denotes the average number of observed substitutions leading to amino acid X at site k; and denotes the average value of the ES statistic corresponding to amino acid X at site k. Averages are taken over 20 parsimonious trees. Standard errors are not shown because they are typically small relative to the averages (mean relative standard error is 2.4%, maximum 13.8%). In italics are the sites and amino acids whose average ES statistic is significantly elevated or depressed at the 5% level, with the Bonferroni correction (one-tailed tests).)
| site (k) | epitope | dN/dS | amino acid (X) | ||
|---|---|---|---|---|---|
| 3 | 1.8 | Ile | 10.2 | 6.6 | |
| Pro | 2.0 | −4.9 | |||
| 92 | E | 1.5 | Thr | 6.0 | 5.2 |
| 106 | 0.7 | Val | 7.0 | 4.7 | |
| 121 | D | 1.4 | Thr | 11.0 | 8.3 |
| 124 | A | 1.6 | Gly | 9.5 | 6.1 |
| 133 | A | 2.0 | Asn | 12.1 | 6.2 |
| 137 | A | 2.3 | Ser | 7.6 | 6.2 |
| His | 1.0 | −5.1 | |||
| 138 | A | 2.2 | Ser | 13.0 | 9.2 |
| Val | 0.0 | −7.5 | |||
| 142 | A | 1.1 | Gly | 8.0 | 6.4 |
| 144 | A | 1.5 | Asp | 9.2 | 7.4 |
| 145 | A | 5.0 | Lys | 32.4 | 25.7 |
| Asn | 11.6 | 4.8 | |||
| Arg | 2.1 | −6.1 | |||
| Asp | 0.0 | −6.5 | |||
| Glu | 0.0 | −7.7 | |||
| 156 | B | 2.8 | Lys | 16.7 | 11.8 |
| Arg | 0.5 | −5.8 | |||
| 159 | B | 2.2 | Asn | 8.3 | 5.0 |
| His | 2.0 | −5.4 | |||
| 167 | D | 1.5 | Ala | 11.0 | 5.9 |
| Ile | 1.0 | −4.3 | |||
| 186 | B | 2.2 | Ile | 19.9 | 17.4 |
| Arg | 0.0 | −5.6 | |||
| Asn | 2.0 | −7.7 | |||
| 190 | B | 1.0 | Val | 7.0 | 6.4 |
| 193 | B | 1.5 | Lys | 6.0 | 5.1 |
| 194 | B | 2.0 | Ile | 13.0 | 8.9 |
| Pro | 0.0 | −8.2 | |||
| 213 | D | 0.6 | Val | 4.9 | 4.6 |
| 220 | 1.7 | Gly | 13.2 | 7.6 | |
| Lys | 1.0 | −8.8 | |||
| 226 | D | 6.7 | Gln | 16.9 | 15.2 |
| Ile | 32.4 | 14.1 | |||
| Leu | 8.2 | 6.7 | |||
| Met | 2.5 | −4.0 | |||
| Phe | 1.0 | −4.3 | |||
| Pro | 1.0 | −10.3 | |||
| Ala | 0.0 | −10.8 | |||
| 229 | D | 3.1 | Ile | 15.7 | 11.2 |
| Lys | 9.4 | −8.2 | |||
| 233 | 0.9 | His | 9.0 | 4.7 | |
| 248 | D | 1.6 | Ile | 13.0 | 8.0 |
| Ala | 0.0 | −4.7 | |||
| 275 | C | 0.9 | Asp | 10.2 | 5.3 |
The presented list of sites with at least one amino acid with an elevated ES statistic partially overlaps with previously published lists of sites with an elevated dN/dS ratio (Bush et al. 1999a; Yang 2000; Suzuki 2006). We do not compare these lists explicitly because the dN/dS and the ES statistics measure different quantities, and we have no a priori expectation for their relationship. Even though we have excluded from our analysis all variants that were denoted as being cultured in chicken eggs, some of the amino acids listed in table 1 (e.g. lysine at sites 145 and 156, serine at site 186) were previously identified as being adaptations of the influenza virus to growth in eggs (Meyer et al. 1993). This is not surprising, as it is known that some mutations first observed in egg cultured variants also exist in circulating human strains (Rocha et al. 1993).
Figure 2 shows the phylogenetic distribution of lysine at site 145, which has the highest value of the ES statistic and probably evolved under directional selection: it was not present in the majority of the virus population until the second half of the 1990s, when it became fixed after emerging repeatedly in different subpopulations.
Figure 2.
Homoplastic substitutions resulting in lysine at site 145. The phylogenetic tree of HA is shown, with the root marked by an asterisk. Branches where the sequence at the descendent node has lysine at site 145 are in black. On this tree, lysine appeared 33 times at site 145. Branch lengths are measured in nucleotide substitutions.
4. Discussion
Our simulation null model of HA sequence evolution is based on a detailed codon-based substitution model that is similar to the widely used Markov chain-based models. As in most such models, the direction of substitutions is independent of the identity of the target amino acid. The simulated data faithfully reproduce most of the coarse characteristics of evolution observed in the true empirical data (see the electronic supplementary material). However, the null model fails to adequately describe finer aspects of non-synonymous evolution. Our analysis has focused on the discrepancies between the data and the model in the probabilities of different amino acid substitutions at a site.
(a) Protein conservation
Owing to functional constraints, the protein shape is generally more conserved in the course of evolution than in the underlying amino acid sequence (Lesk & Chothia 1980). For the same reason, we might expect that substitutions between biochemically dissimilar amino acids would occur less frequently than between similar amino acids (Miyata & Yasunaga 1980).
Our results indicate that non-synonymous substitutions at the second codon positions of HA occurred less frequently than expected under the mGY substitution model, which does not account for differences in the rates of non-synonymous substitutions between the three codon positions (see the electronic supplementary material). This mismatch indicates that different codon positions evolve, on average, under different selective pressures, probably due to the structure of the genetic code. Indeed, non-synonymous changes at the second amino acid position tend to change the properties of the encoded amino acid more radically than non-synonymous changes at the first or third positions (e.g. Haig & Hurst 1991; Urbina et al. 2006).
The mGY+λ model provides a better overall fit to the data (see the electronic supplementary material), as it controls for the differences in probabilities of non-synonymous substitutions between the three codon positions. However, even under this model, amino acid substitutions are significantly more conservative with regard to the physicochemical properties than expected from their mutational opportunities (see the electronic supplementary material). The conservation is lower in epitopic than in non-epitopic regions of HA (see the electronic supplementary material). This conforms with previous observations (Ina & Gojobori 1994; Bush et al. 1999b) that epitopes are less constrained than the rest of the HA protein, but also suggests that the mGY+λ model is not sufficient, even after fitting site-specific dN/dS values and codon-position corrections, to capture the amino acid level patterns of HA protein evolution.
(b) Directional selection
The direction of amino acid substitutions has site-specific biases. Specifically, in both epitopic and non-epitopic sites, homoplasies are more frequent than expected: amino acid substitutions at a site tend to repeatedly give rise to identical amino acids, whereas other specific amino acids are avoided (figure 1; table 1; see the electronic supplementary material). These deviations are not caused by the difference in the number of non-synonymous substitutions between the data and the simulations, since the simulations approximately preserve this number (see the electronic supplementary material).
Although we can pinpoint the amino acids that arose more frequently than expected (table 1), it is possible that some of these amino acids were driven to high frequencies by hitch-hiking along with other selected amino acids rather than due to direct selection. It is difficult to assess the degree to which hitch-hiking affects our site-by-site statistics. However, since the hitch-hiking hypothesis implies that selective sweeps occurred at least at some sites, it is unlikely that hitch-hiking accounts for all significant results presented in table 1.
Therefore, the differences between model and data have to do with natural selection affecting the variability of amino acids at a site. These differences could be due to either negative (selective constraint) or positive selection. Under a conceivable negative selection scenario, if just a few amino acids are permitted at an amino acid site, substitutions between these amino acids predominate (Bazykin et al. 2007), leading to more observed cases of homoplastic evolution in the data than in the simulation. On the other hand, positive selection could also lead to repeated substitutions towards identical novel amino acids, either due to repeated changes of the selective landscape or the independent response of different evolving lineages to the similar selective pressure.
Several arguments suggest that the observed statistical results are primarily due to the action of positive, rather than negative, selection. First, homoplastic substitutions at rates higher than expected on the basis of their mutational opportunities, as shown by the significant elevation of the ES statistic, are unlikely to be due to negative selection. Second, most of the sites with high ES statistics listed in table 1, and all of the sites for which the results are most significant (indicated in italic in table 1), have dN/dS values greater than 1, suggesting that positive selection is the predominant regime at those sites. Finally, most of these sites are located in the epitopes (table 1), for which the importance of positive selection is well known (Bush et al. 1999a; Yang 2000). Therefore, it is reasonable to assume that most amino acids with the positive ES statistic listed in table 1 were positively selected, at the sites specified.
Our results, therefore, suggest that the shape of the fitness landscape at each site has a major effect on the evolution of HA by positive selection, in contrast to the unrealistic assumption of the identical fitness of target amino acids made by most Markov chain models. Other authors recently came to similar conclusions (Kosakovsky Pond et al. in press). Positive selection is directional, in the sense that the substitution probabilities into different amino acids are non-uniform. Frequently, directional selection leads to repeated (homoplastic) substitutions into the same amino acid at a site and/or substitutions into amino acids that are unlikely to be based on mutation probabilities alone. Ascribing the selection coefficients to individual amino acids at a site is a subject for future research.
Acknowledgments
S.K. gratefully acknowledges support by the Burroughs Wellcome Fund Training programme in Biological Dynamics (no 1001782) and the DARPA grant HR0011-05-1-0057. G.A.B. was partially supported by the Molecular and Cellular Biology programme of the Russian Academy of Sciences. J.B.P. and S.K. were funded by grants from the Burroughs Wellcome Fund and the James S. McDonnell Foundation. J.D. gratefully acknowledges support by the Natural Sciences and Engineering Research Council of Canada and the DARPA grant HR0011-05-1-0057.
Supplementary Material
Estimation of simulation parameters. Protein conservation statistics. Robustness of results. Detailed characterization of the behaviour of the mGY+\lambda model. Homoplasy statistic results
Tables containing the evolutionary parameter values inferred from 20 parsimonious trees
References
- Bazykin G.A, Kondrashov F.A, Brudno M, Poliakov A, Dubchak I, Kondrashov A.S. Extensive parallelism in protein evolution. Biol Direct. 2007;2:20. doi: 10.1186/1745-6150-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush R.M, Bender C.A, Subbarao K, Cox N.J, Fitch W.M. Predicting the evolution of human influenza A. Science. 1999a;286:1921–1925. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- Bush R.M, Fitch W.M, Bender C.A, Cox N.J. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 1999b;16:1457–1465. doi: 10.1093/oxfordjournals.molbev.a026057. [DOI] [PubMed] [Google Scholar]
- Bush R.M, Smith C.B, Cox N.J, Fitch W.M. Effects of passage history and sampling bias on phylogenetic reconstruction of human influenza A evolution. Proc. Natl Acad. Sci. USA. 2000;97:6974–6980. doi: 10.1073/pnas.97.13.6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.B. Genetics and the making of Homo sapiens. Nature. 2003;422:849–857. doi: 10.1038/nature01495. [DOI] [PubMed] [Google Scholar]
- Chen L, Perlina A, Lee C.J. Positive selection detection in 40,000 human immunodeficiency virus (HIV) type 1 sequences automatically identifies drug resistance and positive fitness mutations in HIV protease and reverse transcriptase. J. Virol. 2004;78:3722–3732. doi: 10.1128/JVI.78.7.3722-3732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmic M.W, Mindell D.P, Goldstein R.A. Modeling evolution at the protein level using an adjustable amino acid fitness model. Pac. Symp. Biocomput. 2000;5:18–29. doi: 10.1142/9789814447331_0003. [DOI] [PubMed] [Google Scholar]
- Fitch W.M, Leiter J.M.E, Li X, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc. Natl Acad. Sci. USA. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W.M, Bush R.M, Bender C.A, Cox N.J. Long term trends in the evolution of H(3)NA1 human influenza type A. Proc. Natl Acad. Sci. USA. 1997;94:7712–7718. doi: 10.1073/pnas.94.15.7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornasari M.S, Parisi G, Echave J. Site-specific amino acid replacement matrices from structurally constrained protein evolution simulations. Mol. Biol. Evol. 2002;19:352–356. doi: 10.1093/oxfordjournals.molbev.a004089. [DOI] [PubMed] [Google Scholar]
- Forsberg R, Christiansen F.B. A codon-based model of host-specific selection in parasites, with an application to the influenza A virus, Mol. Biol. Evol. 2003;20:1252–1259. doi: 10.1093/molbev/msg149. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Guindon S, Rodrigo A.G, Dyer K.A, Huelsenbeck J.P. Modeling the site-specific variation of selection patterns along lineages. Proc. Natl Acad. Sci. USA. 2004;101:12 957–12 962. doi: 10.1073/pnas.0402177101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D, Hurst L.D. A quantitative measure of error minimization in the genetic code. J. Mol. Evol. 1991;33:412–417. doi: 10.1007/BF02103132. [DOI] [PubMed] [Google Scholar]
- Halpern A.L, Bruno W.J. Evolutionary distances for protein-coding sequences: modeling site-specific residue frequencies. Mol. Biol. Evol. 1998;15:910–917. doi: 10.1093/oxfordjournals.molbev.a025995. [DOI] [PubMed] [Google Scholar]
- Holmes E.C, et al. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 2005;3:e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ina Y, Gojobori T. Statistical analysis of nucleotide sequences of the hemagglutinin gene of human influenza A viruses. Proc. Natl Acad. Sci. USA. 1994;91:8388–8392. doi: 10.1073/pnas.91.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977;267:275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. Cambridge University Press; Cambridge, UK: 1983. The neutral theory of molecular evolution. [Google Scholar]
- Kosakovsky Pond, S. L., Poon, A. F. Y., Brown, A. J. L. & Frost, S. D. W. In press. A maximum likelihood method for detecting directional evolution in protein sequences and its application to influenza A virus, Mol. Biol. Evol. (doi:10.1093/molbev/msn123) [DOI] [PMC free article] [PubMed]
- Koshi J.M, Goldstein R.A. Models of natural mutations including site heterogeneity. Proteins. 1998;32:289–295. doi: 10.1002/(SICI)1097-0134(19980815)32:3<289::AID-PROT4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Koshi J.M, Mindell D.P, Goldstein R.A. Using physical-chemistry-based substitution models in phylogenetic analysis of HIV-1 subtypes. Mol. Biol. Evol. 1999;16:173–179. doi: 10.1093/oxfordjournals.molbev.a026100. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-sites heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Lesk A.M, Chothia C. How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globins. J. Mol. Biol. 1980;136:225–270. doi: 10.1016/0022-2836(80)90373-3. [DOI] [PubMed] [Google Scholar]
- Lindstrom S.E, Cox N.J, Klimov A. Genetic analysis of human H2N2 and early H3N2 influenza viruses, 1957–1972: evidence for genetic divergence and multiple reassortment events. Virology. 2004;328:101–119. doi: 10.1016/j.virol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Mayrose I, Doron-Faigenboim A, Bacharach E, Pupko T. Towards realistic codon models: among site variability and dependency of synonymous and non-synononymous rates. Bioinformatics. 2007;23:i319–i327. doi: 10.1093/bioinformatics/btm176. [DOI] [PubMed] [Google Scholar]
- Meyer W.J, Wood J.M, Major D, Robertson J.S, Webster R.G, Katz J.M. Influence of host cell-mediated variation on the international surveillance of influenza A (H3N2) viruses. Virology. 1993;196:130–137. doi: 10.1006/viro.1993.1461. [DOI] [PubMed] [Google Scholar]
- Miyata T, Yasunaga T. Molecular evolution of mRNA: a method for estimating evolutionary rates of synonymous and amino acid substitutions from homologous nucleotide sequences and its application. J. Mol. Evol. 1980;16:23–36. doi: 10.1007/BF01732067. [DOI] [PubMed] [Google Scholar]
- Muse S.V, Gaut B.S. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome, Mol. Biol. Evol. 1994;11:715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- Nelson M.I, Holmes E.C. The evolution of epidemic influenza. Nat. Rev. Genet. 2007;8:196–205. doi: 10.1038/nrg2053. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Mol. Biol. Evol. 2003;20:1231–1239. doi: 10.1093/molbev/msg147. [DOI] [PubMed] [Google Scholar]
- Nixon K.C. The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics. 1999;15:407–414. doi: 10.1111/j.1096-0031.1999.tb00277.x. [DOI] [PubMed] [Google Scholar]
- Plotkin J, Dushoff J. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza A virus. Proc. Natl Acad. Sci. USA. 2003;100:7152–7157. doi: 10.1073/pnas.1132114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin J, Dushoff J, Levin S.A. Hemagglutinin sequence clusters and the antigenic evolution of influenza A virus. Proc. Natl Acad. Sci. USA. 2002;99:6263–6268. doi: 10.1073/pnas.082110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.M, Jones D.T, Kishino H, Goldman N, Thorne J. Protein evolution with dependence among codons due to tertiary structure. Mol. Biol. Evol. 2007;20:1692–1704. doi: 10.1093/molbev/msg184. [DOI] [PubMed] [Google Scholar]
- Rocha E.P, Xu X, Hall H.E, Allen J.R, Regnery H.L, Cox N.J. Comparison of 10 influenza A (H1N1 and H3N2) haemagglutinin sequences obtained directly from clinical specimens to those of MDCK cell- and egg-grown viruses. J. Gen. Virol. 1993;74:2513–2518. doi: 10.1099/0022-1317-74-11-2513. [DOI] [PubMed] [Google Scholar]
- Rodrigue N, Lartillot N, Bryant D, Philippe H. Site interdependence attributed to tertiary structure in amino acid sequence evolution. Gene. 2005;347:207–217. doi: 10.1016/j.gene.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Sainudiin R, Wong W.S.W, Yogeeswaran K, Nasrallah J.B, Yang Z, Nielsen R. Detecting site-specific physicochemical selective pressures: applications to the class I HLA of the human major histocompatibility complex and the SRK of the plant sporophytic self-incompatibility system. J. Mol. Evol. 2005;60:315–326. doi: 10.1007/s00239-004-0153-1. [DOI] [PubMed] [Google Scholar]
- Seoighe C, et al. A model of directional selection applied to the evolution of drug resistance in HIV-1. Mol. Biol. Evol. 2007;24:1025–1031. doi: 10.1093/molbev/msm021. [DOI] [PubMed] [Google Scholar]
- Shih A.C.-C, Hsiao T.-C, Ho M.-S, Li W.-H. Simultaneous amino acid substitutions at antigenic sites drive influenza A hemagglutinin evolution. Proc. Natl Acad. Sci. USA. 2007;104:6283–6288. doi: 10.1073/pnas.0701396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E.H. Measurement of diversity. Nature. 1949;163:688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- Suzuki Y. Natural selection on the influenza A virus genome. Mol. Biol. Evol. 2006;23:1902–1911. doi: 10.1093/molbev/msl050. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L. 2002 PAUP*. Phylogenetic analysis using parsimony (*and other methods), v. 4. Sunderland, MA: Sinauer Associates.
- Thompson J.D, Higgins D.G, Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne J.L, Goldman N, Jones D.T. Combining protein evolution and secondary structure. Mol. Biol. Evol. 1996;13:666–673. doi: 10.1093/oxfordjournals.molbev.a025627. [DOI] [PubMed] [Google Scholar]
- Urbina D, Tang B, Higgs P.G. The response of amino acid frequencies to directional mutation pressure in mitochondrial genome sequences is related to physical properties of the amino acids and to the structure of the genetic code. J. Mol. Evol. 2006;62:340–361. doi: 10.1007/s00239-005-0051-1. [DOI] [PubMed] [Google Scholar]
- Wilson I.A, Cox N.J. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 1990;8:737–771. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- Wolf Y.I, Viboud C, Holmes E.C, Koonin E.V, Lipman D.J. Long intervals of stasis punctuated by bursts of positive selection in the seasonal evolution of influenza A virus. Biol. Direct. 2006;1:34–53. doi: 10.1186/1745-6150-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W.S.W, Sainudiin R, Nielsen R. Identification of physicochemical selective pressure on protein encoding nucleotide sequences. BMC Bioinform. 2006;7:148. doi: 10.1186/1471-2105-7-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood estimation on large phylogenies and analysis of adaptive evolution in human influenza A. J. Mol. Evol. 2000;51:423–432. doi: 10.1007/s002390010105. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Mutation–selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol. Biol. Evol. 2008;25:568–579. doi: 10.1093/molbev/msm284. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Hasegawa M. Models of amino acid substitution and applications to mitochondrial protein evolution. Mol. Biol. Evol. 1998;15:1600–1611. doi: 10.1093/oxfordjournals.molbev.a025888. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen A.-M. K. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M, Kumar S, Hughes A.L. Sequence convergence in the peptide-binding region of primate and rodent MHC class Ib molecules. Mol. Biol. Evol. 1997;14:1035–1041. doi: 10.1093/oxfordjournals.molbev.a025709. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kumar S. Detection of convergent and parallel evolution at the amino acid sequence level. Mol. Biol. Evol. 1997;14:527–536. doi: 10.1093/oxfordjournals.molbev.a025789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Estimation of simulation parameters. Protein conservation statistics. Robustness of results. Detailed characterization of the behaviour of the mGY+\lambda model. Homoplasy statistic results
Tables containing the evolutionary parameter values inferred from 20 parsimonious trees