Abstract
Lipoprotein Lipase (LPL) plays a pivotal role in lipid metabolism by hydrolyzing triglyceride (TG) rich lipoprotein particles. Abnormalities in normal LPL function are associated with the risk of coronary artery disease (CAD). A number of genetic variants have been identified in the LPL gene that affects different functions of the LPL protein. A common HindIII polymorphism in intron 8 (T/G) of the LPL gene has been found to be associated with altered plasma TG and HDL-cholesterol, and CAD risk in several studies, but its functional significance is unknown. It has been shown that certain intronic sequence contain regulatory elements that are important for transcription and translational regulation of a gene. In this study we tested the hypothesis that this polymorphism affects the binding site of a transcription factor that regulates the transcription of LPL gene. Electrophoretic mobility shift assays revealed that the HindIII site binds to a transcription factor and that the mutant allele has lower binding affinity than the wild type allele. Transcription assays containing the entire intron 8 sequence along with full-length human LPL promoter were carried out in COS-1 and human vascular smooth muscle cells. The mutant allele was associated with significantly decreased luciferase expression level compared to the wild type allele in both the muscle (3.394 ± 0.022 vs. 4.184 ± 0.028; P=4.7 × 10−6) and COS-1 (11.603 ± 0.409 vs. 14.373 ± 1.096; P<0.0001) cells. In conclusion, this study demonstrates for the first time that the polymorphic HindIII site in the LPL gene is functional because it affects the binding of a transcription factor and it also has an impact on LPL expression.
Keywords: Lipoprotein lipase, HindIII polymorphism, electrophoretic mobility shift assay, luciferase reporter gene assay, coronary artery disease
1. Introduction
Lipoprotein lipase (LPL) plays a pivotal role in lipid metabolism by hydrolyzing triglyceride (TG) -rich lipoproteins. Many diseases, including coronary artery disease (CAD), atherosclerosis and obesity seem to be directly or indirectly related to abnormalities in LPL function [1–3]. A large number of variants have been identified in the LPL gene, of which 80% occur in coding regions while the rest are in noncoding regions [4–5]. Most of these functional LPL variants are rare, as they are mostly restricted to families with LPL deficiency or are found only in isolated geographic regions [3].
Several common LPL genetic variants have been widely studied, and among them HindIII in intron 8 (T→G at position 481), and S447X in exon 9 are of particular interest because of their common occurrence (25% and 9% of the less common alleles, respectively in most populations) and their associations with plasma lipid profile [6–11] and susceptibility to CAD [10–15] in several studies, although some inconsistent results have also been reported [16–18]. Since the HindIII polymorphism is located in the middle of intron 8, it is not considered to be functional but rather in linkage disequilibrium with a putative functional variant. Some groups, including ours, have attempted to find such a functional variant and identified coding SNPs, including D9N, N291S and S447X [6, 19–21]. Among these, only the S447X was common with a frequency of about 9% in Caucasians. The 447X allele results in a premature truncation of LPL by two amino acids and thus has been considered functional. However, the in vitro data on the effect of the S447X mutation on LPL function is equivocal. Compared to the wild type, the LPL activity associated with the X447 mutation was higher in one, lower in another, but comparable in three studies [reviewed in 22]. In association studies, although the X447 allele has been reported to be associated with a favorable lipid profile in some studies [7–8, 22] the results have not been confirmed in other studies [15]. Similarly, the association of the S447X polymorphism with CAD is inconsistent [10–18]. In a meta analysis the association of the S447X polymorphism with TG/HDL-C and CAD was found to be small compared to three other LPL amino acid polymorphisms examined [23] or it was gender-specific [24]. Most likely, the S447X polymorphism has some direct effect on plasma TG and HDL-C, but this does not explain all the effect associated with the HindIII polymorphism.
Because of these findings, a logical next step is to postulate that either the HindIII polymorphism is functional by itself or the regulatory elements in the 5’ or 3’ regions contain functional mutations for which HindIII acts as a marker, a notion which has been favored by several other investigators as well [9–10, 20, 23]. The rationale that HindIII could be functional is supported by an early study suggesting the presence of regulatory elements important for LPL expression in introns 8 or 9 of the LPL gene [25]. The goal of this study was to test the hypothesis that the HindIII polymorphic site is functional.
2. Materials and Methods
2.1. Plasmid DNA Constructs for Luciferase Assay
Initially, the entire intron 8 was sequenced from 14 homozygous subjects for the HindIII wild type allele (TT genotype). One sample that carried no other sequence variation was chosen for subcloning and site-directed mutagenesis. A DNA fragment of 1,180 bp containing the entire LPL intron 8 fragment (1,029 bp) plus 151 bp flanking sequence harboring the wild type (T) HindIII site was amplified using intron 8 PCR primers containing the XbaI restriction site: Forward primer (LPLXBAI-F 5’-3’) TCTAGACTTTAGCTGGTCAGACTGGTG; Reverse primer: (LPLXBAI-R) AGATCTTTCACAAATACCGCAGGTG. The wild type PCR product was then ligated to the pCR II-TOPO cloning vector (Invitrogen, Carlsbad, CA, USA) using the supplier's standard procedure. The full-length LPL promoter (1,537 bp) was inserted into the BamHI restriction site upstream of the intron 8 insert. The size and orientation of the DNA insert and fidelity of DNA polymerase was confirmed by restriction analysis and DNA sequencing. Site-directed mutagenesis of this wild type clone was performed using Stratagene’s QuikChange® Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA) to create the mutant type HindIII/G site as per the manufacturer’s protocol. The fragments containing 5' LPL promoter and intron 8 insertion of either wild (T) or mutant (G) HindIII polymorphic site were then inserted upstream of the firefly Luc reporter gene into the promoter-less pGL3-basic vector (Promega, Madison, WI, USA) using appropriate restriction enzymes followed by ligation with T4DNA ligase (New England Biolabs, Inc., Beverly, MA, USA), as described elsewhere [26]. Positive clones with full-length insertions of both promoter and intron 8 sequences were confirmed by restriction analysis. Identification of wild (T) and mutant (G) HindIII alleles in the intron 8 region was confirmed by both restriction analysis and DNA sequencing.
2.2. Transient Transfection and Dual-Luciferase Assay
The wild (T) and mutant (G) type chimeric-firefly Luc constructs (4µg) were used to transiently transfect human vascular smooth muscle (Lonza [Cambrex], Walkersville, MD) and COS-1 cells along with an internal control vector, Renilla Luciferase (Rluc) vector (pRL-CMV) to assess their ability to drive transcription of the luciferase gene using LipofectAMINE (Gibco/BRL). Human vascular smooth muscle cell line was maintained in SmGM-2 smooth muscle media with 5% fetal bovine serum. COS-1 cells were maintained in DMEM with 10% fetal bovine serum, 2mM glutamine, 100µ/ml penicillin and 100 mg/ml streptomycin at 37 °C under 5% CO2. After 48 hours of transfection, cells were washed twice with PBS and collected for the measurement of luciferase activity to determine the level of transcription. The transfected cells were lysed in lysis buffer (Promega, Madison, WI, USA) and the lysates used for the measurement of luciferase activity using Dual-luciferase Reporter (DLR) assay system (Promega, Madison, WI, USA) by TD 20/20 luminometer (Turner Design, Sunnyvale, CA, USA), as described elsewhere [26]. We co-transfected the LPL-reporter gene constructs with Rluc vector, which served as internal control to normalize difference in transfection efficiencies, while pGL3-basic vector with no insert was served as a negative control. Actual Luc activity was calculated as the ratio of firefly to Renilla Luc activity for each experiment.
2.3. Electrophoretic Mobility Shift Assay (EMSA)
Two double-stranded 30-mer oligonucleotides (wild type—CTA TAG GAT TTA AAG CtT TTA TAC TAA ATG; mutant—CTA TAG GAT TTA AAG CgT TTA TAC TAA ATG) corresponding to each variant (wild type or mutant) were prepared (Operon, Alameda, CA). Non-radioactive and radioactive EMSAs were used. For non-radioactive assay, 1.75 pmol of each oligo or commercially available nuclear factor consensus oligos (TFIID, AP2, AP1, SP1, OCT1 and CREB) were incubated with 2 µl (~20 ng) TATA binding protein (TBP) (Promega or Santa Cruz Biotechnologies) in 2 µl TBP 4x buffer [40% glycerol, 80 mM Tris-HCl (pH 8.0), 320 mM KCl, 40 mM MgCl2, 8 mM DTT], and 6 µl ddH2O at room temperature for 15 minutes. 1 µl of 10x loading buffer [250 mM Tris-HCl, pH 7.5, 40% glycerol, 0.2% bromophenol blue] was added to the reaction and the mixture was loaded on 6% polyacrylamide retardation gel (Novax). The gel was run at 25 °C constant temperature in 0.5x TBE buffer with constant voltage of 250V for 15 minutes. The gel was then stained with a 1:10000 diluted SYBR Gold (BioProbes) for 10 minutes and photographed. For the radioactive competitor assay, the wild type oligonucleotide was 5-end-labeled with γ-32P ATP and purified using the QIAquick Purification Kit (QIagen, Valencia, CA). Same concentrated, non-radioactive competitor DNA (wild type or mutant oligos) were added 1x, 3x, 5x, 10x, 20x, and 50x; excess volumes of the labelled probe. The mixture of unlabelled and labelled oligos were incubated with 2 µg aorta smooth muscle cell nuclear extracts (Geneka, Toronto, Canada) for 20 minutes at room temperature in biding buffer [1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50mM NaCl, 10 MM Tris-HCl (pH=7.5), 20% glycerol]. The protein complexes were then separated on 6% non-denaturing polyacrylamide gel at 120 volts for 2 hours, and the gel was dried and autoradiaographed overnight.
2.4. Statistical Analysis
A matrix-based algorithm [27] was used to identify putative regulating elements in intron 8 of the LPL gene. The luciferase activity of each construct was calculated as a mean ± SD value of three experiments in triplicate after adjusting the transfection efficiency by normalizing them with the Rluc control value. Differential luciferase expression between the wild type and mutant type was determined using Student’s T-test.
3. Results
3.1. Identification of Regulatory Elements in Intron 8
We performed a computer-based analysis to identify putative regulatory elements in intron 8 of the LPL gene using a matrix-based algorithm. Intron 8 harbors seven putative regulatory elements, including the consensus sequence of TATA, which is affected by the HindIII polymorphism (Table 1). The presence of consensus sequences for well known vertebrate-encoded transcription factors in intron 8 (Sp1, GATA, C/EBP, and TATA) indicates that intron 8 may participate in the LPL transcription.
Table 1.
Putative Regulatory Elements in Intron 8 of the Human LPL Gene.
| Element | Consensus Sequence | Nucleotide Start Position of the Element* |
|---|---|---|
| Lmo2 | CAGG | 62* |
| Sp1 | GGCG | 196 |
| MZF-1 | GGGG | 204, 664 |
| GATA | GATA | 372, 770 |
| TATA | TAAA | 481 (HindIII site), 489 |
| C/EBPβ | GCAA | 526 |
| Ap-1 | TGAC | 600 |
The number of consensus nucleotide from the start of intron 8 sequence.
3.2. Effect of the HindIII Polymorphism on Reporter Gene Expression
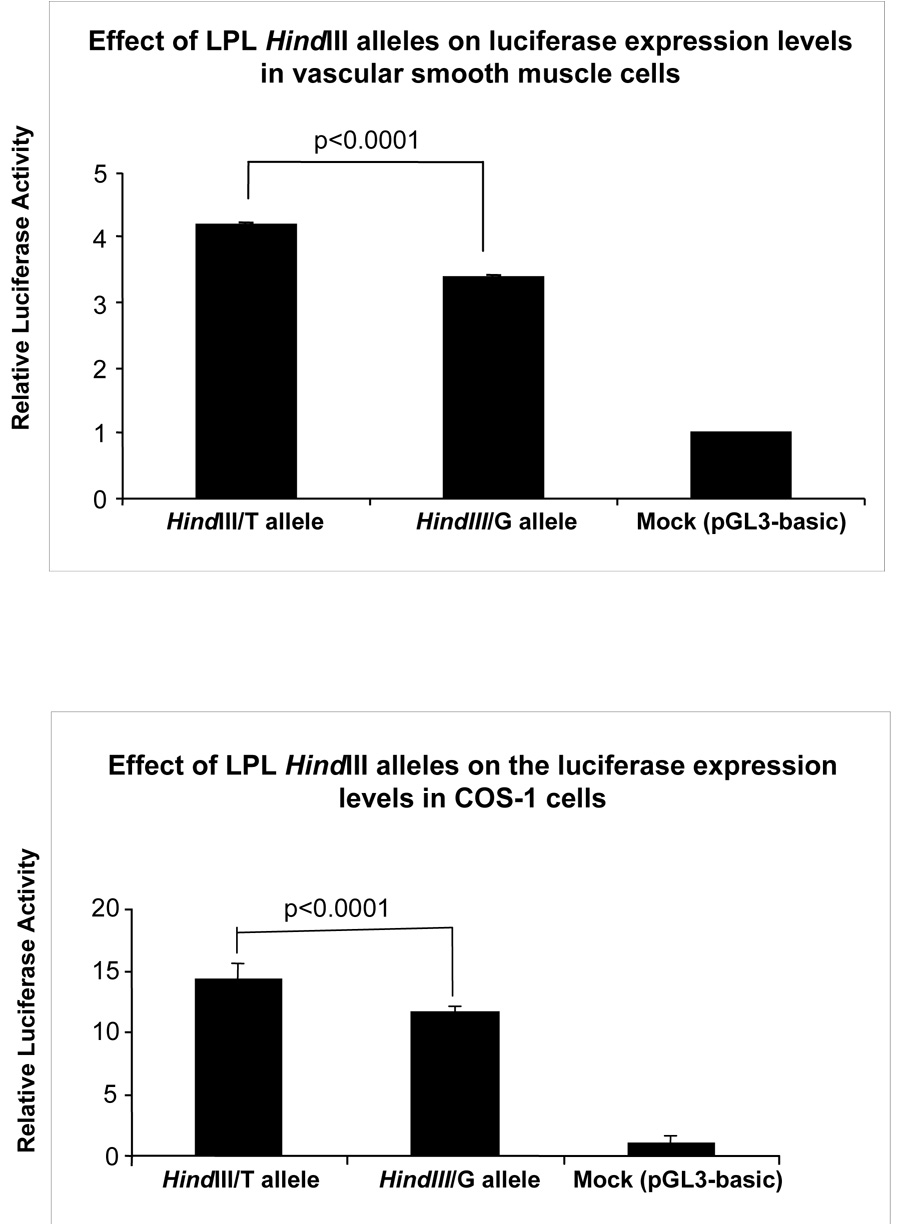
Wild (T) and mutant (G) type constructs were tested for their effects on promoter activity by cotransfecting COS-1 and human vascular smooth muscle cells along with the control vector (pR1-CMV), which was used to adjust the transfection efficiency within different sets and experiments. The smooth muscle cells were used for cell-specific expression, while COS-1 cells were used as a control cell line for comparison purpose. The promoter activity of each vector was determined by the dual luciferase assay system that revealed about 23% lower promoter activity associated with the mutant type allele (G) as compared to the wild type allele (T) in both vascular smooth muscle cells (3.394 ± 0.022 vs. 4.184 ± 0.028; P=4.7 × 10−6) (Figure 1a) and COS-1 cells (11.603 ± 0.409 vs. 14.373 ± 1.096; P<0.0001) (Figure 1b).
Figure 1.
Effect of the HindIII mutation on reporter gene expression in vascular smooth muscle cells and COS-1 cells. The effect of HindIII mutation on promoter activities was measured as the mean of the firefly Luc levels, which was adjusted with the Renilla Luc levels. The results presented are from three independent clones for each construct in triplicate, and each error bar represents the standard error.
3.3. Electrophorectic Mobility Shift Assay (EMSA)
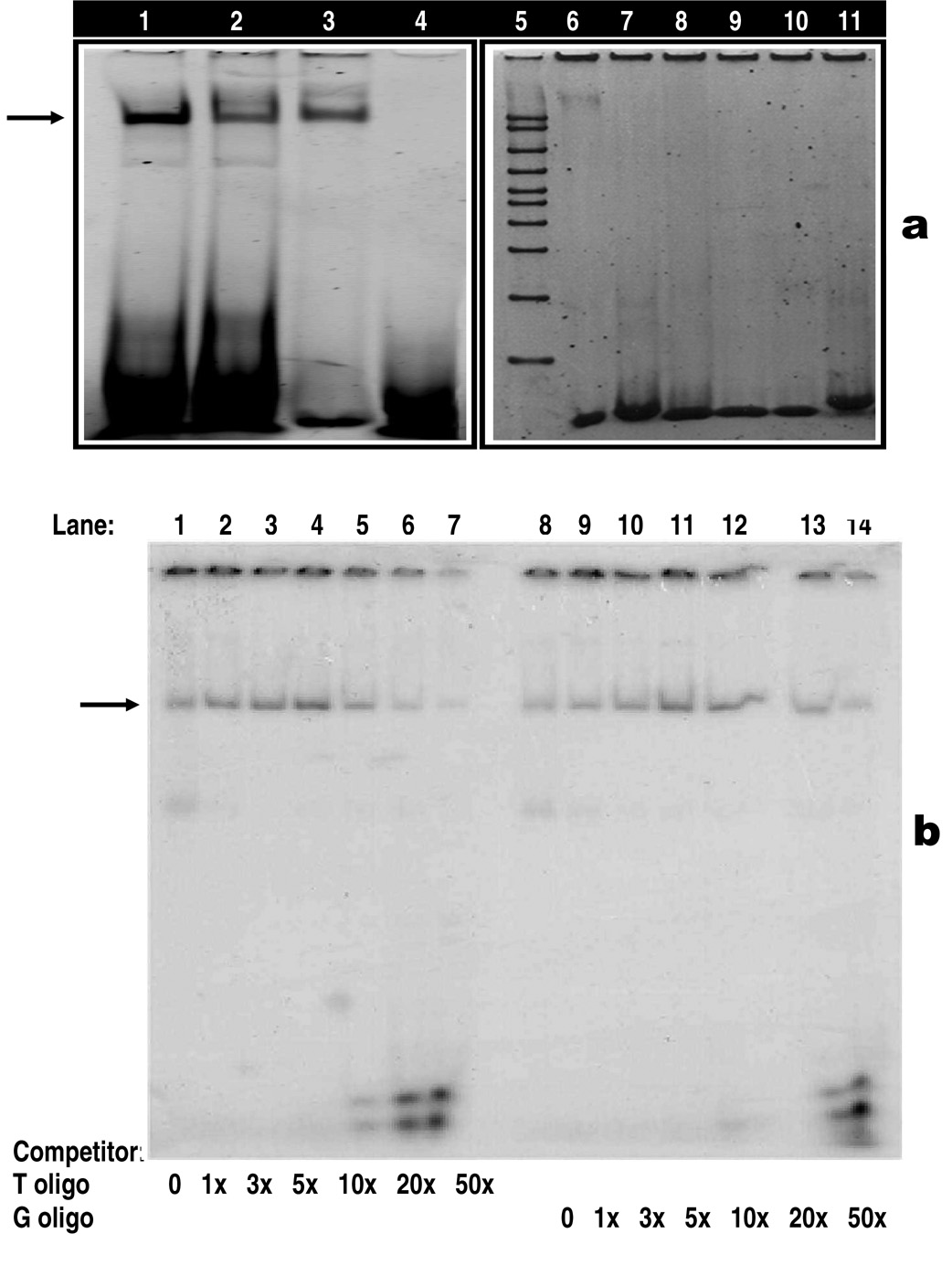
To confirm our hypothesis that the HindIII site in intron 8 is involved in binding with a transcription factor, we performed both non-radioactive and radioactive competitor EMSAs. We postulated that since the HindIII site is part of a putative TATA box, it will likely have a binding with TATA binding protein (TBP). Our non-radioactive assay showed that a subunit of TFIID or TBP that recognizes TFIID consensus sequence (lanes 3 and 6 on Figure 2a), is bound to the HindIII wild type and mutant type oligos. The results also showed that the TBP has higher affinity for the HindIII wild type (T oligo, lane 1) than the mutant type (G oligo, lane 2) (Figure 2a). The binding between the HindIII oligos and TBP was specific as TBP did not bind to consensus sequences for other nuclear factors such as AP2, SP1, AP1, OCT1 and CREB (lanes 4, 7 to 11).
Figure 2.
a: Non-radioactive electrophoretic mobility shift assay (EMSA). Arrow head indicates the specific DNA-protein binding (lanes 1–3, 6) or no DNA-protein binding (lanes 7–11). Binding of HindIII wild type (lane 1) and mutant type (lane 2) with purified human TATA binding protein (TBP). Lanes 1 and 2 contain identical amounts of HindIII oligos and TBP, but the affinity of TBP is higher for the HindIII wild type (lane 1) than the mutant type (lane 2). Lanes 3 and 6 are positive controls showing the binding of TBP with TFIID consensus oligo. Purified TBP used in Lane 3 was obtained from Promega, whereas purified TBP used in lane 6 comes from Santa Cruz Biotechnologies and this may explain the difference in the intensity between the two bands. Lanes 4, 7–11 are negative controls showing no binding of TBP with consensus oligos, including AP2 (lanes 4 and 7), SP1 (lane 8), AP1 (lane 9), OCT1 (lane 10) and CREB (lane 11). Lane 5 contains 50 – 2000 bp marker.
b: Radioactive competitor EMSA for LPL/HindIII T→G polymorphism. Each sample contains a mixture of 5 µg of nuclear extract derived from aorta smooth muscle cells and a 30-mer 32P-labeled LPL/HindIII oligonucleotide containing the T allele. The arrowhead indicates the specific DNA-protein complex associated with the LPL/HindIII T→G polymorphic site. Competition assay performed by adding excess cold oligonucleotides containing either the T allele (lanes 2–7) or the G allele (lanes 9–14). Lanes 1 and 8: no competitor, lanes 2–7 have increasing amounts of T oligo competitor (1x, 3x, 5x, 10x, 20x and 50x, respectively); lanes 9–14 have increasing amounts of G oligo competitors (1x, 3x, 5x, 10x, 20x and 50x, respectively).
In order to further distinguish the binding affinity between the T and G alleles, we performed competition mobility shift assay by using the non-labelled oligos (both wild type and mutant) to compete with radioactive-labelled oligo. As LPL is abundantly expressed in smooth muscles, we used aorta smooth muscle cell nuclear extract in this experiment. The results show that the HindIII/T or G oligonucleotides formed a DNA-protein complex with aorta smooth muscle cell nuclear extract (Figure 2b). By using increasing amounts of unlabeled competitor oligos (T or G allele) to compete with the labelled T oligo, we found that the T allele had a higher binding affinity compared with the G allele (Figure 2b, DNA-protein complex are less visible in lanes 5–7 compared to the ones in lanes 12–14). Identical allele-dependent differences in binding were observed on repeating EMSA three times. The EMSA data indicate that the HindIII sequence binds regulatory protein and that the wild type T allele has a higher affinity for binding compared with the mutant G allele.
4. Discussion
The objective of this study was to test the hypothesis that the HindIII polymorphic site in intron 8 of the LPL gene is functional. Our computer-based analysis on the presence of regulatory elements in intron 8 as well as functional studies of EMSA and reporter gene assays, strongly suggest that the HindIII polymorphic site is functional. We identified 7 putative regulatory elements in intron 8, including the consensus sequence of TATA that is affected by the HindIII polymorphism. To confirm our hypothesis that the HindIII site in intron 8 is involved in binding with a transcription factor, we constructed wild type and mutant type oligos (T→G) and examined their binding with purified human TATA binding protein (TBP or TFIID). We postulated that since the HindIII site is part of putative TATA box, it will more likely bind with TBP. The TBP not only bound with the HindIII wild type (T) and mutant type (G) oligos, there was a differential binding with the two alleles such that the binding affinity for TBP was higher with the wild type allele than the mutant type. Furthermore, the binding between the HindIII oligos and TBP was specific, as TBP did not bind to consensus sequences for other nuclear factors (see Fig. 2; lanes 4, 7–11). Our competition mobility shift assays with the two HindIII oligos in aorta smooth muscle cell nuclear extracts also confirmed the finding that the HindIII sequence binds regulatory factors (most likely TBP) and that this binding is affected by the HindIII sequence variation such that the mutant type allele (G) has reduced binding compared to the wild type allele. Finding a functional TATA box in the middle of an intron is unusual and to our knowledge has not been reported in the literature before. We suspect that this motif somehow contributes to the regulation of the LPL gene. Additional molecular work is necessary to work out the exact function of this intronic motif.
Further evidence that the HindIII polymorphic site is functional comes from our luciferase reporter gene assay performed in human smooth muscle cells and COS-1 cells. In both cell types, the mutant G allele was associated with a modest 20% lower transcription activity than the wild type T allele. This modest effect (20% difference in promoter activity) is predicted from a common variant and is similar to the effect size of 18–24% difference in promote activity reported earlier for a common LPL promoter variant (−93 T/G) [28]. These data strongly suggest that human LPL promoter controls LPL expression in conjunction with regulatory elements present in intron 8 and confirm the original observation made by Enerbäck et al. [25]. Both positive and negative regulatory elements have been identified not only in the 5’ region [29], but also in the intragenic and 3’ regions [25] of the LPL gene. Previato et al. [29] localized a single positive element (−368 to −35) and a negative element (−724 to −565) in the 5’ region by assessing the LPL transcription in 3T3-L1 adipocyte or HepG2 cells. Using 3T3-F442A preadipocyte cells, Enerbäck et al. [25] identified two major positive elements at −430 to −400 and −196 to −168, and two major negative elements at −4 kb to −827 and −239 to −196. The construct containing the −4 kb to −827 deletion was associated with an 8-fold drop in promoter activity. Using DNaseI hypersensitivity assay, which detects sites of altered chromatin structure that correlate with transcriptional regulation, Enerbäck et al. [25] identified two enhancer regions present in introns 8 or 9 and in the 3’ UTR of the LPL gene, which counteracted the negative effect of the −4 kb to −827 construct. The identification of regulatory elements in the LPL intronic regions is analogous to that described for several other genes which harbor regulatory elements in their introns [30–35].
The lower transcription activity and lower binding of TBP associated with the HindIII mutant G allele may appear counterintuitive because this allele is associated with protection from CAD risk and/or with favorable lipid profile possibly due to higher LPL activity. However, we do not have the LPL activity data available in this study. Previously one study has shown a non-significantly higher LPL activity among HindIII mutant carriers (n=15) than the homozygotes of the wild type (n=16) in postheparin plasma (9.21 ± 4.6 vs. 8.11 ± 4.16) [9]. However, the postheparin plasma LPL activity studies may not fully reveal the impact of the HindIII polymorphism on LPL activity as exemplified by the inconsistent findings with the S447X polymorphism. To date eight studies have analyzed the impact of the S447X polymorphism on postheparin LPL activity and only three studies found significant association [22, 36–37] but the other five did not [15, 38–41]. Even one study that found a significant association with a haplotype carrying the 447X mutant allele suggested that other variants in the 3’UTR of LPL are responsible for the increase in LPL activity rather than the S447X variant [37]. Thus the inclusion of the LPL activity data in this study might have not provided a conclusive answer about the functional impact of the HindIII polymorphism on LPL activity because the postheparin plasma reflects the total LPL activity and not tissue-specific activity. Likewise the in vitro measurement of LPL catalytic activity or mass may also be inconclusive as reflected in equivocal findings with the S447X polymorphism [22].
A possibility is that the effect of the HindIII polymorphism on LPL expression is influenced by other tightly linked functional variants that also affect LPL expression. In addition to the presence of potential transcriptional regulatory elements in the 3’ region, translational regulatory elements in the 3’ region of the LPL gene have also been identified [42–44]. A recent study suggests that regulatory polymorphisms are complex and sensitive to modifiers with multiple trans-acting or environmental factors affecting their function [45]. Multiple cis-acting sites may affect the contribution of a variant to the expression of a gene and if they are not examined together then in vitro experiments would not represent the true in vivo effect of a variant. If the HindIII site indeed increases LPL activity by increasing LPL expression in conjunction with other unknown cis-acting regulatory sequences in or around the LPL gene then they need to be examined together. Our constructs contained only the LPL promoter and the entire intron 8 sequence with a caveat that additional unknown cis-acting regulatory sequences may be required to understand the full spectrum of the LPL expression. Furthermore, LPL gene expression is sensitive to hormonal regulation of insulin. For example, it has been shown that LPL mRNA increase significantly in brown adipose tissue after insulin injection to fasting rats [46]. On the other hand, LPL activity is reduced under insulin resistance state because the ability of insulin to up-regulate LPL gene is impaired in insulin resistant individuals, leading to hypertriglyceridemia [47]. It is conceivable that insulin differentially affects the allele-specific effect of the HindIII polymorphism in individuals with different state of insulin sensitivity. Notwithstanding its true in vivo effect, our consistent in vitro results in two different cell systems (muscle and COS-1) along with EMSA results provide compelling evidence that the HindIII polymorphic site is functional.
In conclusion, we provide new information about the functional significance of the HindIII polymorphism and our data in conjunction with previous genetic association data reinforce the concept that this is a common functional variant with a modest effect for a common trait.
Acknowledgement
The study was supported in part by the National Heart, Lung and Blood Institute (NHLBI) grant HL 070169. We thank Mr. Wei-jun Xu, and Ms. Sangita Suresh for their excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goldberg IJ. Lipoprotein lipase and lipolysis: central roles in lipoprotein metabolism and atherogenesis. J Lipid Res. 1996;37:693–707. [PubMed] [Google Scholar]
- 2.Mead JR, Cryer A, Ramji DP. Lipoprotein lipase, a key role in atherosclerosis? FEBS Lett. 1999;462:1–6. doi: 10.1016/s0014-5793(99)01495-7. [DOI] [PubMed] [Google Scholar]
- 3.Brunzell JD, Deeb SS. Familial lipoprotein lipase deficiency, apo C-II deficiency, and hepatic lipase deficiency. In: Scriver CR, Beundet AL, Sly WS, Valle D, editors. The Metabolic & Molecular Bases of Inherited Disease. 8th Ed. New York: McGraw-Hill; 2001. pp. 2789–2816. [Google Scholar]
- 4.Murthy V, Julien P, Gagne C. Molecular pathobiology of the human lipoprotein lipase gene. Pharmacol Ther. 1996;70:101–135. doi: 10.1016/0163-7258(96)00005-8. [DOI] [PubMed] [Google Scholar]
- 5.Lalouel J-M, Wilson DE, Iverius P-H. Lipoprotein Lipase and hepatic triglyceride lipase: molecular and genetic aspects. Curr Opin Lipidol. 1992;3:86–95. [Google Scholar]
- 6.Razzaghi H, Aston CE, Hamman RF, Kamboh MI. Genetic screening of the lipoprotein lipase gene for mutations associated with high triglyceride/low HDL-cholesterol levels. Hum Genet. 2000;107:257–267. doi: 10.1007/s004390000367. [DOI] [PubMed] [Google Scholar]
- 7.Kuivenhoven JA, Groenemeyer BE, Boer JM, Reymer PW, Berghuis R, Bruin T, Jansen H, Seidell JC, Kastelein JJ. Ser447stop mutation in lipoprotein lipase is associated with elevated HDL cholesterol levels in normolipidemic males. Arterioscler Thromb Vasc Biol. 1997;17:595–599. doi: 10.1161/01.atv.17.3.595. [DOI] [PubMed] [Google Scholar]
- 8.Morabia A, Cayanis E, Costanza MC, Ross BM, Bernstein MS, Flaherty MS, Alvin GB, Das K, Morris MA, Penchaszadeh GK, Zhang P, Gilliam TC. Association between lipoprotein lipase (LPL) gene and blood lipids: a common variant for a common trait? Genet Epidemiol. 2003;24:309–321. doi: 10.1002/gepi.10229. [DOI] [PubMed] [Google Scholar]
- 9.Vohl MC, Lamarche B, Moorjani S, Prud'homme D, Nadeau A, Bouchard C, Lupien PJ, Despres JP. The lipoprotein lipase HindIII polymorphism modulates plasma triglyceride levels in visceral obesity. Arterioscler Thromb Vasc Biol. 1995;15:714–720. doi: 10.1161/01.atv.15.5.714. [DOI] [PubMed] [Google Scholar]
- 10.Mattu RK, Needham EW, Morgan R, Rees A, Hackshaw AK, Stocks J, Elwood PC, Galton DJ. DNA variants at the LPL gene locus associate with angiographically defined severity of atherosclerosis and serum lipoprotein levels in a Welsh population. Arterioscler Thromb. 1994;14:1090–1097. doi: 10.1161/01.atv.14.7.1090. [DOI] [PubMed] [Google Scholar]
- 11.Gerdes C, Gerdes LU, Hansen PS, Faergeman O. Polymorphisms in the lipoprotein lipase gene and their associations with plasma lipid concentrations in 40-year-old Danish men. Circulation. 1995;92:1765–1769. doi: 10.1161/01.cir.92.7.1765. [DOI] [PubMed] [Google Scholar]
- 12.Wittrup HH, Tybjaerg-Hansen A, Nordestgaard BG. Lipoprotein lipase mutations, plasma lipids and lipoproteins, and risk of ischemic heart disease. A meta-analysis. Circulation. 1999;99:2901–2907. doi: 10.1161/01.cir.99.22.2901. [DOI] [PubMed] [Google Scholar]
- 13.Hokanson JE. Functional variants in the lipoprotein lipase gene and risk cardiovascular disease. Curr Opin Lipidol. 1999;10:393–399. doi: 10.1097/00041433-199910000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Taylor KD, Scheuner MT, Yang H, Wang Y, Haritunians T, Fischel-Ghodsian N, Shah PK, Forrester JS, Knatterud G, Rotter JI. Lipoprotein lipase locus and progression of atherosclerosis in coronary-artery bypass grafts. Genet Med. 2004;6:481–486. doi: 10.1097/01.gim.0000144012.18935.48. [DOI] [PubMed] [Google Scholar]
- 15.Peacock RE, Hamsten A, Nilsson-Ehle P, Humphries SE. Associations between lipoprotein lipase gene polymorphisms and plasma correlations of lipids, lipoproteins and lipase activities in young myocardial infarction survivors and age-matched healthy individuals from Sweden. Atherosclerosis. 1992;97:171–185. doi: 10.1016/0021-9150(92)90130-9. [DOI] [PubMed] [Google Scholar]
- 16.Fidani L, Hatzitolios AI, Goulas A, Savopoulos C, Basayannis C, Kotsis A. Cholesteryl ester transfer protein TaqI B and lipoprotein lipase Ser447Ter gene polymorphisms are not associated with ischaemic stroke in Greek patients. Neurosci Lett. 2005;384:102–105. doi: 10.1016/j.neulet.2005.04.061. [DOI] [PubMed] [Google Scholar]
- 17.Spence JD, Ban MR, Hegele RA. Lipoprotein lipase (LPL) gene variation and progression of carotid artery plaque. Stroke. 2003;34:1176–1180. doi: 10.1161/01.STR.0000069160.05292.41. [DOI] [PubMed] [Google Scholar]
- 18.Brousseau ME, Goldkamp AL, Collins D, Demissie S, Connolly AC, Cupples LA, Ordovas JM, Bloomfield HE, Robins SJ, Schaefer EJ. Polymorphisms in the gene encoding lipoprotein lipase in men with low HDL-C and coronary heart disease: the Veterans Affairs HDL Intervention Trial. J Lipid Res. 2004;45:1885–1891. doi: 10.1194/jlr.M400152-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Mailly F, Tugrul Y, Reymer PW, Bruin T, Seed M, Groenemeyer BF, Asplund-Carlson A, Vallance D, Winder AF, Miller GJ, Kastelein JJP, Hamsten A, Olivecrona G, Humphries SE, Talmud PJ. A common variant in the gene for lipoprotein lipase (Asp9→Asn). Functional implications and prevalence in normal and hyperlipidemic subjects. Arterioscler Thromb Vasc Biol. 1995;15:468–478. doi: 10.1161/01.atv.15.4.468. [DOI] [PubMed] [Google Scholar]
- 20.Fisher RM, Humphries SE, Talmud PJ. Common variation in the lipoprotein lipase gene: effects on plasma lipids and risk of atherosclerosis. Atherosclerosis. 1997;135:145–159. doi: 10.1016/s0021-9150(97)00199-8. [DOI] [PubMed] [Google Scholar]
- 21.Hata A, Robertson M, Emi M, Lalouel JM. Direct detection and automated sequencing of individual alleles after electrophoretic strand separation: identification of a common nonsense mutation in exon 9 of the human lipoprotein lipase gene. Nucleic Acids Res. 1990;18:5407–5411. doi: 10.1093/nar/18.18.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rip J, Nierman MC, Ross CJ, Jukema JW, Hayden MR, Kastelein JJ, Stroes ES, Kuivenhoven JA. Lipoprotein lipase S447X: a naturally occurring gain-of-function mutation. Arterioscler Thromb Vasc Biol. 2006;26:1236–1245. doi: 10.1161/01.ATV.0000219283.10832.43. [DOI] [PubMed] [Google Scholar]
- 23.Wittrup HH, Tybjaerg-Hansen A, Nordestgaard BG. Lipoprotein lipase mutations, plasma lipids and lipoproteins, and risk of ischemic heart disease. A meta-analysis. Circulation. 1999;99:2901–2907. doi: 10.1161/01.cir.99.22.2901. [DOI] [PubMed] [Google Scholar]
- 24.Wittrup HH, Nordestgaard BG, Steffensen R, Jensen G, Tybjaerg-Hansen A. Effect of gender on phenotypic expression of the S447X mutation in LPL: the Copenhagen City Heart Study. Atherosclerosis. 2002;165:119–126. doi: 10.1016/s0021-9150(02)00183-1. [DOI] [PubMed] [Google Scholar]
- 25.Enerbäck S, Ohlsson BG, Samuelsson L, Bjursell G. Characterization of the human lipoprotein lipase (LPL) promoter: evidence of two cis-regulatory regions, LP-alpha and LP-beta, of importance for the differentiation-linked induction of the LPL gene during adipogenesis. Mol Cell Biol. 1992;12:4622–4633. doi: 10.1128/mcb.12.10.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luedecking EK, DeKosky ST, Mehdi H, Ganguli M, Kamboh MI. Analysis of genetic polymorphisms in the transforming growth factor-beta1 gene and the risk of Alzheimer's disease. Hum Genet. 2000;106:565–569. doi: 10.1007/s004390000313. [DOI] [PubMed] [Google Scholar]
- 27.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall S, Chu G, Miller G, Cruickshank K, Cooper JA, Humphries SE, Talmud PJ. A common mutation in the lipoprotein lipase gene promoter, -93T/G, is associated with lower plasma triglyceride levels and increased promoter activity in vitro. Arterioscler Thromb Vasc Biol. 1997;17:1969–1976. doi: 10.1161/01.atv.17.10.1969. [DOI] [PubMed] [Google Scholar]
- 29.Previato L, Parrott CL, Santamarina-Fojo S, Brewer HB., Jr Transcriptional regulation of the human lipoprotein lipase gene in 3T3-L1adipocytes. J Biol Chem. 1991;266:18958–18963. [PubMed] [Google Scholar]
- 30.Aronow B, Lattier D, Silbiger R, Dusing M, Hutton J, Jones G, Stock J, McNeish J, Potter S, Witte D, et al. Evidence for a complex regulatory array in the first intron of the human adenosine deaminase gene. Genes Dev. 1989;3:1384–13400. doi: 10.1101/gad.3.9.1384. [DOI] [PubMed] [Google Scholar]
- 31.Celis L, Claessens F, Peeters B, Heyns W, Verhoeven G, Rombauts W. Proteins interacting with an androgen-responsive unit in the C3(1) gene intron. Mol Cell Endocrinol. 1993;94:165–172. doi: 10.1016/0303-7207(93)90165-g. [DOI] [PubMed] [Google Scholar]
- 32.Henkel G, Brown MA. PU.1 and GATA: components of a mast cell-specific interleukin 4 intronic enhancer. Proc Natl Acad Sci U S A. 1994;91:7737–7741. doi: 10.1073/pnas.91.16.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang LG, Liu XM, Li ZR, Denstman S, Bloch A. Differential binding of nuclear c-ets-1 protein to an intron I fragment of the c-myb gene in growth versus differentiation. Cell Growth Differ. 1994;5:1243–1251. [PubMed] [Google Scholar]
- 34.van Dijk TB, Caldenhoven E, Raaijmakers JA, Lammers JW, Koenderman L, de Groot RP. The role of transcription factor PU.1 in the activity of the intronic enhancer of the eosinophil-derived neurotoxin (RNS2) gene. Blood. 1998;91:2126–2132. [PubMed] [Google Scholar]
- 35.Yajima I, Sato S, Kimura T, Yasumoto K, Shibahara S, Goding CR, Yamamoto H. An L1 element intronic insertion in the black-eyed white (Mitf[mi-bw]) gene: the loss of a single Mitf isoform responsible for the pigmentary defect and inner ear deafness. Hum Mol Genet. 1999;8:1431–1441. doi: 10.1093/hmg/8.8.1431. [DOI] [PubMed] [Google Scholar]
- 36.Henderson HE, Kastelein JJ, Zwinderman AH, Gagne E, Jukema JW, Reymer PW, Groenemeyer BE, Lie KI, Bruschke AV, Hayden MR, Jansen H. Lipoprotein lipase activity is decreased in a large cohort of patients with coronary artery disease and is associated with changes in lipids and lipoproteins. J Lipid Res. 1999;40:735–743. [PubMed] [Google Scholar]
- 37.Goodarzi MO, Wong H, Quinones MJ, Taylor KD, Guo X, Castellani LW, Antoine HJ, Yang H, Hsueh WA, Rotter JI. The 3' untranslated region of the lipoprotein lipase gene: haplotype structure and association with post-heparin plasma lipase activity. J Clin Endocrinol Metab. 2005;90:4816–4823. doi: 10.1210/jc.2005-0389. [DOI] [PubMed] [Google Scholar]
- 38.Knudsen P, Murtomaki S, Antikainen M, Ehnholm S, Lahdenpera S, Ehnholm C, Taskinen MR. The Asn-291→Ser and Ser-477→Stop mutations of the lipoprotein lipase gene and their significance for lipid metabolism in patients with hypertriglyceridaemia. Eur J Clin Invest. 1997;27:928–935. doi: 10.1046/j.1365-2362.1997.2190767.x. [DOI] [PubMed] [Google Scholar]
- 39.Garenc C, Perusse L, Gagnon J, Chagnon YC, Bergeron J, Despres JP, Province MA, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. Linkage and association studies of the lipoprotein lipase gene with postheparin plasma lipase activities, body fat, and plasma lipid and lipoprotein concentrations: the HERITAGE Family Study. Metabolism. 2000;49:432–439. doi: 10.1016/s0026-0495(00)80004-9. [DOI] [PubMed] [Google Scholar]
- 40.Nierman MC, Rip J, Kuivenhoven JA, van Raalte DH, Hutten BA, Sakai N, Kastelein JJ, Stroes ES. Carriers of the frequent lipoprotein lipase S447X variant exhibit enhanced postprandial apoprotein B-48 clearance. Metabolism. 2005;54:1499–1503. doi: 10.1016/j.metabol.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 41.Nierman MC, Prinsen BH, Rip J, Veldman RJ, Kuivenhoven JA, Kastelein JJ, de Sain-van der Velden MG, Stroes ES. Enhanced Conversion of Triglyceride-Rich Lipoproteins and Increased Low-Density Lipoprotein Removal in LPLS447X Carriers. Arterioscler Thromb Vasc Biol. 2005;25:2410–2415. doi: 10.1161/01.ATV.0000188506.79946.ce. [DOI] [PubMed] [Google Scholar]
- 42.Yukht A, Davis RC, Ong JM, Ranganathan G, Kern PA. Regulation of lipoprotein lipase translation by epinephrine in 3T3-L1 cells. Importance of the 3' untranslated region. J Clin Invest. 1995;96:2438–2444. doi: 10.1172/JCI118301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ranganathan G, Vu D, Kern PA. Translational regulation of lipoprotein lipase by epinephrine involves a trans-acting binding protein interacting with the 3' untranslated region. J Biol Chem. 1997;272:2515–2519. doi: 10.1074/jbc.272.4.2515. [DOI] [PubMed] [Google Scholar]
- 44.Ranganathan G, Li C, Kern PA. The translational regulation of lipoprotein lipase in diabetic rats involves the 3'-untranslated region of the lipoprotein lipase mRNA. J Biol Chem. 2000;275:40986–40991. doi: 10.1074/jbc.M008775200. [DOI] [PubMed] [Google Scholar]
- 45.Cirulli ET, Goldstein DB. in vitro assays fail to predict in vivo effects of regulatory polymorphisms. Hum Mol Genet. 2007;16:1931–1939. doi: 10.1093/hmg/ddm140. [DOI] [PubMed] [Google Scholar]
- 46.Mitchell JR, Jacobsson A, Kirchgessner TG, Schotz MC, Cannon B, Nedergaard J. Regulation of expression of the lipoprotein lipase gene in brown adipose tissue. Am J Physiol. 1992;263:E500–E506. doi: 10.1152/ajpendo.1992.263.3.E500. [DOI] [PubMed] [Google Scholar]
- 47.Maheux P, Azhar S, Kern PA, Chen YD, Reuven GM. Relationship between insulin-mediated glucose disposal and regulation of plasma and adipose tissue lipoprotein lipase. Diabetologia. 1997;40:850–858. doi: 10.1007/s001250050759. [DOI] [PubMed] [Google Scholar]