Abstract
Purpose
Although microarray technology has been widely adopted by the scientific community, analysis of the ensuing data remains challenging. In this paper we present our experience with a complex design microarray experiment on resistance mechanisms of histone deacetylase inhibitors (HDACIs).
Experimental design
To improve our understanding of the underlying mechanism of HDACI resistance in prostate cancer (PCa) cells, we designed a novel “multiple loop, double cube” cDNA microarray experiment. In the experiment of 22 arrays, DU145 and PC3 cells were treated with two different HDACIs (vorinostat, and Valproic Acid, VPA) and incubation periods (48 and 96 hours). Pre-processing included exploratory analyses of the quality of the arrays and intensity-dependent within-array loess normalization. An ANOVA model was used for inference. The results were validated by western blot analysis of known treatment targets.
Results
Treatment of PC3 and DU145 cells with HDACIs caused 2.8–10% (p<.001) differential expression across conditions. 51–73% of these genes were up-regulated and 28–49% down-regulated. The extent of differential expression was associated with: cell line (DU145>PC3), HDACI (VPA≥vorinostat) and duration of treatment (96h>48h). We identified known and new treatment targets involved in cell cycle and apoptosis.
Conclusion
A multiple loop, double cube microarray design can be used to identify HDACI induced changes in gene expression, possibly related to drug resistance.
Background
Histone deacetylase inhibitors (HDACIs) are anti-cancer agents that inhibit the activity of enzymes that acetylate histone tails and other proteins. Acetylation is thought to play a role in cancer development and progression (1). Several HDACIs are in clinical development for solid malignancies. In preclinical studies, these agents have shown antitumor activity in vitro and in vivo in prostate cancer (PCa) models. Phase I and II clinical trials with HDACI resulted in stable disease and partial responses in a wide range of solid malignancies, including PCa (2, 3). Recently, the FDA approved HDACI vorinostat for the treatment of advanced cutaneous T-cell lymphoma and several clinical trials are ongoing in solid malignancies. Since its initial success as a tool for quantitative transcription level analysis in 1995 (4), microarray technology has become instrumental for global and parallel analysis of cellular biology (5). Although microarray technology has been rapidly adopted by the scientific community, experimental design and interpretation of the resulting data remain challenging. The choice of experimental design can be crucial in determining the extent to which biological and technical variation affect the results of a microarray experiment (7). Commonly used experimental designs in dual-colour arrays include the ‘loop design’ and ‘reference design’. In the loop design all samples are hybridized in such a way that the connected arrays form a loop pattern (1–2, 2–3, 3–4, 4–1). In a reference design all samples of interest are co-hybridized with the same reference sample (1-R, 2-R, 3-R) (6).
In this paper we report results of a microarray experiment conducted using a more complex design, as a case study to increase understanding of the technical aspects in experimental design and analysis of high throughput gene expression studies. Prior to our microarray experiment, we tested the sensitivity of PCa cells to HDACIs. PC3 cells were more resistant than DU145 cells to HDACI treatment. To improve understanding of the mechanism underlying this difference in resistance, we designed an experiment to simultaneously compare global gene expression in 16 different biological samples with 22 arrays. To determine the relative contribution of three experimental variables of interest on gene expression, all samples were hybridized according to a tailor-made “multidimensional loop” design. Independent comparison of all 16 samples was made possible with an adjusted ANOVA model for each feature on the array. This manuscript discusses the pros and cons of our design and its feasibility for other researchers.
Methods
Cell Culture and chemicals
The following human PCa cell lines (e.g. LNCaP, DU145, PC3, CWR22R) were obtained from American Type Culture Collection (Rockwell, MD, USA) and cultured in RPMI 1640 medium (Life Technologies, USA) supplemented with 10% FBS. Vorinostat (also known as Suberoyl Anilide Hydroxamic Acid, SAHA) was obtained from Aton Pharma/Merck and valproic acid (VPA) and Trichostatin A (TSA) were obtained from Sigma-Aldrich.
Proliferation assays
DU145 and PC3 were plated in 96-well plates at 4000 cells/well and CWR22R and LNCaP cells were plated at 10.000 cells/well. Per treatment arm 8 wells were used. All cells were allowed to adhere for 24h prior to treatment, except LNCaP which was allowed to adhere for 48h. At the start of the experiment, freshly prepared drug was added to each well and incubated for 48h. The proliferative activity was quantified using the MTT kit of Promega (Cell Titer 96 Non-Radioactive Cell proliferation assay) which is based on reduction of tetrazolium salt MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazoliumbromide] to colored formazan by mitochondrial enzymes present only in living, metabolically active cells (7). In congruence with the manufacturers’ description, cells were incubated for 4h with the Dye solution after treatment with HDACI, followed by overnight incubation with the Stop solution. To acquire a uniform colour, a shaker was used. The absorbance at 570 nm was read and background was subtracted using the 650 nm reading and a blank reading. P-values were calculated using the two-sample t-test assuming equal variances (Student’s t-test). All MTT assays were conducted in triplicate.
Microarray Experiment
Experimental Design
To increase our insight in the mechanism of action of HDACIs on PCa cells and to gain insight in possible resistance mechanisms against these agents, we compared the transcription levels of PC3 and DU145 cell lines after treatment with vorinostat and valproic acid (VPA). For the microarray experiment 1 μM vorinostat and 1 mM of VPA were chosen as the treatment doses. It has been shown that these doses are achievable in humans. 48h or 96h treatment with either 1 μM vorinostat or 1 mM VPA in both DU145 and PC3 cell lines were compared.
In total, we compared 16 conditions with 22 arrays. We used dual colour microarrays to take advantage of the ability to hybridize two samples per array. We preferred direct comparison on the more important questions, while also allowing answering additional questions via indirect comparison. Some samples were hybridized 3 or 4 times to facilitate the comparisons of interest. In a loop design, a larger number of edges between samples is associated with higher variance in the corresponding comparison (6). No assumptions were made concerning (in)dependence of our experimental conditions. Taking into account dye swaps and technical replicates, the microarray set up resulted in a double cube. The directed graph (8) in figure 2b gives a schematic overview of the experimental set up. Each arrow represents one microarray slide comparing expression between two RNAs and is considered a block in the experimental layout (6). The sample on the tail side of the arrow is labelled with cy5 (red). The sample on the head side of the arrow is labelled with cy3 (green). The variables of interest included cell line (DU145, PC3), HDACI (vorinostat, VPA) and duration of treatment (48h, 96h). Control samples within each cube utilize the solvent specific for the HDACI. All samples are labelled and hybridized at least once with cy3 and once with cy5 to correct for dye effects.
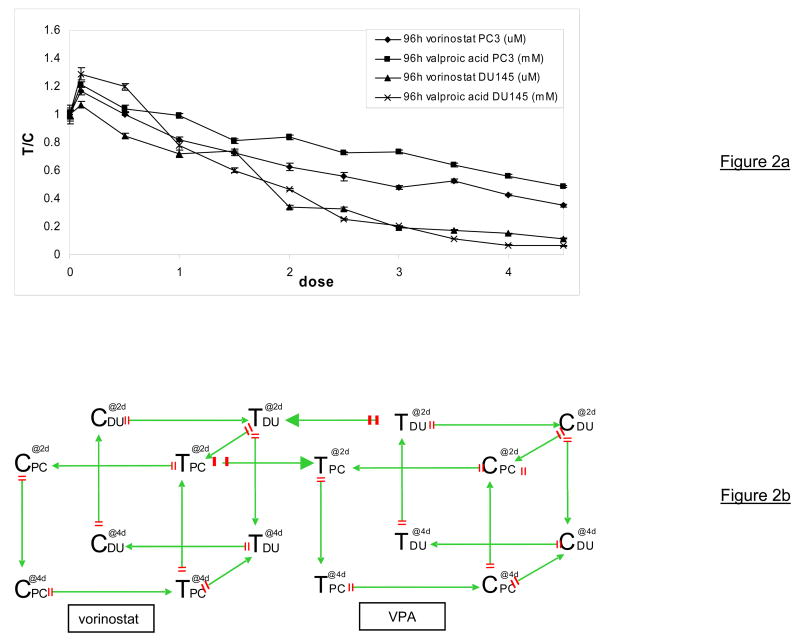
Figure 2.
Figure 2a: MTT assays to test the proliferative activity of PCa cells after 4 days treatment with increasing concentrations of valproic acid or vorinostat. DU145 and PC3 cells were plated in 96-well plates at 2000 cells/well allowed to adhere for 24h prior to treatment. At the start of the experiment the media was removed, replaced by fresh media with or without HDACI and incubated for 96h. Measurements were normalized to control (T/C).
Figure 2b: multiple loop, double cube microarray set up using 22 Agilent human 1A v2 arrays. Arrows represent arrays and direction of hybridization (C control, T treated, PC PC3 cell line, DU DU145 cell line, @2d sample drugged for 48h, @4d sample drugged for 96h, green arrow head Cy3 dye, red arrow tail Cy5 dye, VPA Valproic Acid)
Cell culture
On day 0, DU145 and PC3 cells (low passage) were trypsinized at ~85% confluency and replated at 50.000 cells/100 mm dish. Each treatment was applied in triplicate. Aliquots for each drug concentration and time point were prepared in RPMI on day 0 and stored in −20°C for use in the following four days. FBS was aliquoted separately and stored in −20°C. The concentration DMSO for the vorinostat aliquots was 0.05%. On day 1 the media was removed and replaced with the above described (drug-containing) media. The following four days the media was carefully aspirated each day and replaced with fresh (drug-containing) media. After 48h and 96h dishes were harvested on ice using scrapers and 1 ml TRIzol reagent (Invitrogen) per dish and the pooled samples were stored in −80°C until RNA isolation at the end of the experiment.
RNA isolation
Total RNA was isolated from DU145 and PC-3 cells using a combination of TRIzol ® and Rneasy® Kit (Qiagen) with slight modifications to manufacturer’s protocol. Briefly, cells were harvested in TRIzol®. After addition of chloroform and centrifugation, the aqueous phase was separated (supernatant). This was approximately 50% of the initial TRIzol® volume. Next, an equal volume of 70% ethanol was added to the supernatant. Since the final alcohol concentration now was only 30%, the RNA did not precipitate. The mixture was then passed through RNeasy® column and further purification carried out according to the manufacturer’s instructions. A DNase step was also included to ensure removal of any contaminating DNA.
RNA quality assessment
Total RNA of all samples was quantified by the Johns Hopkins Sidney Kimmel Comprehensive Cancer Center (SKCCC) Microarray Core facility with NanoDrop ND-100 followed by quality assessment with 2100 Bioanalyzer (Agilent Technologies) according to manufacturer’s protocol. rRNA ratio (28S/18S) was 1.7–2.2 and RIN (RNA Integrity Number) value was 9.4–9.9.
Sample amplification and labeling
Sample amplification and labelling procedures were carried out by the SKCCC Microarray Core facility using Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies). One microgram of total RNA was reverse transcribed into first strand and second strand cDNA by MMLV-RT using an oligo dT primer that incorporates a T7 promoter sequence. The cDNA was then used as a template for in vitro transcription in the presence of T7 RNA polymerase and Cyanine labelled CTPs. The labelled cRNA was purified using RNeasy mini kit (Qiagen) and followed by quantification on both concentrations of cRNA and dye labelled. RNA spike-in controls (Agilent Technologies) were added to the RNA samples before amplification and labelled according to manufacturer’s protocol. In total 0.75 microgram of each sample labelled with Cy3 or Cy5 was mixed with control targets (Agilent Technologies).
Target fragmentation and microarray hybridization
Since the size of our microarray experiment was too large to process on one day, it was decided to split the experiment into manageable portions. For target fragmentation, hybridization and scanning, the experiment was split evenly. Fragmentation was carried out by the SKCCC Microarray Core facility by incubating at 60°C for 30 minutes and stopped by adding equal volume of 2x hybridization buffer (Agilent Technologies). Fragmented targets were added on 22 human 1Av2 Microarrays (Agilent Technologies) all from the same batch. Agilent’s Human 1Av2 micorarray consists of 21,073 60-mer probes printed at random on a glass slide. In total there are 1000 replicates (10 × 100 gene targets) randomly printed on the array. In addition 1080 control spots are printed on the array. The microarrays were assembled into a hybridization chamber (Agilent Technologies) and hybridized at 60°C for 17 hours in a hybridization oven with rotation. Hybridized microarrays were washed and dried according to the Agilent microarray processing protocol.
Microarray scanning and data Acquisition
Microarrays were scanned by the SKCCC Microarray Core facility with the Agilent G2565BA microarray scanner under default settings recommended by Agilent Technologies for expression microarrays with 100% PMT and 10 μm resolutions. Data was extracted using Feature Extraction Software v8.1 (Agilent Technologies).
Software
All analyses were performed using the Bioconductor limma or marray packages for R (9). Bioconductor is an open source and open development software project for the analysis and comprehension of genomic data (www.bioconductor.org) and R is a language and environment for statistical computing and graphics (www.r-project.org).
Pre-processing
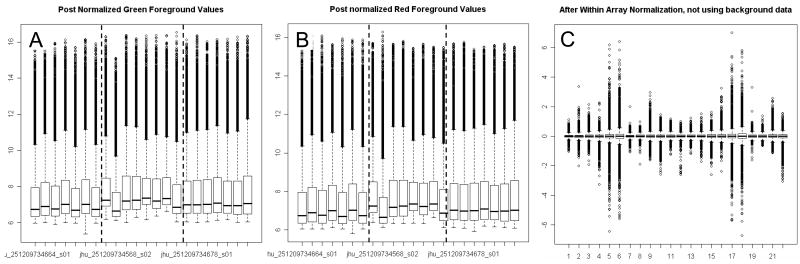
The first step during pre-processing of raw microarray data is quality assessment. Image plots to visualize spatial trends and standard MA plots to visualize intensity trends (10) were created for each array. Eleven probes affected by a small artifact identified in the MA plot of one array were assigned missing values. The need for background subtraction was assessed with plots of the foreground log(red/green) versus background log(red/green) values (11). Following within-array loess normalization of the median foreground intensities, box plots of the loess normalized fold changes (M) from all arrays were made. An across array normalization was considered, but omitted in order not to eliminate the differences between cell lines observed in these boxplots.
Data analysis
Normalized values of the median red and green foreground were calculated from M and A: Rf = A + (M/2) and Gf = A − (M/2). Partitioning of the total variation in the foreground signal into different sources enables comparison of the variance between levels of a factor to the variance within each level of the factor, due to noise. Our Analysis of Variance (ANOVA) model for each probe represents how different factors contribute to the observed signal. All two-way interactions of the experimental factors: treatment, cell line and length of treatment, were included in the model, because we could not assume the effects of each factor were consistent across the levels of other factors. In summary, we assumed:
where yijklm is the log2 of the foreground fluorescent intensity from array (I) i and dye (D) j representing the fixed effects of treatment (T) k, cell line (C) l and length of treatment (L) m. Since between-array normalization was not performed during pre-processing, factors for the effects of dye (D) and array (I) were included in the model.
Quantities of interest were contrast coefficients reflecting comparisons of treatment and controls within cell line and duration of treatment. For significance analysis we used the moderated t-statistic. This has the same interpretation as an ordinary t- statistic except that the standard errors have been moderated across genes, that is pulled towards a common value estimated from the entire set of gene using a Bayesian model. Details are provided in (12), who also describe how to determine p-values from moderated statistics. Lastly, tests of the specified contrasts for every gene were adjusted for multiple comparisons with the Benjamini-Hochberg method.
Western Blots
Cells were plated and treated as described in the cell culture section of the microarray experiment. After 24h, 48h, 72h and 96h cells were washed once with ice cold PBS and harvested on ice using scrapers and the lysis M-Per solution of Pierce (Rockford, IL, USA) containing a cocktail of protease inhibitors (Roche, Steinheim, Germany). After 10 min. incubation on ice, the lysates were centrifuged at 6000 RPM at 4°C for 10 min. The protein-containing supernatant was stored in −80°C until further analysis with the BCA assay (Pierce, Rockford, IL, USA). Laemmli sample buffer at a ratio of 1:2 was mixed to the sample and heated at 95°C for 5 min. The denatured proteins were stored in −80°C until use. Using a 4–15% Tris-SDS-PAGE gel (Biorad) 10–20 μg proteins were separated at 100 V for 1 h. The separated proteins in the gel were electrophoretically transferred onto polyvinylidene difluoride membranes (Bio-Rad, Munich, Germany) at 380 mA for 1 h. Blocking was carried out with TBS plus 5 % non-fat milk (20 mM Tris-HCl, pH 7.6, 137 mM NaCl). After washing, the membrane was incubated overnight with primary antibodies for p21WAF1 (SantaCruz, Biotech, California, USA) and Ac-H3K9 (Cell Signalling) diluted in TBS. Appropriate horseradish peroxidase-conjugated secondary antibodies (Amersham, Buckinghamshire, UK) were incubated for 1h. For detection, autoradiography using ECL was performed according to the manufacturer’s instructions (Amersham, Buckinghamshire, UK).
Results
Cell-specific sensitivity for SAHA and TSA using MTT assays
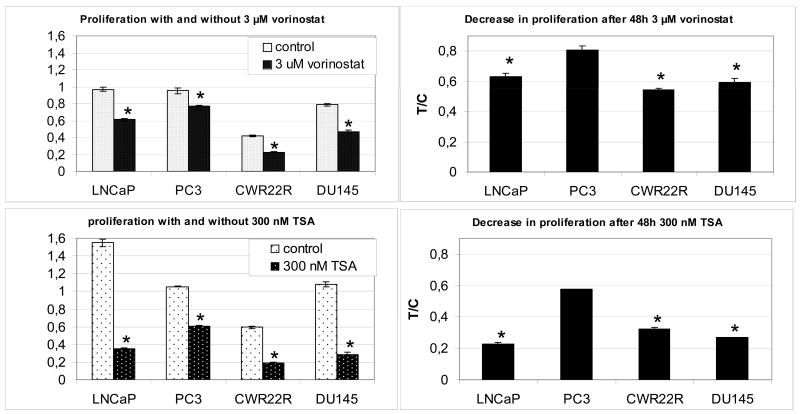
To test the sensitivity of prostate cancer (PCa) cells to HDACIs, a standard MTT proliferation assay was used. Four epithelial PCa cell lines (LNCaP, PC3, CWR22R, DU145) were treated with 3 μM vorinostat or 300 nM TSA (figure 1). The assay revealed that compared to LNCaP, CWR22R and DU145, the proliferation rate of PC3 cells was significantly less affected after 48h treatment with either vorinostat or TSA.
Figure 1.
MTT assay to test the proliferative activity of PCa cells with and without treatment with 3 μM vorinostat or 300 nM TSA. Left two panels indicate proliferation measurements after 48h in control and treated cells. Right panels compare decrease in mean proliferation after treatment in PC3 cells with the proliferation of LNCaP, CWR22R and DU145 (T/C mean treated sample/control sample PCa prostate cancer TSA Trichostatin A, error bars indicate SEM, * indicates p-value < 0.05)
For all other experiments we decided to compare PC3 cells with DU145 cells since the doubling time, morphology, androgen responsiveness and p53 status of PC3 and DU145 is relatively comparable. Figure 2a shows that when DU145 and PC3 cells are treated for 96h, growth of DU145 cells is completely inhibited at the highest concentration of HDACI treatment while proliferation of PC3 cells is only inhibited by 50%.
Microarray experiment
Three independent variables were studied in our experiment: two PCa cell lines (DU145 and PC3) with varying sensitivity to Histone Deacetylase Inhibitors (HDACIs), two structurally different HDACIs (vorinostat (also known as Suberoyl Anilide Hydroxamic Acid, SAHA) and small chain fatty acid Valproic Acid) and two treatment intervals (48 and 96 hours). To determine the relative contribution of these three independent variables of interest on gene expression, all samples were hybridized according to a novel “multidimensional loop” (figure 2b). This set up allowed for independent analysis of each variable without assuming dependency between each variable, and with minimal use of resources.
Pre-processing summary
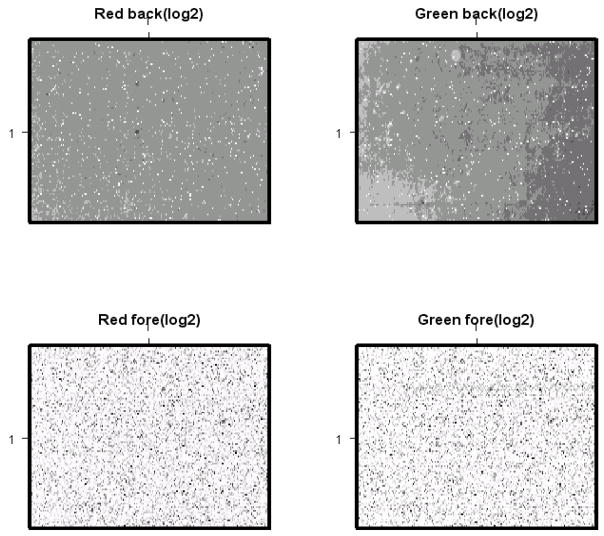
Image plots of raw expression data for each individual microarray (figure 3) revealed some spatial artefacts in the green background signal. Plots of the foreground log2(red/green) versus background log2 (red/green) revealed little correlation (figure 4a+b), indicating that the spatial artifacts seen in the green background did not significantly affect foreground signal (11). Background subtraction was therefore omitted.
Figure 3.
Image plots of unprocessed Red and Green signals: log2 Rb (upper left corner), log2 Gb (upper right corner), log2 Rf (lower left corner) and log2 Gf (lower right corner) of a representative microarray visualize spatial artefacts in log2 Gb. Image plots visualize the intensity of each expression value on the glass slide. Their purpose is to identify spatial defects in the data sets such as scratches (Rb Red background, Gb Green background, Rf Red foreground, Gf Green foreground)
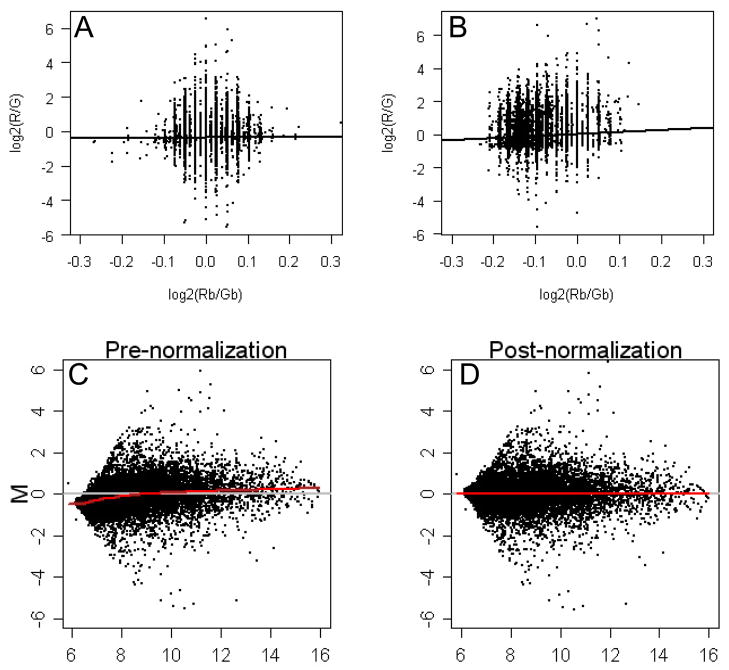
Figure 4.
Figure 4a+b. Correlation plots of raw foreground and background signals of two representative microarrays. The black line shows no correlation between Log2(R/G) (=foreground) and Log2(Rb/Gb) (=background) signals (left = 0.07, right = 0.1)
Figure 4c+d: MA-plots of pre-and post- normalized foreground expression data for a representative microarray. Each dot represents one probe. Red line represents a fitted Loess curve that summarizes any non-linear relationship (M = log2(Red/Green) = ratio of intensity; A = log2(Red+Green)/2 = average intensity)
In MA-plots of foreground signals we noted for all arrays a deviation from zero in the low expression signals (figure 4c). We used loess, as implemented in the limma package (12) to adjust for this systematic technical variation associated with intensity (figure 4d). Boxplots of the Loess normalized red and green foreground signals from all arrays visualized a batch effect (figure 5a+b) which was most likely caused by our splitting target fragmentation, hybridization and scanning of the arrays into three different days. This batch effect was not visible in boxplots of the Loess normalized fold changes (M) (figure 5c), indicating that the batch effect did not favor one channel. Boxplots of the Loess normalized fold changes (M) also indicated that most arrays had similar variability except for all four arrays with both cell lines on the same array. Although across-array normalization could correct for the batch-effect, it might also eliminate true cell line differences. Therefore, an array factor was added to the statistical model to adjust for this effect without performing a separate across array normalization (see methods).
Figure 5.
Figure 5a+b. Boxplots of green (Gf) and red (Rf) foreground signals after Loess normalization visualizing a batch effect in each channel (Rf Red foreground channel, Gf Green foreground channel)
Figure 5c: Boxplots of Loess normalized fold changes (M) for all 22 arrays using foreground data. Increased variability in four arrays having both cell lines co-hybridized is visible: array 5+6 both have vorinostat treated cell lines co-hybridized, array 17+18 both have valproic acid treated cell lines co-hybridized)
Gene expression data
Using a custom-made ANOVA-model (methods), genes with significantly different expression were identified. The comparison of interest in this study was differential expression after treatment. Table 1 displays the differential expression caused by these variables.
Table 1A.
Differential expression after treatment with VPA and vorinostat
| Cell line | Treatment | Treatment | % upregulated (# probes) | % downregulated (# probes) | Total change (# probes) |
|---|---|---|---|---|---|
| DU145 | 1 μM vorinostat | 48h | 53.8% (648) | 46.2% (557) | 5.72% (1205) |
| 96h | 51.3% (916) | 48.7% (869) | 8.47% (1785) | ||
| 1 mM VPA | 48h | 69.7% (1177) | 30.3% (512) | 8.02% (1689) | |
| 96h | 65.5% (1385) | 34.5% (731) | 10.0% (2116) | ||
| PC3 | 1 μM vorinostat | 48h | 67.7% (409) | 32.3% (195) | 2.87% (604) |
| 96h | 63.8% (572) | 36.2% (324) | 4.25% (896) | ||
| 1 mM VPA | 48h | 70.7% (425) | 29.3% (176) | 2.85% (601) | |
| 96h | 72.5% (577) | 27.5% (219) | 3.78% (796) |
Table 1a: Differential expression (p<0.001) after 48h or 96h treatment with 1 μM vorinostat or 1 mM VPA in PC3 or DU145 cell lines
A p-value cut-off of 0.001 was used. No cut-off for fold change was used (see discussion). Genes with a fold change smaller than 1 were reported as down-regulated, genes with a fold change greater than 1 were reported as upregulated. HDACIs induced significant differences in gene expression of 2.85–10.1% of all probes (p<0.001). 51.3–72.5% of the genes were upregulated and 27.5–48.7% downregulated. HDACIs caused less differential expression in PC3 than in DU145 cells (2.85–4.25% vs 5.72–10.0%, respectively). Differential expression was also affected by duration and type of treatment. 96 hour treatment with vorinostat induced an increase of 48% in differential expression in both DU145 and PC3 cells compared to 48 hour treatment. In the VPA treated groups however, 96 hour treatment caused an increase of only 25% in DU145 and 33% in PC3 compared to 48 hour treatment. This HDACI-specific difference is also reflected by comparing the extent of differential expression per timepoint and per cell line. After 96h treatment, 12.4% more expression changes were seen in PC3 cells treated with vorinostat compared to VPA. In the earlier timepoint (48h), no quantitative difference between vorinostat and VPA was seen in the PC3 cell line. In contrast, vorinostat caused 28.7% less expression changes compared to VPA when DU145 cells were treated for 48 hours, while at 96 hour treatment this difference decreased to 15.3%. Table 1b presents the expression data of some commonly changed genes after HDACI treatment (1, 13).
Table 1B.
expression pattern of commonly changed genes
| tVPAda2DU | tVPAda4DU | tVPAda2PC3 | tVPAda4PC3 | tSAHAda2DU | tSAHAda4DU | tSAHAda2PC3 | tSAHAda4PC3 | GeneName | SystematicName |
|---|---|---|---|---|---|---|---|---|---|
| 2.00 | 2.56 | 1.70 | 2.18 | 2.10 | 3.48 | 1.25 | 2.07 | CDKN1A | NM_000389 |
| 2.04 | 2.07 | 1.82 | 1.85 | 1.73 | 2.02 | 1.56 | 1.83 | GLRX | NM_002064 |
| 1.55 | 1.59 | 1.16 | 1.18 | 1.59 | 1.61 | 1.18 | 1.19 | FUCA1 | NM_000147 |
| 1.32 | 1.42 | 1.21 | 1.30 | 1.39 | 1.83 | 1.29 | 1.69 | VEGFC | NM_005429 |
| 0.53 | 0.53 | 0.82 | 0.81 | 0.45 | 0.43 | 0.78 | 0.75 | TYMS | NM_001071 |
| 0.66 | 0.68 | 0.92 | 0.95 | 0.71 | 0.69 | 0.90 | 0.88 | CTPS | NM_001905 |
| 1.01 | 0.94 | 1.09 | 1.02 | 1.02 | 0.99 | 1.02 | 0.99 | RARB | NM_000965 |
| 1.17 | 1.22 | 1.20 | 1.25 | 1.03 | 1.01 | 1.23 | 1.20 | SEMA3A | NM_006080 |
| 0.96 | 0.92 | 1.01 | 0.97 | 0.99 | 0.96 | 0.97 | 0.94 | NRP1 | NM_003873 |
| 1.01 | 0.97 | 1.00 | 0.96 | 1.02 | 1.07 | 0.99 | 1.04 | TNF | NM_000594 |
| 1.00 | 1.02 | 1.03 | 1.05 | 1.01 | 1.01 | 1.00 | 1.00 | IL2 | NM_000586 |
| 1.10 | 1.41 | 1.92 | 2.48 | 1.17 | 1.69 | 1.33 | 1.93 | IL6 | NM_000600 |
| 1.00 | 0.96 | 1.04 | 1.01 | 1.01 | 1.02 | 1.00 | 1.01 | IFNG | NM_000619 |
| 0.95 | 0.97 | 1.02 | 1.04 | 1.02 | 1.00 | 1.04 | 1.02 | AR | NM_000044 |
| 0.99 | 1.03 | 0.97 | 1.01 | 1.08 | 1.09 | 0.96 | 0.96 | KLK3 | NM_001648 |
| 0.49 | 0.59 | 0.83 | 0.99 | 0.52 | 0.59 | 0.80 | 0.92 | HIST2H2AA | NM_003516 |
Validation: Western Blots of differentially expressed genes
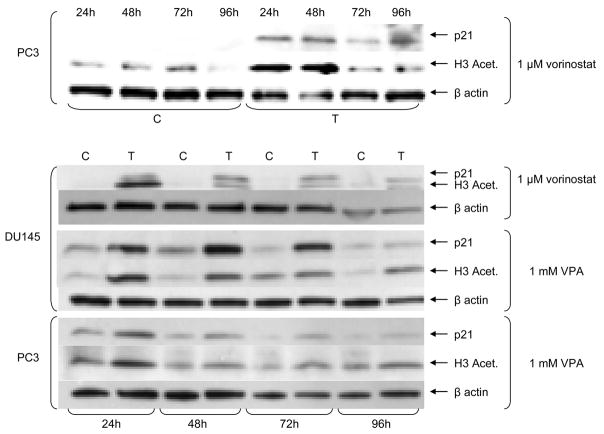
Western blot analysis was used to validate the up-regulation of the classic HDACI-activity marker p21 and the acetylation status of histone 3 after treatment with VPA or vorinostat. Figure 6 shows that the acetylation status of H3 and the protein expression of p21 are increased throughout the course of the experiment. Treatment of DU145 cells compared to PC3 cells resulted in a much higher protein expression of acetylated H3 and p21.
Figure 6.
Western blot validation of p21 expression and acetylation of histone 3 of DU145 and PC3 cells treated 1, 2, 3 and 4 days with 1 μM vorinostat or 1 mM VPA (C control treated with corresponding solvent, T treated with 1 μM vorinostat or 1 mM VPA, VPA valproic Acid)
Discussion
While clinical trials with HDACIs are ongoing, little is known about the mechanisms of action and resistance of these compounds. To improve our understanding of underlying mechanisms of resistance we studied the effect of HDACIs on gene expression profiles of PCa cell lines with different drug sensitivity. We decided to prepare 16 RNA samples of interest that represented three variables: cell line, duration of treatment and type of HDACI. To extract data from these samples to answer our complex questions, we needed bioinformaticists.
In this paper we presented our microarray experiment as a case study to increase understanding of experimental design and analysis in microarray experiments. Each step during data generation and statistical analysis of a microarray experiment may influence the expression data that are generated. Below we will address four issues of importance in the context of microarray experiments: design, biological and technical variation, batch effect and pooling.
One of the first steps in a microarray experiment that may influence the eventual results is its design. Several groups have highlighted the importance of a solid design, but due to the large variety of research questions, general rules are difficult to define. In this study we highlight the importance of a solid design using our own experience. As mentioned above, the two most common set ups in dual colour arrays are the loop design and the reference design. One of the first steps in a microarray experiment that may influence the final results is its design. Several groups have highlighted the importance of a solid design, but due to the large variety of research questions, general rules are difficult to define. In this study we highlight the importance of a solid design using our own experience. As mentioned above, the two most common approaches in dual colour arrays are the loop design and the reference design. The advantages of loop designs include repetitive analysis of samples of interest and the ability to assess dye effect without the use of additional resources. Disadvantage of a loop design include that they provide less information on comparisons between samples that have indirect contact and that the efficiency of these comparisons is distance-dependent (6). In addition, they are often complicated, not robust to chip loss due to technical misadventures and limited in the hypotheses that can be tested. In contrast, reference designs are robust to chip loss and are able to address a much wider range of hypotheses. Furthermore, reference designs are much easier to explain to investigators and are harder to misexecute either in the planning, execution or analysis stage. There are two main disadvantages of a classical reference design: first, all comparisons between the conditions of interest are indirect, as those are never hybridized on the same array; second, unless the reference itself is of interest, half the power of the experimental technique is invested in measurements that are not of direct scientific interest. These two factors generally result in a lower power to identify changes of interest, and in a larger statistical variance on the estimated effects. Recently, it has been proposed that reference designs show primarily advantage in experiments that can not be completed at once (14) (6). In clinical trials for instance, tumour samples might come available throughout the course of the trial. With the use of a (commercially available) reference sample that is co-hybridized with each tumor sample, the measurement can be performed while technical variation and quality can be assessed. For each experiment a careful deliberation on the pros and cons of each set up in the light of the experimental question will aid the eventual data that is generated. In our experiment, we concluded that the use of a reference design would have resulted in the use of ~50% more microarray slides. Instead, we chose for a modified loop design (figure 2b). Because 16 samples needed to be analyzed, a classic loop design would have resulted in too great a distance between some of the samples (6). To achieve a lower residual variance, we decided to hybridize more samples directly.
The choice of experimental design can be crucial in determining the extent to which biological and technical variation impact results of a microarray experiment (7). Depending on the experimental set up, some technical variation can be adjusted by normalisation steps. Within-array normalisation may correct for variation that is array-specific, such as scratches, printing effects and local background. Between-array normalisation aims at correcting for systematic variation that occurs across arrays, such as time effect and technical variation in mRNA pools (15). In general one can identify four factors that contribute to variation of two-channel microarray data (6), varieties V (factors of interest, such as time points of a biological process or treatment), genes G (sequences spotted on the arrays), dyes D (Cy3 and Cy5) and arrays A (overall variation in fluorescent signal from array to array). Depending on the interaction between these factors, up to 24 = 16 causes of variation are possible. The interaction between the factors of interest (V) with genes (G) is of primary interest to study differential gene expressions (6). All other interactions, such as the interaction of dye (D) with genes (G) (16), determine the sources of experimental variability and should be corrected before quantification of differential expression. In our experiment, the use of our multiple loop, double cube design for the analysis of 16 samples, enabled profiling of transcription in PCa cell lines DU145 and PC3 during treatment with two HDACIs vorinostat and VPA. An ANOVA model per gene that included additional factors for Dye (D) and Array (A) retained cell line-specific changes while correcting for between-array variability such as our batch-effect (figure 5a+b).
Larger microarray experiments that aim to answer complex questions can cause additional challenges that need to be adequately addressed. An example of this is the batch effect that is visible in the Gf and Rf boxplots of our data (figure 5a+b). Batch or time effects can occur when not all arrays are hybridized at the same time and/or by the same person. In our case, the experiment included 22 arrays and therefore a batch effect was inevitable. Time-specific changes may affect both signals equally and if a ratio (fold change) is taken during analyses, the time-effect may be missed (figure 5c). For a loop design the separate expression values are used instead of fold change during ANOVA analysis. A correction step for time effects is very important in this set up, because true differential expression will be confounded otherwise. The “normalizeBetweenArrays” function in Limma includes a normalization step to correct for scale differences across arrays. The basic assumption of this procedure is that the distribution of log2 intensities between arrays is similar. Major distribution and scale differences in the intensities are therefore assumed to be due to technical variation. In our experimental set up, this basic assumption could have eliminated cell line-specific differences of interest (figure 5c). Therefore we corrected for the batch effect that was visible in the separate channels (figure 5a+b), by adding a factor for array (A) to the ANOVA model.
While setting up designs for microarray experiments, most investigators face the question of whether to pool replicate samples or to measure each biological replicate separately. Biological replicates are independent preparations of samples taken from the same biosource that have been treated identically. Biological replicates assess the natural variability in a system. The separate measurement of biological replicates versus pooling of replicate samples is under continuing debate (17), (18), (19). The main argument in favour of measuring biological replicates separately is that biological variation may occur from sample to sample. This variation is especially prominent when tissue samples from different patients are being studied. The basic assumption when one decides to pool samples is that the expression data of pooled samples is equal to the average of the same measure taken from each individual sample that contributed to the pool. We assumed that the biological variation within our cell lines is limited. Variation from experiment to experiment will primarily come from technical variation. Based on this assumption, we decided to pool three samples from the same group. To assess the technical variation arising from the microarray technology, each pooled sample was split and multiple technical replicate measurements were carried out. As a result of our experimental set up there were no degrees of freedom available to estimate the biological variance. We did however look at expression between the DMSO control and the water control and did not find any differentially expressed genes (data not shown). When data generation, normalization and statistical analysis have taken place, the data needs to be analyzed and interpreted. To determine which genes are differentially expressed and which gene expressions remained unchanged, arbitrary cut-offs for fold change and/or p-value (or B-statistic) are often used. However, Hughes et al. demonstrated that the basal expression of some genes fluctuates much more than others (20) (21). In addition, Pritchard et al. found a functional relationship between genes whose expression is most variable (22). During the process of data interpretation, it is important to realize that the strength of microarray technology is to generate comprehensive data that precedes the formulation of hypotheses. In contrast to the data generation, data interpretation is often guided by pre-existing ideas in microarray experiments (5). In this study we found several differentially expressed genes that have been linked to HDACI treatment in previous studies although some previously published genes showed not to be differentially expressed in our data set (table 1b). Likely reason for these differences in expression profiles include differences in type of HDACI, cell line used, duration of treatment and dose. Further studies are necessary to determine the importance of these differences.
Not to lose sight of the purpose of our multiple loop double cube microarray experiment, earlier in vitro and in vivo experiments with VPA by our group and others (23) have shown that PC3 cells are relatively resistant to VPA and other HDACIs. In this paper, we describe that treatment of PCa cell lines PC3 and DU145 with HDACIs causes distinct changes in expression profiles. The changes in expression levels in the relatively resistant PC3 were significantly lower than that of DU145. Our microarray design will help us discern the contribution of duration of treatment and type of HDACI to the resistance pattern seen in PC3. Next steps in our analysis will include bioinformatics tools that aid interpretation of microarray data such as Gene Set Enrichment Analysis and clustering algorithms.
Conclusion
In this study, one experiment was used to compare differential expression profiles of 16 different biological samples that represented three factors of interest. We conclude that our multiple loop, double cube microarray design can be used to identify molecular pathways that may be relevant in mechanisms of resistance to HDACIs.
Acknowledgments
Morgan A Marks for helpful discussions and assistance with R and Bioconductor
Conover Talbot
Wayne Yu
Catharine van Tussenbroek Fonds
Jo Keurfonds
Leiden International Studie Fund
Aegon
Supported by the Prostate Cancer Foundation and NCI SPORE grant 58236
Footnotes
Statement of Clinical Relevance
We report on a novel complex design “multiple loop, double cube” cDNA microarray experiment to study gene expression profiles important in resistance to Histone Deacetylase inhibitors (HDACIs). We were able to simultaneously compare global gene expression in 16 different biological samples using 22 arrays. HDACIs are increasingly used in patients with hematological and solid tumors and have shown promising results in clinical trials. Recently, the FDA approved HDACI vorinostat for the treatment of advanced cutaneous T-cell lymphoma. In this manuscript we discuss the pros and cons of our microarray design and its feasibility for other researchers to study mechanisms of resistance. Although microarray technology has been rapidly adopted by the scientific community, experimental design and interpretation of the resulting data remain challenging. In this manuscript we provide more insight in the technical challenges of experimental design and analysis of high throughput gene expression studies, which will be of help to clinical researchers setting up microarray studies and will equip them for more fluent collaborations with bioinformaticists.
References
- 1.Kortenhorst MS, Carducci MA, Shabbeer S. Acetylation and histone deacetylase inhibitors in cancer. Cell Oncol. 2006;28:191–222. doi: 10.1155/2006/760183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reid T, Valone F, Lipera W, et al. Phase II trial of the histone deacetylase inhibitor pivaloyloxymethyl butyrate (Pivanex, AN-9) in advanced non-small cell lung cancer. Lung Cancer. 2004;45:381–6. doi: 10.1016/j.lungcan.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Kelly WK, O’Connor OA, Krug LM, et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J Clin Oncol. 2005;23:3923–31. doi: 10.1200/JCO.2005.14.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–70. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 5.Hoheisel JD. Microarray technology: beyond transcript profiling and genotype analysis. Nat Rev Genet. 2006;7:200–10. doi: 10.1038/nrg1809. [DOI] [PubMed] [Google Scholar]
- 6.Kerr MK, Churchill GA. Experimental design for gene expression microarrays. Biostatistics. 2001;2:183–201. doi: 10.1093/biostatistics/2.2.183. [DOI] [PubMed] [Google Scholar]
- 7.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 8.Yang YH, Speed T. Design issues for cDNA microarray experiments. Nat Rev Genet. 2002;3:579–88. doi: 10.1038/nrg863. [DOI] [PubMed] [Google Scholar]
- 9.R DCT. R: A language and environment for statistical computing. Vienna: Austria; 2004. [Google Scholar]
- 10.Dudoit S, Yang YH, Speed TP, Callow MJ. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statistica Sinica. 2002;12:111–40. [Google Scholar]
- 11.Scharpf RB, Iacobuzio-Donahue CA, Sneddon JB, Parmigiani G. When should one subtract background fluorescence in 2-color microarrays? Biostatistics. 2007;8:695–707. doi: 10.1093/biostatistics/kxl041. [DOI] [PubMed] [Google Scholar]
- 12.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 13.Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther. 2003;2:151–63. [PubMed] [Google Scholar]
- 14.Vinciotti V, Khanin R, D’Alimonte D, et al. An experimental evaluation of a loop versus a reference design for two-channel microarrays. Bioinformatics. 2005;21:492–501. doi: 10.1093/bioinformatics/bti022. [DOI] [PubMed] [Google Scholar]
- 15.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–73. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 16.Martin-Magniette ML, Aubert J, Cabannes E, Daudin JJ. Evaluation of the gene-specific dye bias in cDNA microarray experiments. Bioinformatics. 2005;21:1995–2000. doi: 10.1093/bioinformatics/bti302. [DOI] [PubMed] [Google Scholar]
- 17.Kendziorski CM, Zhang Y, Lan H, Attie AD. The efficiency of pooling mRNA in microarray experiments. Biostatistics. 2003;4:465–77. doi: 10.1093/biostatistics/4.3.465. [DOI] [PubMed] [Google Scholar]
- 18.Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci U S A. 2005;102:4252–7. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 20.Hughes TR, Marton MJ, Jones AR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–26. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 21.Hughes TR, Roberts CJ, Dai H, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–7. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 22.Pritchard CC, Hsu L, Delrow J, Nelson PS. Project normal: defining normal variance in mouse gene expression. Proc Natl Acad Sci U S A. 2001;98:13266–71. doi: 10.1073/pnas.221465998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia Q, Sung J, Chowdhury W, et al. Chronic administration of valproic acid inhibits prostate cancer cell growth in vitro and in vivo. Cancer Res. 2006;66:7237–44. doi: 10.1158/0008-5472.CAN-05-0487. [DOI] [PubMed] [Google Scholar]