Abstract
Lentiviruses chronically infect a broad range of mammalian species and have been transmitted from primates to humans, giving rise to multiple outbreaks of HIV infection over the past century. Although the circumstances surrounding these recent zoonoses are becoming clearer, the nature and timescale of interaction between lentiviruses and primates remains unknown. Here, we report the discovery of an endogenous lentivirus in the genome of the gray mouse lemur (Microcebus murinus), a strepsirrhine primate from Madagascar, demonstrating that lentiviruses are capable of invading the primate germ line. Phylogenetic analysis places gray mouse lemur prosimian immunodeficiency virus (pSIVgml) basal to all known primate lentiviruses and, consistent with this, its genomic organization is intermediate between the nonprimate lentiviruses and their more derived primate counterparts. Thus, pSIVgml represents the first unambiguous example of a viral transitional form, revealing the acquisition and loss of genomic features during lentiviral evolution. Furthermore, because terrestrial mammal populations in Madagascar and Africa are likely to have been isolated from one another for at least 14 million years, the presence of pSIVgml in the gray mouse lemur genome indicates that lentiviruses must have been infecting primates for at least this period of time, or have been transmitted between Malagasy and African primate populations by a vector species capable of traversing the Mozambique channel. The discovery of pSIVgml illustrates the utility of endogenous sequences for the study of contemporary retroviruses and indicates that primate lentiviruses may be considerably older and more broadly distributed than previously thought.
Keywords: lemur, retrovirus, immunodeficiency, ERV, Madagascar
Lentiviruses are complex retroviruses that cause chronic infections in a broad range of mammalian species including primates, ungulates, and felids. Lentiviruses that circulate among free-ranging populations of African apes and monkeys have occasionally been transmitted to humans, resulting in pandemic spread of HIV-1 (group M) as well as the epidemic spread of HIV-1 (group O) and two major HIV-2 lineages (1). Through phylogenetic analysis of lentiviral sequence data, the circumstances surrounding these relatively recent events have been reconstructed in detail (2–6). However, because of very high viral mutation rates, contemporary sequence data have limited power to reveal the distant evolutionary history of viruses (7). Consequently, the long-term evolutionary history of lentiviruses, including their origin within primates, the timescale of their interaction with mammals, and the pattern of viral gene acquisition and loss, remain unknown (8).
Retroviral genome invasions—whereby retroviruses infect germ line cells, causing integrated proviral sequences to be inherited as host alleles termed ‘endogenous retroviruses’ (ERVs)—have occurred throughout the evolution of vertebrates and continue to the present day (9–11). Because ERV sequences evolve relatively slowly, and can be dated by a variety of means, they provide a unique molecular ‘fossil record’ that can reveal the long-term dynamics of retroviral evolution (12, 13). Although genome invasion by lentiviruses appears to be relatively uncommon compared with other retroviral groups, a recent study has established that it can occur (14). It is therefore likely that a paleovirological record of lentiviral evolution may be reconstructed through targeted screening of vertebrate genomes. In this study, we report the discovery of an endogenous lentivirus in the genome of a strepsirrhine primate, the gray mouse lemur (Microcebus murinus). We investigate the phylogenetic, genomic, and biogeographic characteristics of this virus and discuss the implications of our results for lentivirus evolution.
Results
Host Genome Screening and Construction of pSIVgml Consensus.
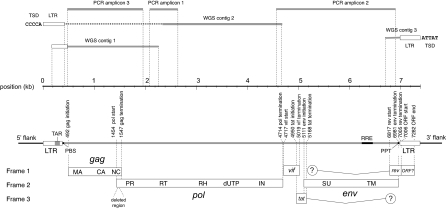
We screened complete, low coverage, and trace archive genome sequence data from a total of 21 primate species (see Table S1) using the tBLASTn program and peptide sequences derived from the Pol proteins of representative lentiviruses. Screening identified several highly significant matches to lentiviral peptides in the low coverage genome sequence of M. murinus. Matching sequences were extracted and found to group robustly with representative lentiviruses in preliminary phylogenetic analysis, confirming their lentiviral origin. Through an iterative process of screening M. murinus genome data and comparing extracted sequences to representative lentivirus genomes, we obtained three contigs that together contained the 5′ and 3′ long terminal repeat (LTR) sequences and nearly complete gag and pol coding domains of an endogenous lentivirus (Fig. 1 and Fig. S1), which we named gray mouse lemur prosimian immunodeficiency virus (pSIVgml).
Fig. 1.
Consensus pSIVgml genome. Horizontal lines above the scale-bar indicate the relative coordinates of the contigs derived from whole genome shotgun (WGS) sequences and PCR amplicons that were used to construct the consensus genome. WGS contig 2 contained a region of indeterminate sequence, indicated by a dashed line. Distinct 5 base pair target site duplication (TSD) sequences located at the 5′ and 3′ flanks of proviral insertions are shown, demonstrating the presence of at least two distinct proviruses. The schematic below the scale shows the locations of ORFs and genomic features within the consensus proviral genome sequence. Circles with inset question marks indicate the possible presence of short, spliced Rev and Tat domains toward the 5′ and 3′ ends of the env gene, respectively; the general lack of sequence similarity of these regions within lentiviruses meant that it was not possible to determine whether or not pSIVgml encodes such domains. LTR, long-terminal repeat; TAR, transactivation responsive element; PBS, primer binding site; MA, matrix; CA, capsid; NC, nucleocapsid; PR, protease; RT, reverse transcriptase; RH, RNaseH; dUTP, dUTPase; IN, integrase; SU, surface glycoprotein; TM, transmembrane; RRE, Rev responsive element.
Through comparison of target site duplication (TSD) and genomic DNA sequences flanking viral LTRs, we established that the currently available M. murinus genome sequence contains at least 10 distinct pSIVgml insertions, including two proviruses and eight ‘solo LTRs’ (Fig. S1). Solo LTRs are formed through homologous recombination between the two (initially identical) LTRs found flanking an intact endogenous provirus, resulting in the deletion of internal viral coding regions and leaving behind a single LTR (15, 16). At one genomic locus, both full-length and solo LTR versions of pSIVgml were identified (see Fig. S1), indicating that solo LTR formation had occurred on one chromosome, but not on the other. Because published M. murinus whole genome shotgun (WGS) data are estimated to represent ≈30% of the complete genome, total pSIVgml copy number in the M. murinus genome is likely to be in the order of six full-length insertions and 24 solo LTRs.
To confirm the presence of lentiviral insertions in the M. murinus germ line, we used PCR to amplify pSIVgml sequences from two genomic DNA samples derived from distinct mouse lemur individuals. PCR amplicons were obtained from both samples and spanned regions of the proviral genome not present in WGS data, allowing construction of a consensus pSIVgml genome (Fig. 1). The consensus genome sequence spanned 7,393 nucleotides and exhibited a typical lentiviral genome structure. In addition to gag, pol, and env coding domains, the pSIVgml genome encodes the trans-regulatory accessory proteins Tat and Rev and their associated RNA secondary structural elements TAR and RRE (Fig. 1 and Figs. S2 and S3). Comparison to representative lentivirus genomes revealed that the pSIVgml pol gene contains a dUTPase, a feature that has previously only been described in nonprimate lentiviruses. A vif gene containing the characteristic and highly conserved SQV/Y motif (17) was identified in the same genomic location as other lentiviral vif genes. A putative ORF was identified in the same genomic location as the nef gene encoded by the lentiviruses found in simian primates, although searches using hidden Markov models (HMM) (18) revealed no detectable similarity to previously described Nef proteins. The consensus genome contained a total of four in-frame stop codons and one frameshifting indel, and a region encoding the C-terminal domain of capsid (CA), the N-terminal part of the nucleocapsid (NC) protein, and the gag-pol junction was deleted.
Phylogenetic Analysis.
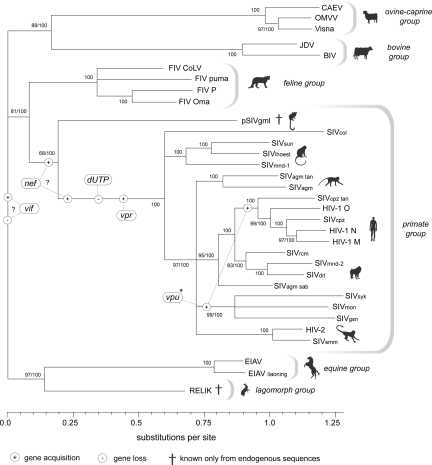
Conserved regions of the gag and pol coding domains were used to construct an alignment of pSIVgml and representative lentivirus sequences (see Methods). Phylogenies constructed using this alignment grouped pSIVgml basal to the known SIV isolates, and intermediate between the feline and primate lentiviruses, under both Bayesian and maximum likelihood (ML) criteria. This phylogenetic positioning is consistent with the transitional structure of the pSIVgml genome, which encodes features that are characteristic of nonprimate lentiviruses (such as the dUTPase), while some derived features characteristic of primate lentiviruses (such as vpr and vpx genes located between pol and env) are absent. Additionally, the phylogeny disclosed strong support (ML bootstrap >80, Bayesian posterior probability = 100) for sister clade relationships among lentiviral groups (primate/feline, ovine/caprine/bovine, and equine/lagomorph), although the precise rooting could not be determined (Fig. 2).
Fig. 2.
Phylogenetic relationships among lentiviruses. The inferred timing of accessory gene acquisition and loss events, based on the principle of parsimony, and assuming no recombination between groups, is indicated. Gene acquisition/loss events that are uncertain with regard to timing or directionality are indicated by question marks. Bootstrap scores and Bayesian posterior probabilities are indicated to the left and right of the forward slash respectively, while nodes with only 100 indicated showed maximal support under both measures. See Methods for taxa definitions and sequence accession numbers. *Subsequent to its origin in the SIVsyk/SIVmon/SIVgsn lineage, the vpu gene was acquired in the HIV-1/SIVcpz lineage via recombination (33). Some primate lentiviruses have an additional gene, vpx (data not shown).
Estimating the Age of the pSIVgml Genome Invasion.
Four lines of evidence indicate that pSIVgml represents an established germ line infection. First, identical insertions were present in two distinct animals. Second, we identified at least nine solo LTR sequences (Fig. S1), which typically form over many host generations (15, 16). Third, in silico screening of M. murinus WGS data for loci that contained pSIVgml insertions revealed no evidence for heterozygous insertion sites, where one allele contains the proviral insertion and the other the preintegration site. This indicates that all insertions are at high allele frequencies (and possibly fixed) within the host population. Last, we compared the 3′ LTR sequence of a full-length provirus with the solo LTR identified at the same genomic locus, based on an unambiguously shared 3′ flanking site (Fig. S1). Because no viral replication could have occurred since the common ancestor of these elements, the date of their divergence can be estimated by using the host's neutral rate of genomic evolution. The divergence between these sequences was 0.0169 substitutions per site. By applying the estimated rates of mouse and human neutral evolution (2.2 × 10−9 and 4.5 × 10−9 substitutions per site per year, respectively) as conservative lower and upper bounds, we obtained an estimated age of 1.9–3.8 million years. It should be noted that this estimate refers only to the divergence of two pSIVgml insertions (i.e., it does not necessarily reflect the age of the pSIVgml germ line invasion, or the extent of the lemur-lentivirus association) and is sensitive to the effects of gene conversion and recombination (19, 20), which could confound the calculation.
Discussion
The discovery of pSIVgml—the first endogenous primate lentivirus—unequivocally demonstrates that lentiviruses are capable of invading primate genomes. Moreover, it illustrates the utility of endogenous sequences for the study of modern retroviruses, including lentiviruses.
In contrast to contemporary, exogenous retroviral sequences, endogenous sequences can reveal the dynamics of host and retrovirus interaction across a timescale of millions of years, providing a robust evolutionary framework in which to investigate retroviral biology and pathogenesis. Because endogenous sequences can independently be assumed to be ancestral to contemporary exogenous viruses, they provide robust outgroups for phylogenetic analyses and for sequence comparisons of modern genomes and proteins. They will therefore prove useful for a wide range of analytical procedures, including classifying new isolates, identifying and characterizing circulating recombinant forms, and dating recent cross-species transmissions. Furthermore, reconstruction and expression of endogenous genes will allow the phenotypic properties of ancestral lentiviral proteins to be explored in vitro (21, 22), potentially providing unique insights into the biology and evolution of contemporary strains.
Our analysis helps to resolve the order of acquisition and loss of genes present within contemporary lentiviral lineages (Fig. 2). Both phylogenetic and genomic analyses indicate that pSIVgml is intermediate between the primate and nonprimate lentiviruses, and thus represents the first unambiguous example of a viral ‘transitional form’. The discovery of pSIVgml thus establishes that the absence of a dUTPase and the presence of vpr (and possibly nef) genes are not prerequisites for the infection of primate hosts. Subsequent screening of mammalian genomes will likely identify additional endogenous lentiviruses, which may provide further insight into the evolution of lentiviral accessory genes.
Lentiviruses have generally been assumed to be a relatively ‘modern’ retrovirus group. However, the recent discovery of rabbit endogenous lentivirus type K (RELIK) demonstrates an association with mammals extending over at least seven million years (14). Because RELIK and pSIVgml share a distantly related ancestor, the relationship between lentiviruses and mammals may be considerably more ancient than this. Although the relatively intact nature of the insertions identified in this study suggests that pSIVgml circulated as an ancestral exogenous lentivirus in Madagascar before entering the M. murinus germ line, it remains uncertain for how long.
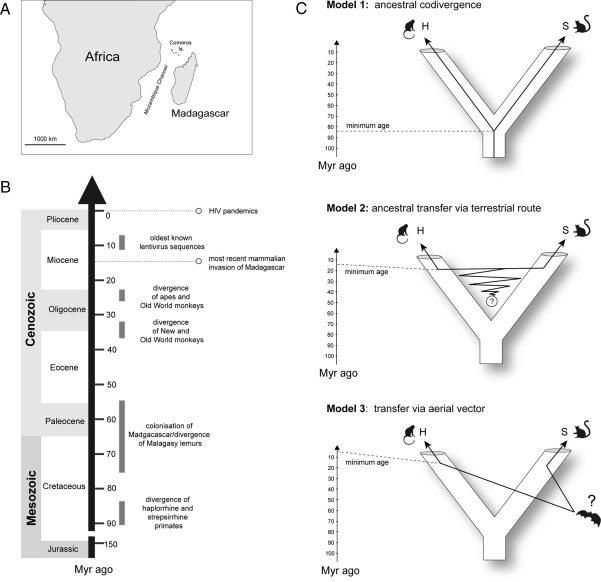
Madagascar has remained at approximately its present position for the past ≈130 million years, and is separated from the African landmass by a deep ocean channel ≈400 km wide at its narrowest point (Fig. 3A) (23). This extended ecological isolation is reflected in a unique and relatively limited mammalian fauna. Strepsirrhine primates were likely the first terrestrial mammalian group to colonize the island ≈75 Myr ago (24), and African and Malagasy primate populations have therefore remained ecologically isolated from one another for the greater part of their evolution. This raises the question of how lentiviruses have come to be present in both primate populations. We propose three competing evolutionary models (Fig. 3C): (1) ancestral codivergence; (2) ancestral terrestrial transfer during periods of intermittent access between the two regions; and (3) transfer mediated by an aerial vector.
Fig. 3.
Models of viral origin. (A) Map showing geographic location of Madagascar relative to Africa. (B) Evolutionary timeline. Mammalian invasions of Madagascar are believed to have involved only five orders of terrestrial mammals—primates, rodents, carnivores, afrosoricida, and artiodactyls. Molecular and fossil data suggest the most recent colonization occurred 14 million years ago (23). The oldest known lentiviral sequences have been dated to between 7 and 11 million years old (14). (C) Proposed models of primate lentivirus evolution. Three evolutionary scenarios can be proposed to account for the presence of lentiviruses in both streprsirrhine (S) and haplorrhine (H) primates; (1) ancestral codivergence of primate lentiviruses into haplorrhine and strepsirrhine lineages in broad concert with their hosts; (2) transfer of lentiviruses between Malagasy strepsirrhines and African haplorrhines subsequent to their divergence, mediated either by a nonprimate vector or via temporary contact between Malagasy and African primate populations during intermittent periods of land-bridge or dry way-point access between the two regions (23); (3) transfer mediated by an aerial vector species.
The phylogenetic position of pSIVgml (Fig. 2) is compatible with ancestral codivergence of primate lentiviruses and their hosts into strepsirrhine and haplorrhine lineages (model 1, Fig. 3C). This model entails an ancestral association of primates and lentiviruses, potentially dating back 85 Myr to the haplorrhine-strepsirrhine split (25), and implies that undetected lentiviruses currently circulate in New World and Asian primates (or have independently gone extinct in every non-African haplorrhine primate lineage).
However, the apparent codivergence of haplorrhine and strepsirrhine lentivirus lineages does not necessarily require the common ancestor of primate lentiviruses to be as old as that of their hosts; it could also reflect the geographic isolation of African and Malagasy primates subsequent to cross-species transmission (26, 27). The periodic colonization of Madagascar by groups of terrestrial mammals subsequent to the arrival of strepsirrhine primates indicates that limited opportunities for terrestrial transfer of lentiviruses between the two regions could have arisen throughout the Cenozoic Era (23), either by direct primate-to-primate contact or via a nonprimate vector (model 2, Fig. 3C). Although this model allows for a more recent lentivirus origin, it nevertheless requires an association dating back at least 14 million years to when the most recent terrestrial mammal invasion occurred (23) (Fig. 3B).
Alternatively, lentiviruses could have been vectored between Malagasy and African primate populations more recently by an aerial vector species capable of crossing the intervening ocean (model 3, Fig. 3C). However, this scenario would require at least two cross-order transmission events. Currently, lentiviruses identified from distinct mammalian orders are approximately equidistant in phylogenies (Fig. 2), providing no evidence that cross-order transfer occurs readily.
Irrespective of which of the above scenarios is correct, the discovery of pSIVgml indicates that a wide range of primate species should be considered in studies of primate lentivirus biology. For example, the independent evolution of TRIM5CypA mediated restriction in both New World and Asian monkeys could have been selected by as yet undiscovered lentiviruses circulating in these species (28). Broad surveillance should be implemented to locate reservoirs of lentiviruses transmissible to humans, and to investigate whether vector-mediated transfer is a potential source of new infections. Endogenous sequences may facilitate such endeavors, because reconstruction and expression of endogenous lentiviral proteins would allow the generation of sensitive antibodies for serological surveillance. Finally, because it is well established that cross-species transmission of lentiviruses within orders is relatively common, it is possible that a range of diverse lentiviruses currently circulate among Madagascar's 33 lemur species (29), potentially providing unique opportunities to study how interactions between primates and lentiviruses have shaped their evolution.
Materials and Methods
Sequence Data.
The following representative lentivirus genomes used for database screening and phylogenetic reconstruction: CAEV (caprine arthritis-encephalitis virus, M33677); SMLV (small ruminant lentivirus, AY445885); Visna (ovine maedi-visna virus, M60610); JDV (Jembrana disease virus, U21603); BIV (bovine immunodeficiency virus, M32690), HIV-1 O (L20587); HIV-1 N (DQ017383); HIV-1 M (K03455); HIV-2 (M30502); EIAV (equine infectious anemia virus, M16575); EIAV liaoning (AF327877); and RELIK (rabbit endogenous lentivirus type K, consensus genome sequence available on request from the authors of this report). For the following simian immunodeficiency viruses (SIVs) and feline immunodeficiency viruses (FIVs), host species, and sequence accession numbers are shown in parentheses: FIV Petaluma (domestic cat, M25381); FIV CoLV (cougar, EF455615); FIV puma (puma, U03982); FIV Oma (Pallas' cat, U56928); SIVagm (African green monkey, M30931); SIVagm sab (African green monkey, sabaeus subspecies, U04005), SIVagm tan (African green monkey, tantalus subspecies, U58991); SIVcol (guereza colobus, AF301156); SIVsun (sun-tailed macaque, AF131870); SIVlhoest (lhoest's monkey, AF188116); SIVrcm (red-capped mangabey, AF349681); SIVmnd (mandrill, M27470); SIVmnd-2 (mandrill, strain 5440, AY159322); SIVdrl (drill monkey, AY159321); SIVsyk (Sykes' monkey, L06042); SIVmon (Mona's monkey, AJ580407); SIVgsn (greater spot-nosed monkey, AF468659); SIVsmm (sooty mangabey, X14307); SIVcpz (chimpanzee, X52154); and SIVcpz tan (chimpanzee, schweinfurthii subspecies, DQ374658). The pSIVgml consensus sequence is available on request from the authors.
PCR and Sequencing.
M. murinus genomic DNA samples were obtained from CORRIEL, the Children's Hospital Oakland Research Institute (CHORI), and a collection held at Stanford University. PCR products were generated from 5–50 ng of lemur DNA in reactions containing 1× PCR buffer, 2.5 mM MgCl2, 150 μM dNTP, 10 pmol of forward and reverse primer (Integrated DNA Technologies), and 2.5 U TaqDNA polymerase (Invitrogen). Amplifications were performed on an ABI 9700 thermal cycler (Applied Biosystems) under the following conditions: 1 cycle of 95°C for 2 min, 35 cycles of 94°C for 15 sec, 55°C for 20 sec, and 72°C for 2 min, and 1 cycle of 72°C for 10 min. Sequencing reactions were performed by using forward and reverse sequencing primers and BigDye v3.1 Terminators (ABI) according to manufacturer's instructions, cleaned up by using BigDye XTerminator Purification Kits (ABI), and analyzed on an ABI 3130XL Genetic Analyzer.
Phylogenetic Analysis.
Phylogenetic trees were constructed based on an alignment of 853 amino acids, spanning conserved regions of the matrix and capsid proteins and the majority of Pol. The underlying nucleotide sequences were used to infer phylogenetic relationships, under both maximum likelihood (ML) and Bayesian MCMC inference, with the programs RAXML (30) and MrBayes 3.0 (31), respectively. ML phylogenies were estimated from 100 distinct random stepwise addition trees, with the topology and substitution rate parameters optimized by using the RAXML heuristic, to an accuracy of 0.1 Log likelihood units. All 100 distinct starting trees converged on a final topology with equal likelihood. Support for the ML trees was assessed via 1,000 nonparametric bootstrap replicates, each starting from a single random stepwise addition tree. Bayesian analyses were run for 5,000,000 steps, sampling trees every 1,000 steps, and discarding the first 500 trees. Convergence of MCMC was indicated by an effective sample size >600 as calculated in Tracer v 1.3 (32). In addition, a second Bayesian MCMC analysis was run to ensure adequate mixing.
Supplementary Material
Acknowledgments.
We thank the Mammalian Genome Project at the Broad Institute for generation and public release of gray mouse lemur genome sequence data. We also thank Pieter J. de Jong [Children's Hospital and Research Center Oakland Institute (CHORI)], and David de Gusta (Stanford) for providing lemur DNA and tissue samples, and Beatrice Hahn, Angela McLean, and Greg Towers for helpful discussion and comments on the manuscript. R.J.G., M.W., and R.W.S. were funded by National Institute of Allergy and Infectious Diseases Grant AI068581. A.K. was funded by a fellowship from the James Martin 21st Century School.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ461356, FJ461357, and FJ461358).
See Commentary on page 20051.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807873105/DCSupplemental.
References
- 1.Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: Scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 2.Korber B, et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 3.Salemi M, et al. Mosaic genomes of the six major primate lentivirus lineages revealed by phylogenetic analyses. J Virol. 2003;77:7202–7213. doi: 10.1128/JVI.77.13.7202-7213.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemey P, et al. Tracing the origin and history of the HIV-2 epidemic. Proc Natl Acad Sci USA. 2003;100:6588–6592. doi: 10.1073/pnas.0936469100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keele BF, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Worobey M, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes EC. Molecular clocks and the puzzle of RNA virus origins. J Virol. 2003;77:3893–3897. doi: 10.1128/JVI.77.7.3893-3897.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharp PM, et al. Origins and evolution of AIDS viruses: Estimating the time-scale. Biochem Soc Trans. 2000;28:275–282. doi: 10.1042/bst0280275. [DOI] [PubMed] [Google Scholar]
- 9.Weiss RA. The discovery of endogenous retroviruses. Retrovirology. 2006;3:67. doi: 10.1186/1742-4690-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarlinton RE, Meers J, Young PR. Retroviral invasion of the koala genome. Nature. 2006;442:79–81. doi: 10.1038/nature04841. [DOI] [PubMed] [Google Scholar]
- 11.Boeke JD, Stoye JP. Retrotransposons, Endogenous retroviruses, and the evolution of retroelements. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. New York: CSHL Press; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 12.Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26:291–315. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- 13.Katzourakis A, Rambaut A, Pybus OG. The evolutionary dynamics of endogenous retroviruses. Trends Microbiol. 2005;13:463–468. doi: 10.1016/j.tim.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc Natl Acad Sci USA. 2007;104:6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belshaw R, et al. Rate of recombinational deletion among human endogenous retroviruses. J Virol. 2007;81:9437–9442. doi: 10.1128/JVI.02216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katzourakis A, Pereira V, Tristem M. Effects of recombination rate on human endogenous retrovirus fixation and persistence. J Virol. 2007;81:10712–10717. doi: 10.1128/JVI.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oberste MS, Gonda MA. Conservation of amino-acid sequence motifs in lentivirus Vif proteins. Virus Genes. 1992;6:95–102. doi: 10.1007/BF01703760. [DOI] [PubMed] [Google Scholar]
- 18.Llorens C, Futami R, Bezemer D, Moya A. The Gypsy Database (GyDB) of mobile genetic elements. Nucleic Acids Res. 2008;36:D38–46. doi: 10.1093/nar/gkm697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson WE, Coffin JM. Constructing primate phylogenies from ancient retrovirus sequences. Proc Natl Acad Sci USA. 1999;96:10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes JF, Coffin JM. Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome. Genetics. 2005;171:1183–1194. doi: 10.1534/genetics.105.043976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science. 2007;316:1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- 23.Tattersall I. Historical biogeography of the strepsirrhine primates of Madagascar. Folia Primatol (Basel) 2006;77:477–487. doi: 10.1159/000095393. [DOI] [PubMed] [Google Scholar]
- 24.Horvath JE, et al. Development and application of a phylogenomic toolkit: Resolving the evolutionary history of Madagascar's lemurs. Genome Res. 2008;18:489–499. doi: 10.1101/gr.7265208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bininda-Emonds OR, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 26.Wertheim JO, Worobey M. A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes. PLoS Pathog. 2007;3:e95. doi: 10.1371/journal.ppat.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charleston MA, Robertson DL. Preferential host switching by primate lentiviruses can account for phylogenetic similarity with the primate phylogeny. Syst Biol. 2002;51:528–535. doi: 10.1080/10635150290069940. [DOI] [PubMed] [Google Scholar]
- 28.Stoye JP, Yap MW. Chance favors a prepared genome. Proc Natl Acad Sci USA. 2008;105:3177–3178. doi: 10.1073/pnas.0800667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sondgeroth K, et al. Assessing flavivirus, lentivirus, and herpesvirus exposure in free-ranging ring-tailed lemurs in southwestern Madagascar. J Wildl Dis. 2007;43:40–47. doi: 10.7589/0090-3558-43.1.40. [DOI] [PubMed] [Google Scholar]
- 30.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 31.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 32.Rambaut A, Drummond AJ. Tracer v1.4. 2007 Available from http://beast.bio.ed.ac.uk/Tracer.
- 33.Sharp PM, Shaw GM, Hahn BH. Simian immunodeficiency virus infection of chimpanzees. J Virol. 2005;79:3891–3902. doi: 10.1128/JVI.79.7.3891-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.