Summary
wingless (wg)/Wnt family genes encode secreted glycoproteins essential for the development of virtually all metazoans. In short germ insects, including the red flour beetle, Tribolium castaneum, the segment-polarity function of wg is conserved [1]. Wnt signalling is also implicated in posterior patterning and germband elongation [2–4], but despite its expression in the posterior growth zone, Wnt1/wg alone is not responsible for these functions; [1–3]. Tribolium contains additional Wnt family genes of unknown function that are also expressed in the growth zone [5]. After depleting one of these, Tc-WntD/8, we found a small percentage of embryos lacking abdominal segments. Additional removal of Tc-Wnt1 significantly enhanced this phenotype, suggesting functional redundancy. Seeking alternative methods to deplete Wnt signal, we performed RNAi with other components of the Wnt pathway including wntless (wls) and porcupine (porc), which process Wnt ligands, and pangolin (pan), which transduces the signal to the nucleus. Tc-wls RNAi caused segmentation defects similar to Tc-Wnt1, but not Tc-WntD/8 RNAi, indicating that the effects of Tc-WntD/8 depletion are Tc-wls-independent. In contrast, depletion of Tc-porc and Tc-pan resulted in embryos resembling those of double Tc-Wnt1,Tc-WntD/8 RNAi, suggesting Tc-porc is essential for the function of both ligands and that they signal through the canonical pathway. Our results provide the first evidence of functional redundancy between Wnt ligands in posterior patterning in short germ insects. This Wnt function appears to be conserved in other arthropods [6] and vertebrates [7–9].
Results and Discussion
The Wnt1/Wingless (Wg) protein is a secreted glycoprotein involved in cell-to-cell communication. In the presence of Wg, Armadillo/β-catenin (Arm) is stabilized and translocated to the nucleus where it activates target genes by associating with transcription factor Pangolin/Tcf (Pan). In intermediate and short germ insects, a few anterior segments are specified at the blastoderm stage and the rest are added sequentially from a posterior growth zone during embryonic germband extension [10]. Wnt signaling appears to be involved in this process, since depletion of arm in Gryllus results in embryos lacking all abdominal segments [2] and similar depletion of pan in Oncopeltus dramatically truncates segmentation [3]. However, depletion of wg/Wnt1 in Gryllus, Oncopeltus or Tribolium does not reduce the number of segments initially formed [1–3, and Figure 1C–D’]. Thus, if Wnt signaling plays a role in the posterior growth zone, we must look for the signaling molecule among the other Wnt family members.
Figure 1. Cuticular and embryonic RNAi phenotypes.
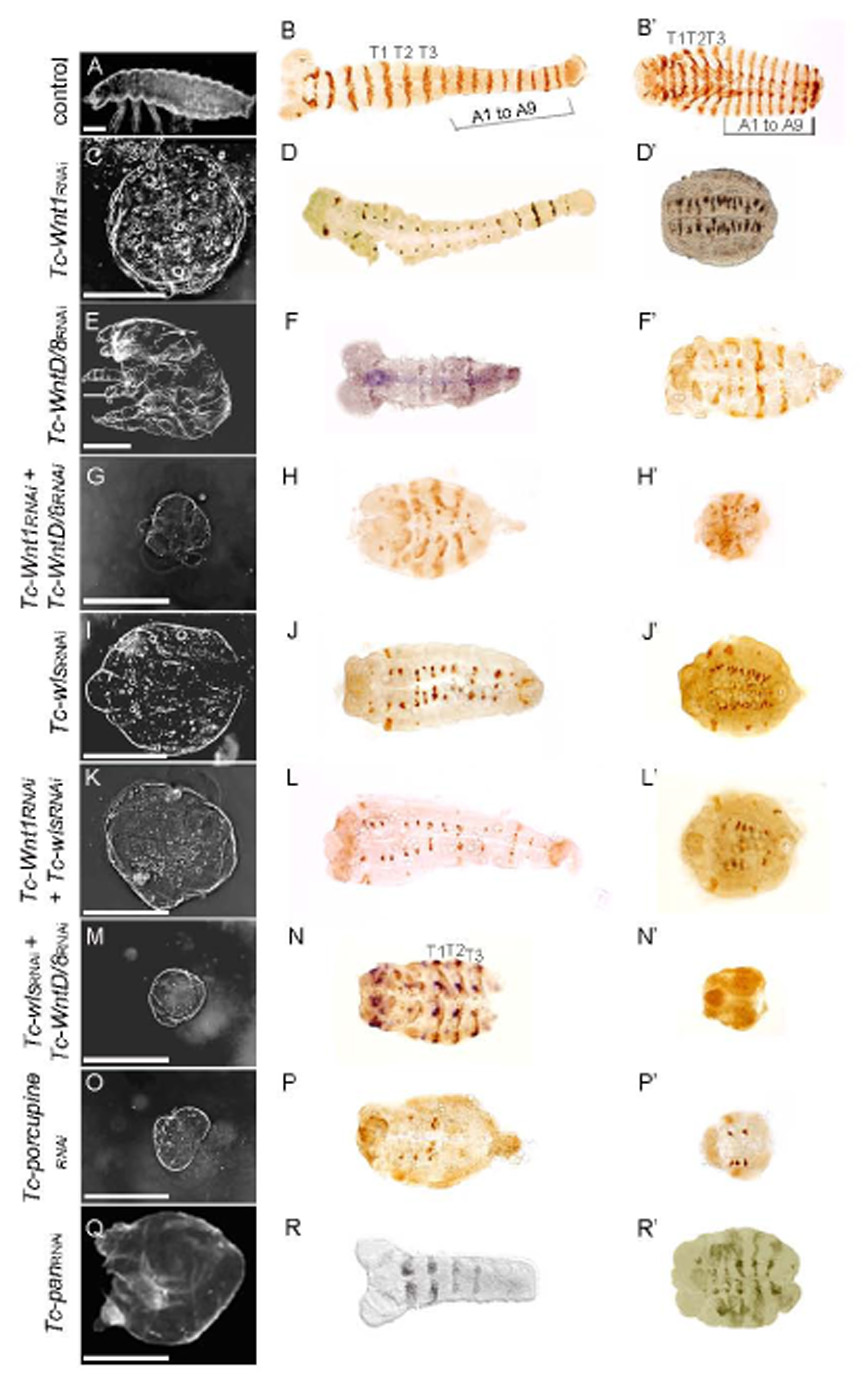
When possible to determine, embryos are oriented with anterior to the left.
(A–B’) Wild-type embryos. (A) Wild-type embryonic cuticle. (B–B’) Developmental stage assessed by Tc-En staining during elongation (B) and retraction (B’). Wild type En expression is visible in all thoracic and abdominal segments.
(C–D’) Tc-Wnt1 RNAi embryos. Cuticles are small spheres without signs of segmentation (C). During elongation all segments are formed although En expression is fading (D), after germband retraction the embryos are highly compacted but still contain Tc-En-expressing cell derivatives in all segments (D’).
(E–F’) Tc-WntD/8 RNAi embryos. Cuticles have two pairs of legs but no abdominal segments. (E). In more severely affected embryos, abdominal segments failed to form (F) and three thoracic segments are observed after germband retraction (F’).
(G–H’) Tc-Wnt1, Tc-WntD/8 double RNAi embryos. Cuticular phenotypes (G) are smaller than those produced by either single RNAi. Tc-En expression is visible in three thoracic segments (H). After germband retraction, the embryos are very compacted and show as few segments as Tc-WntD/8 RNAi embryos, as assessed by Tc-En (H’).
(I–J’) Tc-wls RNAi embryos. Cuticles (I) resemble those from Tc-Wnt1 RNAi (C). Tc-En expression fades during germband elongation (J), and after germband retraction derivatives of all segments are highly compacted (J’), as in Tc-Wnt1 RNAi embryos.
(K–L’) Tc-Wnt1,Tc-wls double RNAi. Cuticles (K) resemble those from Tc-Wnt1 or Tc-wls single RNAi. Tc-En expression fades during germband elongation (L), and after germband retraction derivatives of all segments are highly compacted (L’), as in Tc-Wnt1 or Tc-wls single RNAi germbands.
(M–N’) Tc-wls,Tc-WntD/8 double RNAi embryos. Cuticular phenotypes (M) are more severe than those produced by either single RNAi. Tc-Eve (brown) and Tc-Wnt1 (purple) mRNA expression are visible in three thoracic segments (N). After germband retraction, the embryos are very small and show rudimentary heads and three thoracic segments as assessed by Tc-En (N’).
(O–P’) Tc-porc RNAi embryos. Cuticular phenotypes (O) resemble those produced by Tc-wls/Tc-WntD/8 double RNAi (M). Abdominal segments do not form (P) and after germband retraction, the embryos are very small and show rudimentary heads and three thoracic segments as assessed by Tc-En (P’), similar to Tc-wls,Tc-WntD/8 double RNAi embryos.
(Q–R’) Tc-pan RNAi embryos. Cuticles are small spheres (Q) that resemble Tc-Wnt1 embryos (C). Tc-En staining shows that abdominal segments failed to form during germband elongation (R) and after germband retraction Tc-En is expressed in the remaining segments (R’), like Tc-WntD/8 RNAi embryos.
Scale bars represent 0.1mm.
Functional analysis of Tribolium Wnt genes during embryogenesis
Functional analysis using RNAi to knockdown each of the nine Tribolium Wnt genes [5] failed to produce segmentation phenotypes in over 900 cuticles examined per gene, except for Tc-Wnt1 and Tc-WntD/8 (mRNA depletion confirmed by in situ hybridization). Tc-Wnt1 and Tc-WntD/8 RNAi revealed strikingly different phenotypes. Female reproduction in Tribolium appeared to be highly sensitive to depletion of Tc-Wnt1; female pupae or adults injected with high levels of dsRNA did not produce eggs. When the amount of dsRNA was lowered, only a few embryos were produced, and they displayed a range of phenotypes [1] (Table S1, Figure S1B and S1C). The most severely affected Tc-Wnt1 RNAi embryos produced small spheres of cuticle with no recognizable appendages or segmental grooves (Figure 1C). In these embryos, expression of Tc-Engrailed (Tc-En), a Tc-Wnt1 target, initiated normally during early development, but faded from the ventral epidermis as each segment matured (Figure 1D). Later, during germband retraction, segmental grooves failed to form. Tc-En expression in the CNS demonstrated that derivatives of each segment were present but highly compacted [1] (Figure 1D’). Less severe Tc-Wnt1 RNAi embryos displayed some signs of segmentation and rudimentary appendages (Figure S1B and S1C).
In contrast, Tc-WntD/8 RNAi produced segmentation defects quite different from Tc-Wnt1 RNAi. Female pupae injected with Tc-WntD/8 dsRNA (Table 1 and Table 2) did produce eggs, albeit at reduced number, suggesting that Tc-WntD/8 signalling is also required during oogenesis. Of the eggs produced after Tc-WntD/8 depletion, 61% were empty and a considerable portion (33%) appeared to be developing normally (Table 1). A small number of cuticles (4.4%, Table 1) displayed severe segmentation defects. Although anterior segments including the first and sometimes the second thoracic segment appeared normal, these embryos lacked all posterior segments (Figure 1E, Figure S1D), reminiscent of the arm and pan RNAi mutant phenotypes in other short germ insects [2,3]. More weakly affected cuticles (1.6%, Table 1) contained all gnathal and thoracic segments, but lacked most of the abdominal segments (Figure S1E and S1F). Thus, 6% of the Tc-WntD/8 RNAi progeny showed some degree of posterior truncation.
Table 1.
cuticular RNAi phenotypes
| Severe % | Hypomorphic % | Wild-type % | Empty % | Total (number of individuals examined) | |
|---|---|---|---|---|---|
| WntD/8 | 4.4* | 1.6† | 33 | 61 | 137 |
| Wls | 26** | 0 | 12 | 62 | 1306 |
| WntD/8,Wls | 50*** | 2†† | 0 | 48 | 178 |
truncated embryos lacking T3 and abdominal segments
small spherical embryos lacking limbs
extremely small spherical embryos lacking limbs.
Truncated embryos lacking abdominal segments
small spherical embryos occasionally displaying rudimentary limbs and a few segmental grooves.
Table 2.
germband RNAi phenotypes
| Severe % | Hypomorphic % | Wild-type % | Unstained % | Total (number of individuals examined) | |
|---|---|---|---|---|---|
| WntD/8 | 0.9* | 6.1† | 52 | 41 | 230 |
| Wls | 20** | 0 | 22 | 58 | 419 |
| WntD/8,Wls | 34*** | 1.5† | 9.5 | 55 | 187 |
| Porc | 34*** | 0 | 7.0 | 59 | 321 |
truncated germbands lacking T3 and abdominal segments
overcompacted germbands containing the normal number of segments
truncated, overcompacted germbands lacking T3 and abdominal segments.
Full-length germbands with narrow abdominal segments.
To determine how these terminal cuticular phenotypes developed, we followed Tc-En and Tc-Eve staining in Tc-WntD/8 RNAi embryos. In a collection of 0–72 hr eggs, 52% of the embryos (at various stages of development) appeared normal and 41 % were unstained (Table 2). The unstained eggs may not have contained embryos or they may have contained very young embryos not yet expressing Tc-En or Tc-Eve. A very small percentage of the eggs (0.9%, Table 2) contained severely affected germbands in which abdominal segments had failed to form (Figure 1F and F’). These embryos are likely to produce truncated cuticles, but their numbers are insufficient to account for all such cuticles. Interestingly, 6.1% of the embryos displayed an apparently intermediate phenotype in which the normal number of segments formed during germband elongation, but the abdominal segments were significantly narrower than in wild type (Table 2, Figure S2D and S2E). A few older embryos in this class were broken in two (inset in Figure S2F), suggesting that these embryos account for the remaining truncated cuticles. This phenotype is very intriguing and may provide insight in the mechanism underlying Tc-WntD/8 function in the posterior growth zone. It could be caused by cell death, slower rates of cell division or loss of cell proliferation. Interestingly, a cell proliferation role for Wnt signalling has been shown in Drosophila [11].
Given these germband phenotypes, we conclude that the truncated cuticular phenotypes could result from either the loss of the posterior region of the embryo during germband retraction, or the inability to form the posterior segments in the first place. Although the number of affected embryos is very low, in situ hybridization with the WntD/8 probe indicated the RNAi had successfully depleted the Tc-WntD/8 mRNA in all embryos. The low penetrance of the TcWntD/8 truncation phenotype led us to hypothesize that the function of Tc-WntD/8 is partially redundant with that of one or more of the other Wnts.
Effects of depleting multiple Wnts in the posterior growth zone
To determine which, if any, of the other Wnt genes expressed in the posterior growth zone (Tc-Wnt1, Tc-Wnt5 and Tc-WntA) [5] might function redundantly with Tc-WntD/8, we performed simultaneous RNAi with multiple Wnts. Tc-Wnt5,Tc-WntA double RNAi failed to produce any mutant phenotypes in more than 500 embryos examined (Table S1) (for this and subsequent experiments, a sample of embryos was stained with the appropriate probes to demonstrate that the target mRNA(s) had been successfully depleted). RNAi combining Tc-WntD/8 with Tc-Wnt5 or Tc-WntA produced the same range and frequency of phenotypes observed in Tc-WntD/8 single RNAi (approximately 6% of the cuticles were truncated, Table S1). Similar results were obtained when both Tc-WntA and Tc-Wnt5 dsRNA were combined with Tc-WntD/8 dsRNA (triple RNAi). Since Tc-Wnt1 RNAi produced a class of highly compacted but not truncated embryos, we tested combinations of Tc-Wnt1 with Tc-Wnt5 and/or Tc-WntA. In these RNAi experiments, adult females were injected with a reduced concentration of Tc-Wnt1 dsRNA to avoid complete sterility (approximately 100 embryos examined per RNAi experiment). All combinations produced the same range and frequency of phenotypes produced by Tc-Wnt1 single RNAi (Figure S1A–C and Table S1), including some highly compacted, almost asegmental embryos, but no trunctated embryos. In these experiments testing multiple Wnts in combination with Tc-Wnt1 or Tc-WntD/8, we did not observe an increase in frequency or severity of either single RNAi phenotype. Thus, we found no obvious evidence for redundancy of Tc-Wnt5 and/or Tc-WntA in posterior patterning. However, given the low frequencies of the most severe phenotypes, we can not rule out the possibility of low-level redundancy from Tc-Wnt5 and/or Tc-WntA.
Finally, we performed Tc-Wnt1,Tc-WntD/8 double RNAi to determine how depletion of both might affect patterning in the posterior growth zone. The resulting cuticular phenotypes ranged from wild-type to extremely small spherical cuticles that were significantly smaller than those obtained after Tc-Wnt1 RNAi alone (Figure 1G and Table S1). Additional phenotypes including cuticles which displayed random combinations of hypomorphic to severe effects of depleting both genes were also observed (Table S1). To determine the developmental profile of the embryos that produced the extremely small cuticles, we followed Tc-En expression during embryogenesis. In addition to the Tc-En expression patterns expected from RNAi with either gene alone (data not shown), we observed embryos in various stages of germband retraction (Figure 1H and H’) that lacked abdominal segments (similar to Tc-WntD/8 RNAi embryos, Figure 1F’) and were highly compacted by the end of germband retraction (similar to Tc-Wnt1 RNAi embryos, Figure D’). Thus, the extremely severe germband and cuticular phenotypes produced by Tc-Wnt1,Tc-WntD/8 double RNAi appeared to result from the additive effect of depleting both genes.
Interestingly, truncated embryos were observed more often after Tc-Wnt1,Tc-WntD/8 double RNAi than after Tc-Wnt8 single RNAi (15% versus 6%, Table S1), suggesting that these Wnts may function redundantly in the posterior growth zone. However, since all RNAi experiments involving Tc-Wnt1 proved to be somewhat problematic in that it was difficult to obtain a high percentage of eggs displaying the most severe phenotype, we sought an alternative method to block Wnt signaling. Thus, we examined other components of the Wnt pathway including Wntless (wls) and porcupine (porc), which process Wnt ligands and pangolin (pan), which transduces the signal to the nucleus.
Effects of depleting Wntless
Wls [12], also known as Evenness interrupted [13] or Sprinter [14], has been reported to control Wnt ligand secretion from Wnt-producing cells [12–14]. Wls is involved in wg/Wnt1 secretion in Drosophila, in MOM-2-governed polarization of blastomeres in C. elegans and in Wnt3a-mediated communication between cultured human cells [12–13]. Based on these results, the authors speculate that Wls is required for the function of all members of the Wnt family [12–13]. Hence, to investigate the effect of depleting all Wnt signal, we injected female pupae with Tc-wls dsRNA, assuming this would block the secretion of all Wnt ligands in the resulting embryos. In contrast to injection of Tc-Wnt1, injection of Tc-wls dsRNA into female pupae did not appear to be detrimental to oogenesis, since plenty of eggs were available for analysis (Table 1). Thus, we speculate that Tc-Wnt1 has a wntless-independent function in oogenesis.
In Tribolium, we found Tc-wls to be expressed ubiquitously during embryogenesis (data not shown). The majority of eggs produced by Tc-wls RNAi were empty (62%) and a small percentage (12%) were wild-type. The remaining embryos (26%) showed severe segmentation defects (Figure 1I and Figure S1G–S1J) similar to those produced by Tc-Wnt1 RNAi (Figure 1C), but none showed defects similar to those produced by Tc-WntD/8 RNAi. Tc-wls RNAi cuticles were small round structures, lacking any signs of segmentation (Figure 1I). In Tc-wls RNAi embryos, Tc-Eve expression was normal during early germband extension (Figure S2H). Tc-En expression initiated normally, but faded as the segments matured (Figure 1J). Tc-Wnt1 expression also faded during germband elongation in Tc-wls RNAi embryos (Figure S2G). After germband retraction, the embryos were highly compacted but contained derivatives of all segments as assessed by Tc-En (Figure 1J’) and Tc-Eve (Figure S2I) expression in the CNS. Furthermore, Tc-wls and the most severely affected Tc-Wnt1 RNAi embryos were dorsally open, similar to the “dorsal-open” phenotype described for Drosophila wg/Wnt1 [15]. One month after Tc-wls dsRNA injection, hypomorphic phenotypes were observed (Figure S1G–J, not considered in Table 1). Similar to Tc-Wnt1 RNAi embryos, Tc-wls RNAi hypomorphic embryos either completely lacked appendages or their appendages were defective (Figure S1G–S1J). These striking similarities in the defects produced by depletion of either Tc-Wnt1 or Tc-wls led us to conclude that the Tc-wls RNAi phenotype is most likely due to effects on the secretion of Wnt1, as in Drosophila [12, 14]. To confirm these data, we also tested the effect of Tc-Wnt1/Tc-wls double RNAi (Figure 1K). Adult females were injected with a reduced concentration of Tc-Wnt1 dsRNA to avoid effects on oogenesis. In these embryos, Tc-En expression faded during germband elongation (Figure 1L) and CNS derivatives from all segments were present after germband retraction (Figure 1L’), as seen in either Tc-Wnt1 or Tc-wls single RNAi embryos. Interestingly, Tc-wls RNAi either alone or in combination with Tc-Wnt1, did not produce any truncated embryos, and although the truncated Tc-WntD/8 RNAi phenotype is rare (7% total expected at the germband stage), we did not find any during careful examination of 419 Tc-wls RNAi embryos (Table 2). Thus, depletion of Tc-wls affected the segment-polarity function of Tc-Wnt1 but not the growth zone function of Tc-WntD/8. Further studies are necessary to determine whether Tc-wls is also required for the secretion of Wnt ligands other than Wnt1 (in this case, Wnt5 and WntA). However, these results indicate that if Tc-wls is required for their function, their depletion does not result in truncated embryos.
Tc-wls,Tc-WntD/8 double RNAi
Since Tc-wls RNAi readily produced embryos depleted of Tc-Wnt1 function, we performed Tc-wls,Tc-WntD/8 double RNAi experiments to further examine the interaction between Tc-Wnt1 and Tc-WntD/8. If Tc-wls were required for the function of Tc-Wnt1 but not Tc-WntD/8, then we might expect the combined phenotype of Tc-wls and Tc-WntD/8 RNAi to be more severe than the phenotypes produced by RNAi with either gene alone, similar to Tc-Wnt1,Tc-WntD/8 double RNAi. Indeed, we found a high frequency of Tc-wls,Tc-WntD/8 double RNAi embryos that showed additive effects (Figure 1M–N’, Table 1 and 2). These embryos resembled Tc-wls (and Tc-Wnt1) single RNAi embryos in that their cuticles were dorsally open and lacked both limbs and signs of segmentation (compare Figure 1M with 1C, I). Moreover, after germband retraction, their germbands were highly compacted (Figure 1N’). On the other hand, they resembled Tc-WntD/8 RNAi embryos in that they lacked abdominal segments (Figure 1N, Figure S2J and L). These additive effects resulted in retracted germbands and cuticles that were significantly smaller than either Tc-wls or Tc-WntD/8 single RNAi embryos, and more closely resembled the Tc-Wnt1,Tc-WntD/8 double RNAi embryos (compare Figure 1 M and G).
Importantly, as in Tc-Wnt1,Tc-WntD/8 double RNAi, there seems to be a synergistic effect as well, in that the penetrance of the Tc-WntD/8 truncation phenotype was greatly enhanced in the double RNAi experiments. The percentage of truncated germbands increased from 0.9% in Tc-WntD/8 single RNAi embryos to 34% in the double RNAi embryos (Table 2). In addition, the number of wild-type embryos was greatly reduced (from 52 to 9.5%). The effect was even more dramatic in the cuticlar phenotypes in that 100% of the embryos that developed cuticles (50% of the total, Table 1) displayed the severe phenotype (smaller than Tc-Wnt-1,Tcwls or Tc-WntD/8 single RNAi cuticles) indicating that they were both truncated and highly compacted. These results suggest that Wnt1 and WntD/8 function redundantly in the posterior growth zone. If they were completely redundant neither single RNAi would show the truncation phenotype. In this case, Tc-Wnt1 single RNAi does not produce any truncated embryos, but Tc-wntD/8 single RNAi does (Table S1), albeit at a very low frequency, indicating that posterior patterning was somewhat more sensitive to loss of Tc-WntD/8 than to loss of Tc-wnt1. Moreover, since Tc-wls,Tc-WntD/8 double RNAi eliminated the wild-type phenotype, and maximized the percentage of truncated embryos among those that developed, it is likely that the other Wnt ligands contribute little if any to patterning from the posterior growth zone.
Effects of depleting Tc-porcupine
Since Wls does not appear to be required by all Wnts in Tribolium, at least not Tc-WntD/8, we sought an independent method to eliminate all Wnt signal from the posterior. We chose to deplete the ortholog of porcupine (porc), which acts upstream of wls in Drosphila [12]. Tc-porc RNAi resulted in embryos that produced cuticles lacking limbs and signs of segmentation (Figure 1O). They were smaller than those obtained with Tc-wls RNAi (compare Figure 1O with 1I), more closely resembling the extremely small cuticles obtained with Tc-Wnt1,Tc-WntD/8 or Tcwls,Tc-WntD/8 double RNAi. Examination of Tc-En expression revealed defects in these Tc-porc RNAi embryos that also resemble those of Tc-WntD/8,Tc-wls double RNAi embryos; these germbands contained remnants of all anterior segments, but lacked abdominal segments (Figure 1P) and after germband retraction they were highly compacted (Figure 1P’). Interestingly, 34% of the total Tc-porc RNAi embryos displayed this severe phenotype, exactly the same percentage as obtained for Tc-wls,Tc-WntD/8 double RNAi (Table 2) suggesting that the high penetrance of the truncation phenotype is only obtained when the function of both Tc-Wnt1 and Tc-WntD/8 is depleted. Furthermore, these data indicate that, in contrast to Tc-wls, Tc-porc is involved in the secretion of both Tc-Wnt1 and Tc-WntD/8 ligands.
Effects of depleting Tc-pangolin
Truncated phenotypes produced by depletion of arm or pan in other short germ insects [2, 3] suggest that a Wnt signal from the posterior growth zone is transmitted through the canonical pathway. To determine whether Tc-WntD/8 signals through this pathway, we examined the effects of depleting Tc-pan. Since pan transduces the wg/Wnt1 signal to the nucleus during segmentation [16] we might expect a more severe phenotype combining the effects of depleting both Tc-Wnt1 and Tc-WntD/8. Indeed, the most severely affected Tc-pan RNAi embryos produced small limbless spheres of cuticle (Figure 1Q) similar to Tc-Wnt1, and their germbands contained fewer segments (as assessed by Tc-En expression, Figure 1R–R’, Figure S2N–S2O) similar to Tc-WntD/8. Truncated limbs and only few abdominal segments were commonly found in less severe Tc-pan RNAi embryos (Figure S1O–R). These results suggest that both Wnt1 and WntD/8 ligands may signal through the canonical pathway. It is important to note that although Tc-pan RNAi appeared to affect both Tc-Wnt1 and Tc-WntD/8 function, the Tc-pan RNAi phenotype described above was not quite as severe as the Tc-Wnt1,Tc-WntD/8 or Tc-wls, Tc-WntD/8 double RNAi phenotypes. The Tc-pan RNAi embryos were truncated and limbless, but not highly compacted, suggesting that genes normally repressed by pan (including targets of Tc-Wnt1) are derepressed, as in Drosophila [17–18].
Tc-WntD/8 and Tc-Wnt1 expression in wild-type embryos
Since both Tc-WntD/8 and Tc-Wnt1 genes seemed to function redundantly in posterior patterning, they are likely to share similar domains of expression in the growth zone. Both genes are first expressed at the blastoderm stage [5]. On closely examination, we found Tc-Wnt1 to be expressed in a subterminal position, leaving the posterior pole free of expression (Figure 2A and 2B). In contrast, Tc-WntD/8 was restricted to the posterior pole (Figure 2D and 2E) in a domain that appeared to complement that of Tc-Wnt1. Later, Tc-WntD/8 expression was limited to two small spots, flanking the mesodermal precursors in the condensing germ rudiment (Figure 2G and 2H), which were maintained during germband elongation (Figure 2F). Tc-Wnt1, however, is expressed more broadly in the posterior growth zone during germband elongation, as well as in the developing segments [5,19] (Figure 2C). The redundancy of these two genes may result from their overlapping domains in the posterior growth zone during germband elongation.
Figure 2. Tc-Wnt1 and Tc-WntD/8 mRNA expression in wild-type and Tc-torso RNAi embryos.
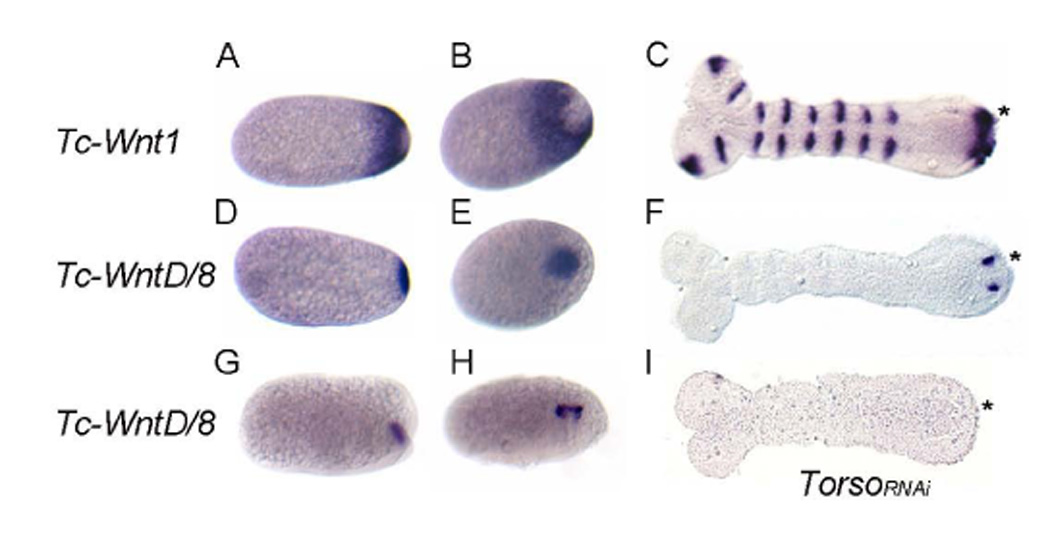
In these embryos, the anterior region is to the left and posterior is to the right.
(A–B) During the blastoderm stage, Tc-Wnt1 is expressed as a ring around the posterior pole, leaving the pole itself free of expression.
(C) During germband elongation Tc-Wnt1 is expressed, broadly in the posterior growth zone.
(D–E) Tc-WntD/8 is expressed at the posterior pole complementary to Tc-Wnt1 expression.
(G–H) Tc-WntD/8 expression resolves into intense spots on either side of the midline in the posterior growth zone of the condensing germ rudiment.
(F) During germband elongation, two distinct spots of Tc-WntD/8 expression are visible in the posterior growth zone (*)
(I) Tc-WntD/8 expression is abolished in Tc-torso RNAi embryos.
Regulation of Tc-WntD/8 in posterior patterning
Torso signaling is important for posterior patterning in Tribolium and regulates several genes expressed in the posterior growth zone, including Tc-Wnt1 [4]. Since, Tc-Torso RNAi embryos lack T3 and all abdominal segments [4], as do a small percentage of TcWntD/8 RNAi embryos, we investigated the effect of Tc-torso depletion on the expression of Tc-WntD/8. In Tc-torso RNAi embryos Tc-WntD/8 expression is completely abolished (Figure 2I), suggesting that posterior patterning by Torso may be realized, at least in part, through Tc-WntD/8.
Conclusions
In Tribolium, the function of Tc-WntD/8 in the posterior growth zone appears to be regulated by Tc-torso and transduced through the canonical Wnt pathway. However, this function does not depend on Tc-wls, providing evidence that Wls is not required for the secretion of all Wnt proteins, as previously thought. Tc-WntD/8 function appears to be redundant with Tc-Wnt1 since the frequency of the truncation phenotype was enhanced when both were removed. Similar truncation phenotypes are produced by depletion of Wnt3 or Wnt8 in vertebrates [7–9] and Wnt8 in the spider A. tepidariorum (McGregor et al. [6], submitted in companion with this article). Thus, Wnt8 may have a conserved function in posterior patterning in vertebrates and arthropods. Wnt3, which has evolved a posterior patterning role in some vertebrates, has not been found in insects [5]. In arthropods, Wnt8 appears to be the major growth zone regulator in a spider [6], while the posterior patterning function of WntD/8 appears to be enhanced by Wnt1 in the short germ insect, Tribolium. A posterior Wnt signal is not required in Drosophila, where all segments are predefined at the blastoderm stage. Instead, Wnt1 appears to have evolved a role in the differentiation of specific posterior structures [11] and WntD/8 function is required for dorsal-ventral patterning [20]. It will be interesting to determine whether Oncopeltus and Gryllus also rely on multiple Wnt ligands (including WntD/8), since there is evidence for Wnt-signaling through the canonical pathway in these intermediate germband insects [2, 3], but the signal does not appear to be provided by Wnt1 alone. Additional investigation will also be necessary to determine whether posterior Wnt signaling regulates cell proliferation and/or other aspects of posterior patterning.
Experimental Procedures
Strain and maintenance
Ga-1 strain of Tribolium castaneum was used for gene expression and functional analysis. The beetles were reared at 30°C in whole wheat flour supplemented with 5% dried yeast.
Isolation of Tribolium Tc-wls and Tc-porc
Tc-wls and Tc-porc were identified by BLAST analysis of the genome. Gene fragments were amplified from cDNA, cloned into TOPO4 vector (Invitrogen) and sequenced at the KSU sequencing facility (http://www.oznet.ksu.edu/pr_dnas/on_campus_form.htm) to confirm their identity.
RNA interference (RNAi)
Template for dsRNA synthesis was amplified by using gene specific primers that have T7 promoter sequences at the end. Double-stranded RNA was synthesized using the T7 megascript kit (Ambion) and purified using the Megaclear kit (Ambion). Different concentrations of dsRNA (0.3–2ug/ul) were mixed with injection buffer (5mM KCl, 0.1 mM KPO4 pH 6.8) prior to injection. Parental or embryonic RNAi was performed and affected embryos were analyzed as previously described [21–22]. For experiments involving Tc-Wnt1, which is required for oogenesis, fecund adult females were injected with 0.3ug/ul of Tc-Wnt1 dsRNA and higher concentrations of dsRNA for the other Wnt genes. For all other experiments, female pupae were injected, allowed to eclosed and mated to wild-type males. Tc-Wnt1 dsRNA was also injected into 2–4 hour embryos which were incubated for 12–36 hr prior to fixation for in situ hybridization. 0–72 h old eggs were collected and split into two samples, one was allowed to develop at 30°C for two days and then submerged in 75% lactic acid for 16 hours at 60°C, the other sample was immediately fixed for in situ hybridization. 500–1000 bp fragments where used as template for digoxigenin-labeled RNA probes [23]. Expression of Engrailed in Tribolium embryos was analysed using the α- Invected antibody 4D9 (Santa Cruz Technology) which cross-reacts with Tc-En [24]. Expression of Tc-Eve was analysed using an anti-Eve (28B) antibody (University of Iowa).
Microscopy and imaging
Cuticles and stained embryos and were viewed with a Nikon Digital Camera DXM 1200F camera on an Olympus BX50 microscope and photographed using Nikon ACT-1 Version 2.62 software. Brightness and contrast of all images were adjusted using Adobe Photoshop CS2 software.
Supplemental Data
The supplemental data for this article contains 2 supplementary figures and 1 supplementary table.
Supplementary Material
Acknowledgements
The work of RB and SJB is supported by NIH grant HD29594. TF was supported by an HMMI undergraduate fellowship. LF was supported by a summer scholarship from the TCJohnson Center for Basic Cancer Research, KSU. We thank Michelle Gordon for technical assistance with the in situ hybridization and Teresa Shippy for insightful discussions during the course of these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ober KA, Jockusch EL. The roles of wingless and decapentaplegic in axis and appendage development in the red flour beetle, Tribolium castaneum. Dev Biol. 2006;294(2):391–405. doi: 10.1016/j.ydbio.2006.02.053. [DOI] [PubMed] [Google Scholar]
- 2.Miyawaki K, Mito T, Sarashina U, Zhang H, Shinmyo Y, Ohuchi H, Noji S. Involvement of Wingless/Armadillo signaling in the posterior sequential segmentation in the cricket, Gryllus bimaculatus (Orthoptera), as revealed by RNAi analysis. Mech Dev. 2004;121:119–130. doi: 10.1016/j.mod.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Angelini DR, Kaufman TC. Functional analyses in the milkweed bug Oncopeltus fasciatus (Hemiptera) support a role for Wnt signaling in body segmentation but not appendage development. Dev Biol. 2005;283:409–423. doi: 10.1016/j.ydbio.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 4.Schoppmeier M, Schroder R. Maternal Torso signaling controls body axis elongation in a shortgerm insect. Current Biology. 2005;15:2131–2136. doi: 10.1016/j.cub.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 5.Bolognesi R, Beermann A, Farzana L, Wittkopp N, Lutz R, Balavoine G, Brown SJ, Schröder R. Tribolium Wnts: evidence for a larger repertoire in insects with overlapping expression patterns that suggest multiple redundant functions in embryogenesis. Development, Genes and Evolution. 2008;218:193–202. doi: 10.1007/s00427-007-0170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGregor AP, Pechmann M, Feitosa NM, Schwager EE, Kruck S, Aranda M, Damen WGM. Wnt8 is required for establishment of the growth zone and generation of opisthosomal segments in the spider Achaearanea tepidariorum. Submitted in companion with this manuscript. [Google Scholar]
- 7.Takada S, et al. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 8.Li HY, et al. FGF8, Wnt8 and Myf5 are target genes of Tbx6 during anteroposterior specification in Xenopus embryo. Dev Biol. 2006;290:470–481. doi: 10.1016/j.ydbio.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 9.Lekven AC, Thorpe CJ, Waxman JS, Moon RT. Zebrafish Wnt8 encodes two Wnt8 proteins on a bicistronic transcript and is required for mesoderm and neuroectoderm patterning. Dev Cell. 2001;1:103–114. doi: 10.1016/s1534-5807(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 10.Davis GK, Patel NH. Short, long and beyond: molecular and embryological approaches to insect segmentation. Annu. Rev. Entomol. 2002;47:669–699. doi: 10.1146/annurev.ento.47.091201.145251. [DOI] [PubMed] [Google Scholar]
- 11.Siegfried E, Perrimon N. Drosophila wingless: a paradigm for the function and mechanism of Wnt signaling. Bioessays. 1994;6:395–404. doi: 10.1002/bies.950160607. [DOI] [PubMed] [Google Scholar]
- 12.Banziger C, Soldini D, Schutt C, Zipperlen P, Hausmann G, Basler K. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell. 2006;125:509–522. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 13.Bartscherer K, Pelte N, Ingelfinger D, Boutros M. Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell. 2006;125:523–533. doi: 10.1016/j.cell.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Goodman RM, Thombre S, Firtina Z, Gray D, Betts D, Roebuck J, Spana EP, Selva EM. Sprinter: a novel transmembrane protein required for Wg secretion and signaling. Development. 2006;133:4901–4911. doi: 10.1242/dev.02674. [DOI] [PubMed] [Google Scholar]
- 15.Morel V, Arias AM. Armadillo/beta-catenin-dependent Wnt signalling is required for the polarisation of epidermal cells during dorsal closure in Drosophila. Development. 2004;131:3273–3283. doi: 10.1242/dev.01217. [DOI] [PubMed] [Google Scholar]
- 16.Schweizer L, Nellen D, Basler K. Requirement for Pangolin/dTCF in Drosophila Wingless signalling. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5846–5851. doi: 10.1073/pnas.1037533100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavalo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signaling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 18.Hays R, Gibori GB, Bejsovec A. Wingless signaling generates pattern through two distinct mechanisms. Development. 1997;124:3727–3736. doi: 10.1242/dev.124.19.3727. [DOI] [PubMed] [Google Scholar]
- 19.Nagy L, Carrol S. Conservation of wingless patterning functions in the short-germ embryos of Tribolium castaneum. Nature. 1994;367:460–463. doi: 10.1038/367460a0. [DOI] [PubMed] [Google Scholar]
- 20.Ganguly A, Jiang J, Ip YT. Drosophila WntD is a target and an inhibitor of the Dorsal/Twist/Snail network in the gastrulating embryo. Development. 2005;132(15):3419–3429. doi: 10.1242/dev.01903. Epub 2005 Jun 29. [DOI] [PubMed] [Google Scholar]
- 21.Bucher G, Scholten J, Klingler M. Parental RNAi in Tribolium (Coleoptera) Curr Biol. 2002;12(3):85–86. doi: 10.1016/s0960-9822(02)00666-8. [DOI] [PubMed] [Google Scholar]
- 22.Shippy TD, Guo J, Brown SJ, Beeman RW, Denell RE. Analysis of maxillopedia expression pattern and larval cuticular phenotype in wild-type and mutant tribolium. Genetics. 2000;155(2):721–731. doi: 10.1093/genetics/155.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown SJ, Hilgenfeld RB, Denell RE. The beetle Tribolium castaneum has a fushi tarazu homolog expressed in stripes during segmentation. Proc Natl Acad Sci U S A. 1994;91(26):12922–12926. doi: 10.1073/pnas.91.26.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown SJ, Parrish JK, Beeman RW, Denell RE. Molecular characterization and embryonic expression of the even-skipped ortholog of Tribolium castaneum. Mech Dev. 1997;1(1–2):165–173. doi: 10.1016/s0925-4773(96)00642-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


