Abstract
In the Asian tropics, a conspicuous radiation of Macaranga plants is inhabited by obligately associated Crematogaster ants tending Coccus (Coccidae) scale insects, forming a tripartite symbiosis. Recent phylogenetic studies have shown that the plants and the ants have been codiversifying over the past 16–20 million years (Myr). The prevalence of coccoids in ant–plant mutualisms suggest that they play an important role in the evolution of ant–plant symbioses. To determine whether the scale insects were involved in the evolutionary origin of the mutualism between Macaranga and Crematogaster, we constructed a cytochrome oxidase I (COI) gene phylogeny of the scale insects collected from myrmecophytic Macaranga and estimated their time of origin based on a COI molecular clock. The minimum age of the associated Coccus was estimated to be half that of the ants, at 7–9 Myr, suggesting that they were latecomers in the evolutionary history of the symbiosis. Crematogaster mitochondrial DNA (mtDNA) lineages did not exhibit specificity towards Coccus mtDNA lineages, and the latter was not found to be specific towards Macaranga taxa, suggesting that patterns of associations in the scale insects are dictated by opportunity rather than by specialized adaptations to host plant traits.
Keywords: Coccus, coevolutionary radiation, mitochondrial DNA cytochrome oxidase I gene phylogeny, myrmecophytic Macaranga, southeast Asia
1. Introduction
Symbiosis has been a pervasive theme in the evolution of living systems. Indeed, much of life comprises composite organisms cobbled together from ancient interactions that evolved into obligate partnerships, exemplified at the cellular level by the mitochondria of eukaryotic cells. At higher levels of biological organization, organisms enlist the help of other species in extracting nutrients from the environment, such as the intestinal flora of animals and the fungi cultivated by leaf-cutter ants (Mueller et al. 2001). The need to defend against exploitation has likewise elicited creative solutions involving members of other species, as seen in ant-defended plants (Davidson & Mckey 1993) and fungus-defended termite eggs (Matsuura et al. 2000). So successful are some of these associations that they have persisted over geological time (e.g. Pellmyr & Leebens-Mack 1999; Machado et al. 2001; Mueller et al. 2001). The need to simultaneously exploit and protect oneself against exploitation has led to the evolution of intricate webs of associations involving multiple species. It is becoming evident that the long-term persistence of such associations is prevalent not only in two-party interactions, but also in more complex systems comprising three or more associates, such as the Attine ant–fungus–parasite (and microbe) system (Currie et al. 2003).
A frequently encountered phenomenon in tropical forests is the association between ants, plants and homopterans (sap-sucking insects). One such tripartite system comprises the obligate association between Macaranga trees, Crematogaster (subgenus Decacrema) ants and Coccus scale insects in southeast Asia (figure 1). The plants are protected against vines and herbivores by the ants that, in turn, gain residence in domatia created by hollow stems and food bodies secreted by the plants (Fiala et al. 1989). The plants do not survive to maturity without their Decacrema ants, and the ants are never found without their Macaranga hosts. In addition, the ants are able to further extract nutrients from the plants indirectly via the exudates of Coccus scale insects feeding on Macaranga sap and cohabiting with the ants in the confines of the Macaranga stem domatia (Heckroth et al. 1998).
Figure 1.
A Crematogaster (Decacrema) ant colony nesting inside a stem of Macaranga beccariana with Coccus symbiotic scale insects on the inner surface.
Recent phylogenetic studies have suggested that Macaranga plants and Decacrema ants have been codiversifying over the past 16–20 million years (Myr; Davies et al. 2001; Itino et al. 2001; Quek et al. 2004, 2007). The host plant assemblage comprises 26 Macaranga species in two closely related clades (Blattner et al. 2001; Davies et al. 2001) and the ants fall into a single mitochondrial DNA (mtDNA) clade consisting of 17 distinct mtDNA lineages (Quek et al. 2007). Although the ants and plants disperse independently of each other, most of the Decacrema mitochondrial lineages are specific to Macaranga species or clades (Itino et al. 2001; Quek et al. 2004, 2007). Coadaptations facilitating species specificity between the ants and plants include (i) species-specific defence systems in the ants (Itino & Itioka 2001; Itino et al. 2003), (ii) host selection using chemical cues by Decacrema founding queens (Inui et al. 2001), (iii) associated life-history traits of ants and plants (Feldhaar et al. 2003), and (iv) specialized stem textures in the plants and ant adaptations to host stem texture (Federle et al. 1997, 2000, 2002).
Scale insects appear to be a common associate of myrmecophytes, such as those in the Neotropical genera Cecropia and Cordia (Gullan 1997). Ward (1991) proposed that one route to the evolutionary origin of ant–plant mutualisms may have involved the tending of scale insects by ants in cavities bored by beetle or lepidopteran larvae. Davidson (1997) additionally proposed that the carbohydrate-rich exudates of homopterans fuel the high-tempo activities of their tending ants. Collectively, these propositions imply an important role for scale insects in the origin and evolution of ant–plant mutualisms.
Extensive sampling of Coccus from Macaranga and other plant genera in southeast Asia has revealed that they are highly specialized on myrmecophytic Macaranga (Heckroth et al. 1998), but have occasionally been found with phytoecious (i.e. dwelling in live plant cavities) Cladomyrma ants (Moog et al. 2005). The Coccus from Macaranga are believed to constitute a monophyletic group (P. J. Gullan 2008, personal communication), but this has yet to be ascertained. An intriguing but thus-far unanswered question is: how do the scale insects colonize the plant? Are they brought in by a founding ant queen, as observed in several phytoecious ant taxa (Moog et al. 2005), or do they arrive of their own accord? Moog et al. (2005) suggest that the former is unlikely in ant–plant–homopteran systems, such as the Decacrema–Macaranga–Coccus alliance, in which a founding queen must first chew a hole into the stem (since the mandibles are needed to secure the homopteran until entry into the domatium). Even more intriguing is to contemplate the question in evolutionary time: how did the tripartite system originate—did Coccus become part of the association only after Decacrema and Macaranga formed the mutualism or was Coccus already in association with Macaranga trees when Decacrema ants entered the scene? Alternatively, were the ants and scales already in association before colonizing Macaranga plants? Is it indeed the case that coccoids are an integral component in the origin of ant–plant mutualisms, as proposed by Ward (1991)? To address these questions, we reconstructed a gene phylogeny of the Macaranga-inhabiting scale insects and estimated a time frame for their diversification using nucleotide sequences from the mitochondrial gene cytochrome oxidase I (COI), the same gene used in dating the ant phylogeny (Quek et al. 2004, 2007). If the scales are brought in by founding queens (i.e. vertically transmitted), then we might expect to observe some influence of the Decacrema ant phylogeny on the Coccus scale phylogeny. And if the scales colonize independently of the ants, then the null expectation is a pattern of association that is independent of the ant phylogeny. Thus, we also characterize the specificity of the Coccus mtDNA lineages reported here to Macaranga clades, and the specificity of Decacrema mtDNA lineages (from Quek et al. 2007) towards the Coccus mtDNA lineages.
2. Material and methods
(a) Sampling
Scale insects were collected from 204 trees representing 22 Macaranga species in 13 locations throughout Borneo and Malaya (listed in the electronic supplementary material). We focused on myrmecophytic Macaranga trees because the Macaranga-inhabiting Coccus are reported to be specific to these (Heckroth et al. 1998). Most of the scale insects were collected from the ant colonies reported in Quek et al. (2007). For out-groups we sequenced (i) three free-living Coccus species (Coccus celatus, Coccus hesperidum and Coccus pseudomagnoliarum) believed to be close relatives of the Coccus on Macaranga (P. J. Gullan & T. Kondo 2008, personal communication), (ii) a member of the genus Pulvinaria within the same subfamily, Coccinae (Hodgson 1994), and (iii) two genera, Eulecanium and Parthenolecanium, in the subfamily Eulecaninae within Coccidae (Hodgson 1994).
(b) DNA extraction, polymerase chain reaction and sequencing
DNA was extracted from single ethanol-preserved scale insects using a ‘salting-out’ protocol (Sunnucks & Hales 1996). A partial COI gene (521 bp) corresponding to positions 1718–2239 in the Drosophila yakuba mtDNA genome was amplified by polymerase chain reaction (PCR) with the primers mtD-6 (5′-GGA TCA CCT GAT ATA GCA TTC CC-3′; Simon et al. 1994) and CI-N4 (5′-CCT GGT AGG ATT AAA ATA TAT AC-3′), and using the following PCR temperature profile: 35 cycles of 95°C for 30 s, 42°C for 30 s and 72°C for 40 s. After amplification, PCR products were purified using Qiagen PCR purification kits. Cycle sequencing reactions for both strands were performed with the BigDye Terminator cycle sequencing ready reaction kit on an ABI 377 automated sequencer. Upon the inspection of the COI sequences, it was discovered that a pseudogene had been amplified in the out-group C. celatus (as indicated by a frame-shift mutation with an ensuing stop codon near the 5′ end) and thus it was removed from analyses.
(c) Phylogenetic analyses
Maximum-likelihood (ML) analyses were performed with Phyml (Guindon & Gascuel 2003), using the general time-reversible substitution model with a proportion of invariant sites and a gamma-distributed rate heterogeneity (GTR+I+G), as selected by hierarchical likelihood ratio tests (LRTs; Huelsenbeck & Rannala 1997) implemented in Modeltest v. 3.06 (Posada & Crandall 1998). Clade support was assessed with 1000 bootstrapped datasets in Phyml. The GTR+I+G model was also used in Bayesian analysis, conducted in MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001), using the default run settings that perform two independent analyses with four Markov chain Monte Carlo chains each (one cold and three heated). The Bayesian analysis was run for five million generations, with a burn-in of four million generations, well after stationarity was reached. Parsimony analyses were performed with PAUP* v. 4.0.b10 (Swofford 2002), using the heuristic search option with 1000 random addition replicates and TBR branch swapping. Clade support was assessed from 1000 bootstrap replicates using 10 random addition replicates.
(d) Partner preference
Specificity of ant lineages to Coccus lineages (using ant lineage designations from Quek et al. 2007) was determined by chi-squared tests of the extent of departure of the observed Coccus lineage's proportion from its expected proportion as indicated by its frequency in all the locations where the ant lineage was sampled. Specificity of Coccus lineages to host plant clades (sections Pachystemon, Pruinosae and Winklerianae) was also determined in a similar manner. Macaranga section Pachystemon contains the majority of the myrmecophytes (21 species) and the closely related section Pruinosae contains five myrmecophytic species; section Winklerianae, more distantly related, has two myrmecophytes (see Davies et al. 2001). Five thousand Monte Carlo simulations of the sampling distribution were also performed to assess the observed sample against the null distribution. Ant clades C and E (n=1 in both clades) were removed from the analyses.
(e) Age estimation
A general conservatism in COI substitution rates in arthropods (Gaunt & Miles 2002) makes this gene useful for dating divergences within arthropod groups (e.g. Degnan et al. 2004). In addition, the pairwise divergence rate of COI was found to cluster approximately 1.5% Myr−1 (uncorrected pairwise distance) in several arthropod groups (Quek et al. 2004); thus we used this rate to estimate divergences within the COI phylogeny. Homogeneity of substitution rate on the ML phylogeny of the scales was tested using the LRT (Huelsenbeck & Rannala 1997) with the GTR+I+G model, implemented in PAUP* v. 4.0b10. We removed all duplicate haplotypes from the LRT. Because the LRT showed significant deviations from rate constancy (p<0.05), branch lengths were subjected to non-parametric rate smoothing (Sanderson 1997), implemented in TreeEdit v. 1.0 (Rambaut & Charleston 2002). Three well-supported nodes of varying genetic divergences (5.8–14.2%) were chosen as fixation points to obtain the relative ages of the other nodes. This range of divergences was chosen to approximate that used for dating divergences in various arthropod groups (table 4 in Quek et al. 2004). This approach produces a range of ages rather than a point estimate, and is thus more conservative. Mean uncorrected pairwise distances between sister taxa were calculated using MEGA v. 2.1 (Kumar et al. 2001).
3. Results and discussion
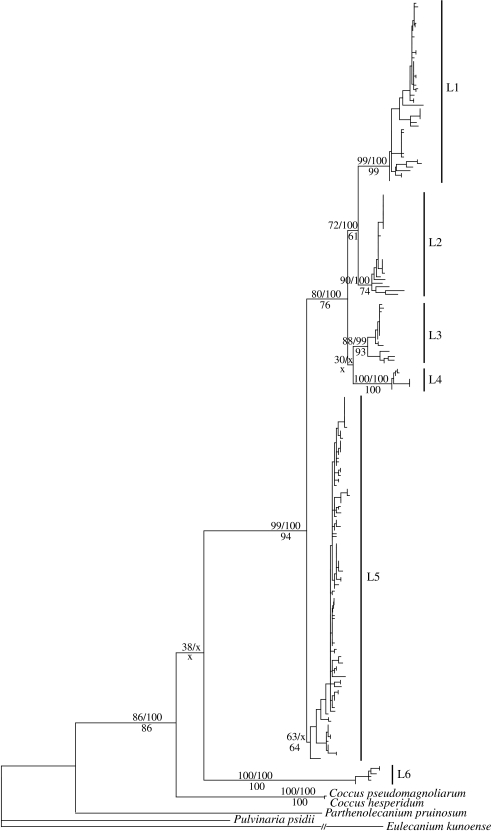
Phylogenetic analyses reveal six well-supported primary mtDNA lineages, L1–L6 (figure 2). The monophyly of the in-group (L1–L6) was recovered in the ML topology, but was poorly supported by ML bootstrapping and not supported by parsimony bootstrapping and Bayesian posterior probability; in parsimony and Bayesian analyses, L6 was placed sister to the two Coccus out-groups, albeit with poor support. The branch uniting L3 and L4 in the ML topology was similarly poorly supported—in Bayesian and parsimony analyses, L3 was placed sister to the node uniting L1 with L2, but again with low support. The membership among lineages varied considerably (from n=5 in L6 to n=87 in L5), probably reflecting the natural abundance of the scale lineages, because our sampling was carried out with no prior knowledge of the scales' distribution and without regard to their morphology.
Figure 2.
Phylogeny of 220 Macaranga-inhabiting Coccus scale insects estimated from 521 bp mtDNA sequences (COI) using ML. The six primary mtDNA lineages of Coccus scales are indicated (L1–L6). The numbers above branches and left of slash indicate ML bootstrap support (1000 replicates), numbers right of slash indicate Bayesian posterior probabilities, and numbers below branches indicate parsimony bootstrap support (1000 replicates). An ‘x’ in the node support values indicates the absence of the node in the parsimony bootstrap or Bayesian consensus topology.
The chi-squared test for the ant lineages' preference for the scale lineages indicated that none of the ant lineages showed significant preferences for particular scale lineages (table 1), and specificity between the scale lineages and Macaranga taxa was also not observed (table 2). By contrast, Quek et al. (2007) revealed that most ant mtDNA lineages showed specificity towards Macaranga taxa. The non-specific association of scales with ants and plants has similarly been documented based on morphospecies of the ants and scales (Heckroth et al. 1998), and is consistent with independent colonization of Macaranga plants by scales and ants. Thus, in contrast to the ants, the scales' association within the system, while obligate, is dictated by opportunity rather than by physiological, behavioural or ecological adaptations to specific ant or host plant taxa.
Table 1.
Chi-squared tests for biased association between ant lineages and scale insect lineages. (The p-values are from 5000 Monte Carlo simulations; n.s., not significant.)
| ant lineage (sample size) | scale insect lineage | expected proportion | observed proportion | p |
|---|---|---|---|---|
| A (3) | L2 | 28/160 (0.18) | 0/3 (0.00) | n.s. |
| L3 | 14/160 (0.09) | 0/3 (0.00) | n.s. | |
| L4 | 7/160 (0.04) | 0/3 (0.00) | n.s. | |
| L5 | 107/160 (0.67) | 2/3 (0.67) | n.s. | |
| L6 | 4/160 (0.03) | 1/3 (0.33) | n.s. | |
| D (12) | L2 | 28/160 (0.18) | 4/12 (0.33) | n.s. |
| L3 | 14/160 (0.09) | 2/12 (0.17) | n.s. | |
| L4 | 7/160 (0.04) | 0/12 (0.00) | n.s. | |
| L5 | 107/160 (0.67) | 6/12 (0.50) | n.s. | |
| L6 | 4/160(0.03) | 0/12 (0.00) | n.s. | |
| F (10) | L2 | 28/160 (0.18) | 0/10 (0.00) | n.s. |
| L3 | 14/160 (0.09) | 4/10 (0.40) | n.s. | |
| L4 | 7/160 (0.04) | 0/10 (0.00) | n.s. | |
| L5 | 107/160(0.67) | 6/10 (0.60) | n.s. | |
| L6 | 4/160 (0.03) | 0/10 (0.00) | n.s. | |
| G (47) | L2 | 28/160 (0.18) | 6/47 (0.13) | n.s. |
| L3 | 14/160 (0.09) | 1/47 (0.02) | n.s. | |
| L4 | 7/160 (0.04) | 2/47 (0.04) | n.s. | |
| L5 | 107/160 (0.67) | 37/47 (0.79) | n.s. | |
| L6 | 4/160 (0.03) | 1/47(0.02) | n.s. | |
| H (61) | L2 | 28/160 (0.18) | 10/61(0.16) | n.s. |
| L3 | 14/160 (0.09) | 2/61 (0.03) | n.s. | |
| L4 | 7/160 (0.04) | 4/61 (0.07) | n.s. | |
| L5 | 107/160 (0.67) | 44/61 (0.72) | n.s. | |
| L6 | 4/160 (0.03) | 1/61 (0.02) | n.s. | |
| K (37) | L1 | 53/60 (0.88) | 35/37 (0.95) | n.s. |
| L2 | 2/60 (0.03) | 1/37 (0.03) | n.s. | |
| L3 | 3/60 (0.05) | 0/37 (0.00) | n.s. | |
| L6 | 2/60 (0.03) | 1/37 (0.03) | n.s. |
Table 2.
Chi-squared tests for biased association between scale insect lineages and host plant section. (The p-values are from 5000 Monte Carlo simulations; n.s., not significant.)
| Coccus lineage (sample size) | Macaranga section tested | expected proportion | observed proportion | p |
|---|---|---|---|---|
| L1 (49) | Pachystemon | 51/60 (0.85) | 45/49 (0.92) | n.s. |
| Pruinosae | 5/60 (0.08) | 4/49 (0.08) | n.s. | |
| L2 (30) | Pachystemon | 174/216 (0.81) | 21/30 (0.70) | n.s. |
| Pruinosae | 38/216 (0.18) | 7/30 (0.23) | n.s. | |
| Winklerianae | 4/216 (0.02) | 2/30 (0.07) | n.s. | |
| L3 (17) | Pachystemon | 174/216 (0.81) | 14/17 (0.82) | n.s. |
| Pruinosae | 38/216 (0.18) | 1/17 (0.06) | n.s. | |
| Winklerianae | 4/216 (0.02) | 2/17 (0.12) | n.s. | |
| L4 (7) | Pachystemon | 123/160 (0.77) | 6/7 (0.86) | n.s. |
| Pruinosae | 33/160 (0.21) | 1/7 (0.14) | n.s. | |
| Winklerianae | 4/160 (0.03) | 0/7 (0.00) | n.s. | |
| L5 (107) | Pachystemon | 123/160 (0.77) | 83/107 (0.78) | n.s. |
| Pruinosae | 33/160 (0.21) | 24/107 (0.22) | n.s. | |
| Winklerianae | 4/160 (0.03) | 0/107 (0.00) | n.s. | |
| L6 (6) | Pachystemon | 174/216 (0.81) | 5/6 (0.83) | n.s. |
| Pruinosae | 38/216 (0.18) | 1/6 (0.17) | n.s. | |
| Winklerianae | 4/216 (0.02) | 0/6 (0.00) | n.s. |
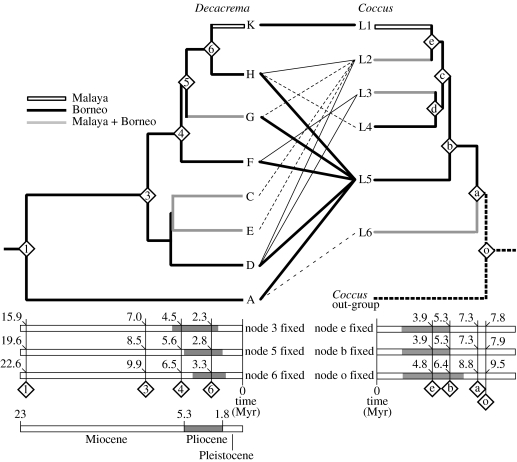
The minimum estimated age of the Coccus inhabiting Macaranga is 7.3–8.8 Myr (table 3; figure 3, node a), while that of the ants is 15.9–22.6 Myr (Quek et al. 2007). The latter estimate is consistent with the origin of ever wet forests in western Malesia at approximately 20 Myr ago (Morley 2000) and also with the origin of myrmecophytic Macaranga, since myrmecophytic Macaranga are confined to the aseasonal ever wet climatic zone there (Davies et al. 2001). Thus, taken at face value, the time-scaled phylogeny suggests that the scales are only approximately half as old as the ants. Certainly, we expect that the true pairwise divergence rate of COI in the scales would vary somewhat around the assumed 1.5% Myr−1. Allowing a similar age for the ants and scales would require halving the scales' rate to approximately 0.6% Myr−1. Such an extensive departure seems unlikely, given that a survey of geologically calibrated COI rates encompassing beetles, cicadas and crustaceans yielded a range between 1.3 and 1.9% Myr−1 (see table 4 in Quek et al. 2004). Furthermore, if we take into account the possibility that the scales collected from Macaranga are not monophyletic (figure 2) and that L6 represents an independent colonization of the Decacrema–Macaranga system by Coccus, then, based on the estimate at node b (for the clade comprising L1–L5 in figure 3), rather than node a, the minimum age estimate is even younger at 5.3–6.4 Myr. However, studies by T. Kondo (2003–2007, unpublished data) indicate that the Macaranga coccids form a morphologically coherent group; thus the hypothesis of independent colonization events seems unlikely. It is also possible that there are older Coccus lineages not detected by our sampling, which might bring our age estimate closer to that of the ants but this seems unlikely, given the broad geographical area sampled. It is also plausible that such older lineages may have become extinct. Alternatively, the mutualism between Macaranga and Decacrema may have originated with a taxon of coccoids that have since become replaced by those in this study. Unfortunately, hypotheses involving the possibility of extinct or replaced taxa are untestable.
Table 3.
Mean uncorrected pairwise distances and inferred ages of nodes on the Coccus chronogram (figure 3) obtained by non-parametric rate smoothing, based on a pairwise divergence rate of 1.5% Myr−1. (Fixed ages are indicated in italics.)
| node | mean pairwise distance (%) | age (node o fixed) | age (node b fixed) | age (node e fixed) |
|---|---|---|---|---|
| o (out-group/L1–L6) | 14.2 | 9.47 | 7.86 | 7.84 |
| a (L6/L1–L5) | 15.4 | 8.78 | 7.28 | 7.27 |
| b (L5/L1–L4) | 7.9 | 6.35 | 5.27 | 5.26 |
| c (L3–L4/L1–L2) | 6.7 | 5.17 | 4.29 | 4.28 |
| d (L3/L4) | 5.6 | 4.91 | 4.07 | 4.06 |
| e (L1/L2) | 5.9 | 4.75 | 3.94 | 3.93 |
Figure 3.
Comparison of the Coccus scale insect chronogram with that of Decacrema ants (modified from Quek et al. 2007). A thick black line connecting an ant lineage to a scale lineage indicates that the scale lineage constitutes 50% or more of the ant's total scale associates; a narrow line indicates 16–50% and dashed line 7–13% (or single sample); scales constituting less than 5% are not shown.
If we accept the data at face value, then we infer that the scale insects were indeed latecomers and, therefore, that the origin of the mutualism between Decacrema and Macaranga did not involve scale insects, in contrast to Ward's (1991) model for the origin of ant–plant mutualisms. If so, the ants may have relied on extrafloral nectaries (EFNs) in place of coccoids or they may have relied only on food bodies provisioned by the plants (as seen in Acacia–ant mutualisms, Janzen 1966). Interestingly, Davies et al. (2001) showed that myrmecophytism evolved only in Macaranga lineages in which EFNs on mature leaf surfaces had been lost and in which leaf margin (as opposed to leaf surface) EFNs became enlarged, suggesting a role for carbohydrate-rich plant exudates, whether in the form of EFNs or homopteran honeydew, in the evolution of myrmecophytism.
The single-locus dataset used here does not lend itself to inferring species boundaries, which can conflict with mitochondrial lineages due to introgression or incomplete lineage sorting (Avise 1994; Sota & Vogler 2001; Shaw 2002; Linnen & Farrell 2007). Indeed, preliminary sequence data from nuclear loci are in disagreement with the mtDNA phylogeny presented here (S. Ueda 2008, unpublished data). However, our main objective in this paper is to infer the antiquity of the Macaranga-associated scale insects. Certainly, introgression and incomplete lineage sorting can introduce errors into our age estimation, but the relative contribution of this error decreases with evolutionary time, especially when measured using fast-evolving genes such as metazoan COI. At the time frame of 7–9 Myr (the older end of the time scale on our phylogeny), the effect of introgression or incomplete lineage sorting should not contribute to any worrying deviation that is not already contained by our bracketing method (where we obtain three time frames based on three fixed nodes).
The obligate and ancient association of Macaranga plants, Decacrema ants and Coccus scale insects shown here appears to be unique in contrast to the tropical American ant–plant systems in which numerous plant and ant genera are involved, and intergeneric host shifts are not uncommon (Davidson & Mckey 1993). Furthermore, other taxa appear to have evolved obligate associations with the Macaranga symbiotic system, such as phytophagous Arhopala butterflies (Maschwitz et al. 1984). The Macaranga-based symbioses thus offer a promising system for the study of coevolutionary radiations of multipartite and obligate associations. Further empirical and theoretical treatment of this complex association may provide a reflection of the evolutionary dynamics of coevolving communities in the rainforests of southeast Asia.
Acknowledgments
We thank P. J. Gullan and T. Kondo for taxonomic advice, provision of out-group specimens and comments on the manuscript, S. J. Davies for plant identification, N. E. Pierce, A. Hamid, U. Ban, T. Fukatsu, T. Yokokawa, S. Usami, Y. Nakamura and S. Matsuda for analytical advice and/or technical support. This research was funded by the Japan Ministry of Education, Culture, Sports, Science and Technology (12640614 and 14540577 to T.I.). We thank Sabah Parks, National Parks Board (Singapore) and Sarawak Forest Department for collecting permission.
Supplementary Material
List of samples showing locality and associated ants and host plants. Host plant species are abbreviated as follows: aet=Macaranga aëtheadenia, ang=M. angulata, ban=M. bancana, bec=M. beccariana, gla=M. glandibracteolata, gri=M. griffithiana, hos=M. hosei, hul=M. hullettii, hyp=M. hypoleuca, ind=M. indistincta, kin=kingii, lam=lamellata, mot=M. motleyana, pea=M. pearsonii, pet=M. petanostyla, pru=M. pruinosa, pse=M. pseudopruinosa, pub=M. puberula, tra=M. trachyphylla, vel=velutina, win=M. winkleri. Ant lineages follow Quek et al. 2007
References
- Avise J.C. Chapman and Hall; New York, NY: 1994. Molecular markers, natural history and evolution. [Google Scholar]
- Blattner F.R, Weising K, Banfer G, Maschwitz U, Fiala B. Molecular analysis of phylogenetic relationships among myrmecophytic Macaranga species (Euphorbiaceae) Mol. Phylogenet. Evol. 2001;19:331–344. doi: 10.1006/mpev.2001.0941. doi:10.1006/mpev.2001.0941 [DOI] [PubMed] [Google Scholar]
- Currie C.R, Wong B, Stuart A.E, Schultz T.R, Rehner S.A, Mueller U.G, Sung G.H, Spatafora J.W, Straus N.A. Ancient tripartite coevolution in the attine ant–microbe symbiosis. Science. 2003;299:386–388. doi: 10.1126/science.1078155. doi:10.1126/science.1078155 [DOI] [PubMed] [Google Scholar]
- Davidson D.W. The role of resource imbalances in the evolutionary ecology of tropical arboreal ants. Biol. J. Linn. Soc. 1997;61:153–181. doi:10.1006/bijl.1996.0128 [Google Scholar]
- Davidson D.W, Mckey D. The evolutionary ecology of symbiotic ant–plant relationships. J. Hym. Res. 1993;2:13–83. [Google Scholar]
- Davies S.J, Lum S.K.Y, Chan R, Wang L.K. Evolution of myrmecophytism in western Malesian Macaranga (Euphorbiaceae) Evolution. 2001;55:1542–1559. doi: 10.1111/j.0014-3820.2001.tb00674.x. doi:10.1554/0014-3820(2001)055[1542:EOMIWM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Degnan P.H, Lazarus A.B, Brock C.D, Wernegreen J.J. Host–symbiont stability and fast evolutionary rates in an ant–bacterium association: cospeciation of Camponotus species and their endosymbionts Candidatus Blochmannia. Syst. Biol. 2004;53:95–110. doi: 10.1080/10635150490264842. doi:10.1080/10635150490264842 [DOI] [PubMed] [Google Scholar]
- Federle W, Mashwitz U, Fiala B, Riederer M, Hölldobler B. Slippery ant–plants and skillful climbers: selection and protection of specific ant partners by epicuticular wax blooms in Macaranga (Euphorbiaceae) Oecologia. 1997;112:217–224. doi: 10.1007/s004420050303. doi:10.1007/s004420050303 [DOI] [PubMed] [Google Scholar]
- Federle W, Rohrseitz K, Holldobler B. Attachment forces of ants measured with a centrifuge: better ‘wax-runners’ have a poorer attachment to a smooth surface. J. Exp. Biol. 2000;203:505–512. doi: 10.1242/jeb.203.3.505. [DOI] [PubMed] [Google Scholar]
- Federle W, Maschwitz U, Hölldobler B. Pruning of host plant neighbours as defence against enemy ant invasions: Crematogaster ant partners of Macaranga protected by ‘wax barriers’ prune less than their congeners. Oecologia. 2002;132:264–270. doi: 10.1007/s00442-002-0947-z. doi:10.1007/s00442-002-0947-z [DOI] [PubMed] [Google Scholar]
- Feldhaar H, Fiala B, Hashim R.B, Maschwitz U. Patterns of the Crematogaster–Macaranga association: the ant partner makes the difference. Insectes Soc. 2003;50:9–19. doi:10.1007/s000400300002 [Google Scholar]
- Fiala B, Maschwitz U, Pong T.Y, Helbig A.J. Studies of a southeast Asian ant–plant association: protection of Macaranga trees by Crematogaster borneensis. Oecologia. 1989;79:463–470. doi: 10.1007/BF00378662. doi:10.1007/BF00378662 [DOI] [PubMed] [Google Scholar]
- Gaunt M.W, Miles M.A. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol. Biol. Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. doi:10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Gullan P.J. Relationships with ants. In: Ben-Dov Y, Hodgson C.J, editors. Soft scale insects—their biology, natural enemies and control. Elsevier Science B. V.; Amsterdam, The Netherlands: 1997. pp. 351–373. [Google Scholar]
- Heckroth H.P, Fiala B, Gullan P.J, Idris A.H, Maschwitz U. The soft scale (Coccidae) associates of Malaysian ant-plants. J. Trop. Ecol. 1998;14:427–443. doi:10.1017/S0266467498000327 [Google Scholar]
- Hodgson C.J. CAB International; Wallingford, UK: 1994. The scale insect family Coccidae: an identification manual to genera. [Google Scholar]
- Huelsenbeck J.P, Rannala B. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science. 1997;276:227–232. doi: 10.1126/science.276.5310.227. doi:10.1126/science.276.5310.227 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Inui Y, Itioka T, Murase K, Yamaoka R, Itino T. Chemical recognition of partner plant species by foundress ant queens in Macaranga–Crematogaster myrmecophytism. J. Chem. Ecol. 2001;27:2029–2040. doi: 10.1023/a:1012290820150. doi:10.1023/A:1012290820150 [DOI] [PubMed] [Google Scholar]
- Itino T, Itioka T. Interspecific variation and ontogenetic change in antiherbivore defense in myrmecophytic Macaranga species. Ecol. Res. 2001;16:765–774. doi:10.1046/j.1440-1703.2001.00432.x [Google Scholar]
- Itino T, Davies S.J, Tada H, Hieda O, Inoguchi M, Itioka T, Yamane S, Inoue T. Cospeciation of ants and plants. Ecol. Res. 2001;16:787–793. doi:10.1046/j.1440-1703.2001.00442.x [Google Scholar]
- Itino T, Itioka T, Davies S.J. Coadaptation and coevolution of Macaranga trees and their symbiotic ants. In: Kikuchi T, Higashi S, Azuma N, editors. Genes, behaviors and evolution of social insects. Hokkaido University Press; Sapporo, Japan: 2003. pp. 283–294. [Google Scholar]
- Janzen D.H. Coevolution of mutualism between ants and acacias in central America. Evolution. 1966;20:249–275. doi: 10.1111/j.1558-5646.1966.tb03364.x. doi:10.2307/2406628 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen I.B, Nei M. MEGA 2.1: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. doi:10.1093/bioinformatics/17.12.1244 [DOI] [PubMed] [Google Scholar]
- Linnen C.R, Farrell B.D. Mitonuclear discordance is caused by rampant mitochondrial introgression in Neodiprion (Hymenoptera: Diprionidae) sawflies. Evolution. 2007;61:1417–1438. doi: 10.1111/j.1558-5646.2007.00114.x. doi:10.1111/j.1558-5646.2007.00114.x [DOI] [PubMed] [Google Scholar]
- Machado C.A, Jousselin E, Kjellberg F, Compton S.G, Herre E.A. Phylogenetic relationships, historical biogeography and character evolution of fig-pollinating wasps. Proc. R. Soc. B. 2001;268:685–694. doi: 10.1098/rspb.2000.1418. doi:10.1098/rspb.2000.1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschwitz U, Schroth M, Hänel H, Pong T.Y. Lycaenids parasitizing symbiotic plant–ant partnerships. Oecologia. 1984;64:78–80. doi: 10.1007/BF00377547. doi:10.1007/BF00377547 [DOI] [PubMed] [Google Scholar]
- Matsuura K, Tanaka C, Nishida T. Symbiosis of a termite and a sclerotium-forming fungus: Sclerotia mimic termite eggs. Ecol. Res. 2000;15:405–414. doi:10.1046/j.1440-1703.2000.00361.x [Google Scholar]
- Moog J, Saw L.G, Hashim R, Maschwitz U. The triple alliance: how a plant-ant, living in an ant-plant, acquires the third partner, a scale insect. Insectes Soc. 2005;52:169–176. doi:10.1007/s00040-005-0791-3 [Google Scholar]
- Morley R.J. Wiley; Chichester, UK: 2000. Origin and evolution of tropical rain forests. [Google Scholar]
- Mueller U.G, Schultz T.R, Currie C.R, Adams R.M, Malloch D. The origin of the attine ant–fungus mutualism. Q. Rev. Biol. 2001;76:169–197. doi: 10.1086/393867. doi:10.1086/393867 [DOI] [PubMed] [Google Scholar]
- Pellmyr O, Leebens-Mack J. Forty million years of mutualism: evidence for Eocene origin of the yucca–yucca moth association. Proc. Natl Acad. Sci. USA. 1999;96:9178–9183. doi: 10.1073/pnas.96.16.9178. doi:10.1073/pnas.96.16.9178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Quek S.-P, Davies S.J, Itino T, Pierce N.E. Codiversification in an ant–plant mutualism: stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) inhabitants of Macaranga (Euphorbiaceae) Evolution. 2004;58:554–570. doi:10.1554/03-361 [PubMed] [Google Scholar]
- Quek S.-P, Davies S.J, Ashton P.S, Itino T, Pierce N.E. The geography of diversification in mutualistic ants: a gene's-eye view into the Neogene history of Sundaland rain forests. Mol. Ecol. 2007;16:2045–2062. doi: 10.1111/j.1365-294X.2007.03294.x. doi:10.1111/j.1365-294X.2007.03294.x [DOI] [PubMed] [Google Scholar]
- Rambaut, A. & Charleston, M. 2002 TreeEdit Phylogenetic tree editor, v. 1.0 α 10. Oxford, UK: Department of Zoology, University of Oxford.
- Sanderson M.J. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol. Biol. Evol. 1997;14:1218–1231. [Google Scholar]
- Shaw K.L. Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: what mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proc. Natl Acad. Sci. USA. 2002;99:16 122–16 127. doi: 10.1073/pnas.242585899. doi:10.1073/pnas.242585899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994;87:651–701. [Google Scholar]
- Sota T, Vogler A.P. Incongruence of mitochondrial and nuclear gene trees in the carabid beetles Ohomopterus. Syst. Biol. 2001;50:39–59. [PubMed] [Google Scholar]
- Sunnucks P, Hales D.F. Numerous transposed sequences of mitochondrial cytochrome oxidase I–II in aphids of the genus Sitobion (Hemiptera: Aphididae) Mol. Biol. Evol. 1996;13:510–524. doi: 10.1093/oxfordjournals.molbev.a025612. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L. 2002 PAUP*: phylogenetic analysis using parsimony, v. 4.0.b10. Sunderland, MA: Sinauer Associates.
- Ward P.S. Phylogenetic analysis of pseudomyrmecine ants associated with domatia-bearing plants. In: Huxley C.R, Cutler D.F, editors. Ant–plant interactions. Oxford University Press; Oxford, UK: 1991. pp. 335–352. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of samples showing locality and associated ants and host plants. Host plant species are abbreviated as follows: aet=Macaranga aëtheadenia, ang=M. angulata, ban=M. bancana, bec=M. beccariana, gla=M. glandibracteolata, gri=M. griffithiana, hos=M. hosei, hul=M. hullettii, hyp=M. hypoleuca, ind=M. indistincta, kin=kingii, lam=lamellata, mot=M. motleyana, pea=M. pearsonii, pet=M. petanostyla, pru=M. pruinosa, pse=M. pseudopruinosa, pub=M. puberula, tra=M. trachyphylla, vel=velutina, win=M. winkleri. Ant lineages follow Quek et al. 2007