Abstract
Cancer/Testis (CT) genes, normally expressed in germ line cells but also activated in a wide range of cancer types, often encode antigens that are immunogenic in cancer patients, and present potential for use as biomarkers and targets for immunotherapy. Using multiple in silico gene expression analysis technologies, including twice the number of expressed sequence tags used in previous studies, we have performed a comprehensive genome-wide survey of expression for a set of 153 previously described CT genes in normal and cancer expression libraries. We find that although they are generally highly expressed in testis, these genes exhibit heterogeneous gene expression profiles, allowing their classification into testis-restricted (39), testis/brain-restricted (14), and a testis-selective (85) group of genes that show additional expression in somatic tissues. The chromosomal distribution of these genes confirmed the previously observed dominance of X chromosome location, with CT-X genes being significantly more testis-restricted than non-X CT. Applying this core classification in a genome-wide survey we identified >30 CT candidate genes; 3 of them, PEPP-2, OTOA, and AKAP4, were confirmed as testis-restricted or testis-selective using RT-PCR, with variable expression frequencies observed in a panel of cancer cell lines. Our classification provides an objective ranking for potential CT genes, which is useful in guiding further identification and characterization of these potentially important diagnostic and therapeutic targets.
Keywords: gene index, prediction
Cancer/Testis (C/T) genes are a heterogeneous group that are normally expressed predominantly in germ cells and in trophoblasts, and yet are aberrantly activated in up to 40% of various types of cancer types (1). A subset of the CT genes has been shown to encode antigens that are immunogenic and elicit humoral and cellular immune responses in cancer patients (2). Because of their restricted expression profile in normal tissues and because the testis is an immunoprivileged site, the CT antigens are emerging as strong candidates for therapeutic cancer vaccines, as revealed by early-phase clinical trials (3–10). Biologically, the CT genes provide a model to better understand complex gene regulation and aberrant gene activation during cancer.
Any gene that exhibits an mRNA expression profile restricted to the testis and neoplastic cells can be termed a CT gene. Existing definitions of CT genes vary in the literature, from genes expressed exclusively in adult testis germ cells and malignant tumors (1, 11) to dominant testicular expression (12), possible additional presence in placenta and ovary and epigenetic regulation (13), or membership of a gene family and localization on the X chromosome (14). Reflecting this lack of a consensus definition, an increasing number of heterogeneous CT candidates have appeared in the literature, with available expression profile information frequently limited to the original defining articles. In some cases, e.g., ACRBP, the original CT-restricted expression in normal tissues could not be confirmed by subsequent experiments (1). Partially due to this lack of a clear and broadly applicable definition, or “type specimen,” for a CT gene, it has become increasingly challenging to identify the CT genes that are most suitable for cancer vaccine development. Moreover, this incoherent classification increases the risk of pursuing unsuitable clinical targets. However, with more expression data becoming available, CT gene transcripts of genes originally thought to have the CT expression profile are being detected in additional tissues (1), resulting in the more stringent “testis-restricted” description being altered to one of “testis-preference.” Based on a compilation from the published literature, the CT database now lists >130 RefSeq nucleotide identifiers as CT genes that belong to 83 gene families (www.cta.lncc.br). An analysis of the human X chromosome has also suggested that as many as 10% of the genes on this chromosome may be CT genes (15). Given this increasing number of CT and CT-like genes, their comprehensive classification based on expression profiles is essential for our understanding of their biological role and regulation of expression.
In an attempt to resolve this and to identify new CT antigens, we have taken an in silico approach to produce a comprehensive survey of CT gene expression profiles by combining expression information from an existing corpus of >8,000 cDNA libraries (16) together with the depth and resolution provided by massively parallel signature sequencing (MPSS) expression libraries (17), cap-analysis of Gene Expression (CAGE) libraries (18), and a survey using semiquantitative reverse-transcription PCR (RT-PCR) on a panel of 22 normal tissues. As a result, we have created a coherent classification of CT genes, and new CT genes have been identified using well-informed, structured prediction and confirmation criteria.
Results and Discussion
CT classification.
CT genes were classified into 3 groups, testis-restricted, testis/brain-restricted and testis-selective, based on their expression profiles obtained from a manually curated corpus of cDNA, MPSS, CAGE expression libraries and RT-PCR (see Dataset S1 for MPSS and CAGE library annotation and http://evocontology.org for the cDNA annotation). By merging expression information using different technology platforms, we were able to leverage their individual strengths—the breadth of tissue coverage associated with the cDNA/EST expression libraries, the high sensitivity of CAGE/MPSS and the ability to custom-tailor PCR primers. Of 153 genes, 39 with transcripts present only in adult testis and no other normal adult tissue except for placenta were classified as testis-restricted; 14 CT genes with additional expression in other adult immuno-restricted sites (all regions of the brain) were classified as testis/brain-restricted, and 85 genes, designated as testis-selective, were ranked by the ratio of testis/placenta expression relative to other expression in normal adult tissues (see Fig. 1 for the expression array, Fig. 2 for the PCR panel of selected testis-restricted CT genes, and Fig. S1 and Dataset S2 for arrays from individual expression sources).
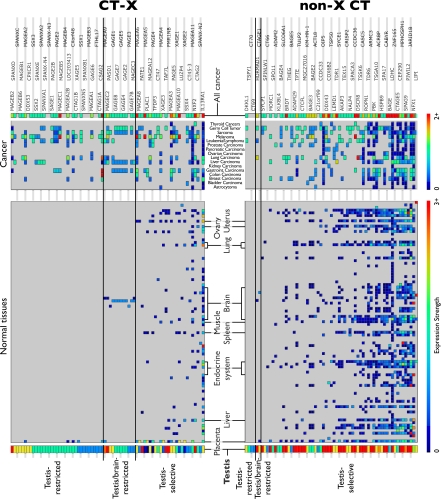
Fig. 1.
Merged expression profiles of CT-X (left array) and non-X CT genes (Right) based on expression data from RT-PCR and cDNA, MPSS and CAGE libraries from tissues sources annotated as normal and “adult” (Lower) or “cancer.” Expression in normal testis, placenta, and selected tissues is marked. Color reflects the support for the expression of a CT genes in a given anatomical site (blue for low combined expression evidence ≥1, red for strong support from at least 3 sources (for the normal tissue panel) with a total score ≥3) or 2 sources (the cancer panel lacking RT-PCR data), respectively. The most abundant expression (red) is seen in testis for most genes, particularly in the non-X CT group. Expression values were normalized on a per-gene basis relative to the combined normal testis/placenta expression confidence (Lower) or the source of the highest cancer expression confidence (Upper). The 3 CT annotation groups (testis-restricted, testis/brain-restricted and testis-selective) are highlighted. See Dataset S3 for the full list of CT classifications.
Fig. 2.
RT-PCR analysis of selected CT genes in the testis-restricted category (MAGEA1, GAGE, SSX2, NY-ESO-1, MAGEC1, and SPANX). Expression profile are shown for a range of 22 normal tissues (Left) and 31 cancer cell lines (Right).
An uneven chromosomal distribution of the CT genes was observed, with 83 of 153 genes (54%) being on the X chromosome, and 70 on non-X chromosomes (Fig. S2). Furthermore, 35 CT-X genes were classified as testis-restricted, whereas only 4 non-X CT genes belong to this group. An additional 12 CT-X genes were found to be testis/brain-restricted, compared with 2 non-X testis/brain-restricted CT genes. CT-X gene family members thus appear to be under more stringent transcriptional restriction in somatic tissues, whereas non-X CT genes are more broadly expressed. This validates the CT gene classification into CT-X and CT non-X groups, with the CT-X group being of particular interest for therapeutic approaches.
Twenty-six CT-X and 59 non-X CT genes belong to the testis-selective category, and 36 of these genes (5 CT-X and 31 non-X CT) had >50% of the expression evidence derived from non-testis or placental libraries, indicating that these might not qualify as CT genes.
Seven CT genes were not identified in any library at all (2 CT-X and 5 non-X CT). An additional 8 CT-X genes (SPANX-N1, PAGE1, CSAG1, SSX5/6/7/9, and CT45-2) were not present in any testis-annotated library. Of these, SSX5 and SSX7 have been shown to be expressed in testis by RT-PCR (19), suggesting a likely discrepancy in mapping short sequence tags to their genomic counterparts, an expected phenomenon for large and highly homologous gene families like SSX. In contrast, the absence of testicular expression of SSX6 and SSX9 was confirmed in that study, indicating that some of the currently recognized CT genes could either be silent or expressed at extremely low levels in testis. The full list with classification and raw expression scores across the merged expression array can be found in Dataset S3.
Associations between different CT gene properties and their assigned classification were analyzed using the APRIORI algorithm. Besides being more likely testis-restricted, CT-X genes were found to be more often members of multigene families than non-X CTs. In addition, Gene Ontology terms showed CT-X genes to be more often in the “molecular function unknown” and “biological process unknown” categories, whereas the non-X CTs are associated with known functions such as meiosis, sexual reproduction, and gametogenesis (see Dataset S4 for all attributes and annotations).
While the description of CT-X genes such as NY-ESO-1 (20), SSX2 (21), and MAGE-A1 (22) match our classification—all are in the testis-restricted category—not all CT genes were found to be as testis-restricted as described in the literature. BAGE, SPO11, LIPI, LDHC, and BRDT, considered to be testis-restricted based on a tissue panel of 13 non-gametogenic normal tissues (1), fall into the testis-selective category in our screen, most likely due to a larger amount of expression sources sampled. Despite the broader coverage we could not confirm an expression of MAGE-A1, MAGE-C1, and NY-ESO-1 at low levels in the pancreas reported in the same study. In agreement with the study in ref. 1, we found IL13RA1, ACRBP, and SPA17 to be expressed in a wide variety of tissues, falling into the lower end of the testis-selective category.
In the present study, we have ranked the testis-selective genes based upon the ratios of their expression evidence in testis and placenta relative to other somatic tissues, rather than using fixed thresholds and the number of somatic tissues in which a CT candidate is allowed as the distinguishing criteria for CT versus non-CT genes (2). Genes without any somatic expression have unique potential for cancer vaccines and other therapeutic approaches to cancer. From past work involving screening of larger sets of genes (23), a cutoff was introduced that defined CT candidate genes as genes with 2-fold higher expression evidence in testis and placenta relative to all other somatic normal tissues. This approach was complementary to our current one and will not require updated thresholds as the number of sampled tissue sources increases.
Intriguingly, a number of CT genes were found to be expressed in no somatic tissues except for brain, suggesting the presence of a distinctive transcriptional control mechanism that functions with tissue specificity in germ cells and in brain. There have been relatively few studies of CT gene expression in different anatomical regions of normal brain and similarly not many in brain tumors (24, 25), except for NXF2, which was shown to be expressed in normal brain (26). Our in silico study has discovered a broader subset of CT genes with brain expression, among them members of the otherwise fully testis-restricted GAGE and MAGE families, found to be expressed in the hippocampus and cerebral cortex. A previous study has similarly identified a group of cancer/testis/brain (CTB) antigens (27). However, despite the bioinformatic evidence, we have not been able to confirm the expression of selected CT genes (MAGEA9, MAGEC2, PASD1, and GAGE) in tissue samples from total brain, cerebellum, caudate nucleus, thalamus, frontal cortex, occipital cortex, pons, or amygdala by RT-PCR (data not shown), and whether these genes are expressed in brain remains to be proven.
Distribution of CT Genes in Cancer Tissues.
Our ranking by the number of different cancer types and anatomical sites of CT genes expressed in cancer-annotated libraries distinguishes CT-“rich” and CT-“poor” tumors based on the in silico analysis obtained from cDNA, CAGE, and MPSS libraries (Fig. 1 and Dataset S5). The broadest distribution of CT genes was found in germ cell tumors, melanomas and lung carcinomas, adenocarcinomas and chondrosarcomas. Breadth of cancer expression was uncorrelated with tissue restriction in normal tissues (r = 0.18 for CT-X genes, r = 0.02 for non-X CT genes using Spearman rank correlation); for instance, the fully testis-restricted CT genes, such as MAGEA2/A2B and CTAG2, were found to be present in a variety of different tumor tissues.
Melanoma, non-small-cell lung cancer, hepatocelluar carcinoma and bladder cancer have been identified as high CT gene expressors, with breast and prostate cancer being moderate and leukemia/lymphoma, renal and colon cancer low expressors (1). Our in silico analysis confirms this distinction, in particular for tumor tissues well represented by the available libraries, showing a broad distribution of CT genes expressed in cancers of skin including melanoma (43% of CT genes with cancer expression were found in at least one melanoma library), lung (37%), and liver (34%). Strong presence of CT expression found in the present study but not by previous RT-PCR studies includes tumors from germ cells (39%), stomach (28%), and cartilage (chondrosarcomas, 26%). One reason for this discrepancy could be the lack of RT-PCR data for certain tumors, e.g., gastric cancer is much rarer than other carcinomas in the Western world, and mesenchymal tumors are also not well represented in many of the RT-PCR studies to date. Our in silico information may thus serve as a guide for future experimental investigations, especially useful for recently described CT genes not yet analyzed in great detail. Discrepancies are also likely to occur due to the potential inclusion of cancer cell line samples in the survey that, unlike normal tissue samples explicitly labeled as normal, are often not diistinguished from primary tumor samples. A third reason for this observed discrepancy could be the bias that resulted from differences in library numbers studied for each tumor type: for instance, ovarian cancer is CT-rich by RT-PCR but not evident from our in silico study, possibly due to the low number of available ovarian cDNA libraries. However, colon cancer, a CT-poor tumor, was correctly shown to have low frequency of CT genes despite the large number of colon libraries in the databases, and this would argue that the difference in library numbers may not have been a significant factor. Last, the in silico finding of high CT expression in germ cell tumor represents a special situation that can be explained by two reasons. One is that a subset of CT genes, particularly the non-X CTs, encode proteins with known specific functions in germ cells, and their expression in germ cell tumors represents the preserved expression of lineage-specific markers—rather than aberrant gene activation, conceptually similar to the expression of thyroglobulin by thyroid cancer or prostate specific antigen by prostate cancer. The other reason would be that the germ cell tumors from which the mRNA expression profiles were derived could have been contaminated by the adjacent or entrapped testicular tissue, which provides the source for CT gene transcripts when the germ cell tumor was actually negative for the CT gene in question.
CT Candidate Prediction.
Prediction of CT candidates based on their expression profiles in cDNA, MPSS, and CAGE libraries resulted in 28 genes supported by 2 expression platforms in the testis- or testis/brain-restricted category, including 10 known CT genes and 18 novel CT candidates (Fig. S3 and Dataset S6). An additional, less stringent screen for CT-X genes identified 47 genes in the same categories, including 34 known CT genes and 13 novel candidates. After manual curation, the list of novel candidates was extended to include the highest scoring testis-selective CT-X candidates, TKTL1 and NXF3, the latter being a known CT gene, a member of the NXF2 CT family (28).
Of 33 novel CT candidate genes, 12 most promising genes were manually selected for experimental validation by RT-PCR based on an evaluation of available gene expression data in human cancer. Of the 5 X- and 7 non-X-chromosomal candidates, 11 transcripts could be amplified, whereas transcripts from VCX2 were not detected in any of the 23 normal tissue RNA samples. Three of the amplified gene transcripts exhibited testis-restricted (AKAP4) or testis-selective (PEPP-2, OTOA) expression (data not shown). RT-PCR products of these genes were also detected in samples from a panel of 30 cancer cell lines.
PEPP-2, an X-linked human homeobox gene, encodes a transcriptional factor with similar cancer/testis restricted expression patterns in both human and mouse (29); it is also a member of a top 50 list of genes under strong positive selection between human and chimpanzee (30). Otoancorin (OTOA) was reported to be specific to sensory epithelia of the inner ear (31), but has also been associated with ovarian and pancreatic cancer due to its homology with mesothelin, a cancer immunotherapy target (32). AKAP4 (CT-X), identified in the 2-platform screen, exhibits weak expression in different cancer cell lines and encodes a kinase anchor protein (33) involved in the cAMP-regulation of motility (34) and was recently suggested as a CT gene in an independent study (35).
All 3 confirmed genes are candidates for immunotherapy based on their restricted expression, and further investigation of their mRNA and protein expression in various tumors is warranted and ongoing. Given the comprehensive nature of our study and the limited number of confirmed novel CT candidates, it seems that the number of true CT genes matching the criterion of stringent testis-restricted expression profile has reached a plateau.
Although it is clear that the CT designation has been inappropriately given to a large number of genes with wide normal tissue expression, it is less evident how precisely the term CT should be applied. There is no difficulty with CT genes whose expression profile have a classic CT pattern; we estimate ≈39 genes presently in this category and ≈90% of them reside on the X chromosome. The challenge for the remaining CT genes, most of which are non-X coded, is that they are expressed in testis and cancer, but are also expressed in a limited number of normal tissues. Should these be designated CT? Perhaps the best solution at this point would be to assemble further information about CT genes and their products, including function, binding partners, evolutionary selection (36), control of gene expression, identification of expressing normal somatic cells, aberrant non-lineage expression in cancer, and immunogenicity, before establishing a uniform classification of CT genes.
Methods
Selection of CT Genes.
A total of 153 CT genes (200 unique RefSeq transcript identifiers) were selected from the CT Antigen DB (http://www.cta.lncc.br) and by manual curation of the literature. Genes were annotated with their most current gene identifiers and merged based on shared National Center for Biotechnology Information RefSeq nucleotide identifiers (Dataset S7). Additional gene identifiers were obtained from RefSeq release 11 (37), IPI version 3.29 (38); genomic coordinates were taken from the University of California, Santa Cruz Genome Browser hg18 human genome build (39). Of these 153 genes, 83 that encode 107 RefSeq transcripts were mapped to the X chromosome (CT-X genes) whereas 70 genes were on autosomes (non-X CT genes). Subcellular localization was based on predictions in the human version of the LOCATE system (40). SEREX information was obtained from the Cancer Immunome Database website (http://ludwig-sun5.unil.ch/CancerImmunomeDB). Ambiguities were resolved by manual curation.
Source of Expression Information.
Gene expression profiles were determined based on 4 different sources: 99 CAGE libraries from the RIKEN FANTOM3 project (18), 47 MPSS libraries (17, 23, 41), a collection of 8401 cDNA expression libraries from the eVOC system (16), and semiquantitative RT-PCR across 22 normal tissue samples. Source materials were annotated with regards to the anatomical site and pathological status of their source tissues. In cases where the anatomical source was unclassifiable, cell type information was used. Bone marrow/blood libraries were designated bone marrow, and all combinations with mucosa (colon, stomach) were merged into “mucosa.” Libraries not explicitly annotated as “normal” were considered as unclassified. Libraries from pooled tissue sources were ignored, and pooled samples were kept as long as the pathological and anatomical status was identical for all donors (see Dataset S1 for annotated libraries).
Pseudoarrays.
Expression information was organized into “pseudoarrays” based on expression information obtained from CAGE-, MPSS-, and cDNA-libraries in the case of cancer expression and merged with RT-PCR results in the case of normal tissue expression. Columns reflect the class of library in which a CT transcript was identified and rows represent individual RefSeq transcripts. Annotation was based on the general library class description (normal, cancer or unclassified) combined with pathological state and anatomical site. To evaluate the relative levels of CT expression we converted expression signals from the 4 sources into “expression evidence”: For CAGE- and MPSS-based expression data, expression evidence was based on detected tags per million (TPM), with matches <3 TPM (≈1 transcript per cell) filtered out. Normalized and subtracted EST libraries prevent quantitaton of expression strength based on EST counts, therefore expression evidence is represented by the number of cDNA libraries in which a given transcript was identified. RT-PCR results were manually binned into 5 groups of expression, ranging from 0 (not expressed) to 4 (strongly expressed). For each expression source, evidence values were normalized on a per-transcript basis by setting the highest expression evidence in normal tissues to a value of 1, reflecting relative changes in expression levels across tissues and pathological states. Pseudoarrays from the 4 expression sources were merged by summing the individual expression evidence scores for a given transcript from each platform. Expression profiles for multiple transcripts associated with the same gene were merged into a single representation, keeping the highest expression score for overlapping annotations. In arrays where annotation was “merged” into single columns based on their class (e.g., all cancer expression information), the highest expression score across all annotated libraries was kept for each gene.
Visualization and Ranking.
Genes were divided into CT-X and non-X CT panels, then individually ranked by their expression properties in normal tissues and classified into the following 3 categories: (i) expression in testis and placenta only (testis-restricted); (ii) expression in testis, placenta and brain-regions only (testis/brain-restricted), and (iii) all other genes (testis-selective). Final ranking within each category was obtained by sorting based on decreasing level of normal tissue specificity as measured by the combined testis and placenta expression evidence divided by all normal expression evidence. All arrays were visualized using MeV 4.0 (www.tm4.org).
Clustering Methods.
Associations between CT annotation and their classification were investigated by recording their assigned class; presence or absence in placenta, brain, testis, and developing ovary; their testis/placenta tissue specificity; their X vs. non-X chromosomal status; membership in a gene family; subcellular localization; and evolutionary status (36) followed by an analysis with the APRIORI algorithm (42), which identifies association rules matching a predefined threshold of support (30%) and confidence (≥ 0.8)
Search Criteria for CT Candidates.
CT candidates were identified using the same in silico expression sources, but with no filters for minimum TPM value and satisfying the following criteria: (i) exhibit expression in testis and at least one cancer-associated tissue at 10 TPM (CAGE, MPSS) or presence in at least one EST/cDNA library with testis and cancer annotation; (ii) not be present above those levels in any other tissue except for placenta, ovary, and brain; and (iii) be supported independently by 2 platforms. Identified candidates were ranked using the same approach used to classify known CT genes. To increase coverage of CT-X genes, a second genome-wide search was conducted requiring support from only a single platform. Candidates were selected for RT-PCR validation by manual curation, removing hypothetical proteins, predicted genes and candidates with multiple publications indicating expression in somatic tissues.
RT-PCR.
RNA preparations were purchased from the normal tissue panels of Clontech and Ambion or prepared from cancer cell lines using the RNAeasy kit (Qiagen) and were used to prepare cDNA for RT-PCR. A total of 1.0 μg of RNA was reverse transcribed into cDNA in a total volume of 20 μL using the Omniscript RT kit (Qiagen) according to the manufacturer's protocol using oligo(dT)18 primers (Invitrogen). The cDNA was diluted 5 times and 3 μL was used in the PCR with primers specific to each analyzed gene in a final volume of 25 μL. Primers used for PCR amplification were designed to have an annealing temperature ≈60 °C using Primer3 software (www.genome.wi.mit.edu/cgi-bin/primer/primer3www.cgi) and were chosen to encompass introns between exon sequences to avoid amplification of genomic DNA. DNase treatment was undertaken before cDNA synthesis to analyze intronless genes. Primers were designed to target all known variants of a gene in RefSeq and their specificity was confirmed by aligning with the National Center for Biotechnology Information sequence databases using BLAST (www.ncbi.nlm.nih.gov/blast/blast.cgi). Primer sequences and amplicon sizes are provided in Dataset S7.
JumpStart REDTaq ReadyMix (Sigma Aldrich) was used for amplification according to the manufacturer's instructions. Samples were amplified with a precycling hold at 95 °C for 3 min, followed by 35 specific cycles of denaturation at 95 °C for 15 seconds, annealing for 30 seconds (10 cycles at 60 °C, 10 cycles at 58 °C and 15 cycles at 56 °C) and extension at 72 °C for 30 seconds followed by a final extension step at 72 °C for 7 min. β-actin was amplified as control. PCR products were separated on 1.5% agarose gels stained with ethidium bromide. For semiquantitative PCR analysis, RT-PCR products were classified into 0 (negative) to 4 (strongest signal) based on the intensity of the product on ethidium bromide-stained gels.
Supplementary Material
Acknowledgments.
We thank Dmitry Kuznetsov for providing access to the SEREX information on CT genes and Erika Ritter (Ludwig Institute for Cancer Research, New York Branch at Memorial Sloan–Kettering Cancer Center, New York) for providing cell lines. This project was supported by the South African National Bioinformatics Network; National Institutes of Health Stanford-South African Informatics Training for Global Health Grant TW-03-008; Atlantic Philanthropies; The Oppenheimer Memorial Trust; a Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government (to Y. H.); and a grant from the Genome Network Project from the Ministry of Education, Culture, Sports, Science and Technology, Japan. This work was conducted as part of the Hilton-Ludwig Cancer Metastasis Initiative, funded by the Conrad N. Hilton Foundation and the Ludwig Institute for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810777105/DCSupplemental.
References
- 1.Scanlan MJ, Simpson AJG, Old LJ. The cancer/testis genes: Review, standardization, and commentary. Cancer Immun. 2004;4:1. [PubMed] [Google Scholar]
- 2.Simpson AJG, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 3.Marchand M, et al. Tumor regressions observed in patients with metastatic melanoma treated with an antigenic peptide encoded by gene MAGE-3 and presented by HLA-A1. Int J Cancer. 1999;80:219–230. doi: 10.1002/(sici)1097-0215(19990118)80:2<219::aid-ijc10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 4.Davis ID, et al. Recombinant NY-ESO-1 protein with iscomatrix adjuvant induces broad integrated antibody and CD4(+) and CD8(+) t cell responses in humans. Proc Natl Acad Sci USA. 2004;101:10697–10702. doi: 10.1073/pnas.0403572101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jäger E, et al. Recombinant vaccinia/fowlpox NY-ESO-1 vaccines induce both humoral and cellular NY-ESO-1-specific immune responses in cancer patients. Proc Natl Acad Sci USA. 2006;103:14453–14458. doi: 10.1073/pnas.0606512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valmori D, et al. Vaccination with NY-ESO-1 protein and CPG in montanide induces integrated antibody/th1 responses and CD8 t cells through cross-priming. Proc Natl Acad Sci USA. 2007;104:8947–8952. doi: 10.1073/pnas.0703395104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uenaka A, et al. T cell immunomonitoring and tumor responses in patients immunized with a complex of cholesterol-bearing hydrophobized pullulan (chp) and NY-ESO-1 protein. Cancer Immun. 2007;7:9. [PMC free article] [PubMed] [Google Scholar]
- 8.Odunsi K, et al. Vaccination with an NY-ESO-1 peptide of HLA class i/ii specificities induces integrated humoral and t cell responses in ovarian cancer. Proc Natl Acad Sci USA. 2007;104:12837–12842. doi: 10.1073/pnas.0703342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atanackovic D, et al. Expression of cancer-testis antigens as possible targets for antigen-specific immunotherapy in head and neck squamous cell carcinoma. Cancer Biol Ther. 2006;5:1218–1225. doi: 10.4161/cbt.5.9.3174. [DOI] [PubMed] [Google Scholar]
- 10.Gnjatic S, et al. NY-ESO-1: Review of an immunogenic tumor antigen. Adv Cancer Res. 2006;95:1–30. doi: 10.1016/S0065-230X(06)95001-5. [DOI] [PubMed] [Google Scholar]
- 11.Scanlan MJ, et al. Identification of cancer/testis genes by database mining and mRNA expression analysis. Int J Cancer. 2002;98:485–492. doi: 10.1002/ijc.10276. [DOI] [PubMed] [Google Scholar]
- 12.Zendman AJW, Ruiter DJ, Muijen GNPV. Cancer/testis-associated genes: Identification, expression profile, and putative function. J Cell Physiol. 2003;194:272–288. doi: 10.1002/jcp.10215. [DOI] [PubMed] [Google Scholar]
- 13.Costa FF, Blanc KL, Brodin B. Concise review: Cancer/testis antigens, stem cells, and cancer. Stem Cells. 2007;25:707–711. doi: 10.1634/stemcells.2006-0469. [DOI] [PubMed] [Google Scholar]
- 14.Kalejs M, Erenpreisa J. Cancer/testis antigens and gametogenesis: A review and “brain-storming” session. Cancer Cell Int. 2005;5:4. doi: 10.1186/1475-2867-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross MT, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelso J, et al. eVOC: A controlled vocabulary for unifying gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jongeneel CV, et al. An atlas of human gene expression from massively parallel signature sequencing (MPSS) Genome Res. 2005;15:1007–1014. doi: 10.1101/gr.4041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 19.Güre AO, Wei IJ, Old LJ, Chen YT. The SSX gene family: Characterization of 9 complete genes. Int J Cancer. 2002;101:448–453. doi: 10.1002/ijc.10634. [DOI] [PubMed] [Google Scholar]
- 20.Chen YT, et al. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Türeci O, et al. The SSX-2 gene, which is involved in the t(x;18) translocation of synovial sarcomas, codes for the human tumor antigen HOM-MEL-40. Cancer Res. 1996;56:4766–4772. [PubMed] [Google Scholar]
- 22.van der Bruggen P, et al. A gene encoding an antigen recognized by cytolytic t lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 23.Chen YT, et al. Identification of cancer/testis-antigen genes by massively parallel signature sequencing. Proc Natl Acad Sci USA. 2005;102:7940–7945. doi: 10.1073/pnas.0502583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahin U, et al. Expression of cancer testis genes in human brain tumors. Clin Cancer Res. 2000;6:3916–3922. [PubMed] [Google Scholar]
- 25.Scarcella DL, et al. Expression of MAGE and GAGE in high-grade brain tumors: A potential target for specific immunotherapy and diagnostic markers. Clin Cancer Res. 1999;5:335–341. [PubMed] [Google Scholar]
- 26.Zhang M, Wang Q, Huang Y. Fragile x mental retardation protein FMRP and the RNA export factor NXF2 associate with and destabilize NXF1 mRNA in neuronal cells. Proc Natl Acad Sci USA. 2007;104:10057–10062. doi: 10.1073/pnas.0700169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: An expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 28.Loriot A, Boon T, Smet CD. Five new human cancer-germline genes identified among 12 genes expressed in spermatogonia. Int J Cancer. 2003;105:371–376. doi: 10.1002/ijc.11104. [DOI] [PubMed] [Google Scholar]
- 29.Wayne CM, MacLean JA, Cornwall G, Wilkinson MF. Two novel human x-linked homeobox genes, hPEPP1 and hPEPP2, selectively expressed in the testis. Gene. 2002;301:1–11. doi: 10.1016/s0378-1119(02)01087-9. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwaenepoel I, et al. Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in autosomal recessive deafness DFNB22. Proc Natl Acad Sci USA. 2002;99:6240–6245. doi: 10.1073/pnas.082515999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muminova ZE, Strong TV, Shaw DR. Characterization of human mesothelin transcripts in ovarian and pancreatic cancer. BMC Cancer. 2004;4:19. doi: 10.1186/1471-2407-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner RM, Johnson LR, Haig-Ladewig L, Gerton GL, Moss SB. An X-linked gene encodes a major human sperm fibrous sheath protein, HAKAP82. genomic organization, protein kinase a-rii binding, and distribution of the precursor in the sperm tail. J Biol Chem. 1998;273:32135–32141. doi: 10.1074/jbc.273.48.32135. [DOI] [PubMed] [Google Scholar]
- 34.Michel JJC, Scott JD. AKAP mediated signal transduction. Annu Rev Pharmacol Toxicol. 2002;42:235–257. doi: 10.1146/annurev.pharmtox.42.083101.135801. [DOI] [PubMed] [Google Scholar]
- 35.Chiriva-Internati M, et al. AKAP-4: A novel cancer testis antigen for multiple myeloma. Br J Haematol. 2008;140:465–468. doi: 10.1111/j.1365-2141.2007.06940.x. [DOI] [PubMed] [Google Scholar]
- 36.Stevenson BJ, et al. Rapid evolution of cancer/testis genes on the X chromosome. BMC Genomics. 2007;8:129. doi: 10.1186/1471-2164-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheeler DL, et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31:28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kersey PJ, et al. The International Protein Index: An integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 39.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fink JL, et al. LOCATE: A mouse protein subcellular localization database. Nucleic Acids Res. 2006;34:D213–7. doi: 10.1093/nar/gkj069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grigoriadis A, et al. Establishment of the epithelial-specific transcriptome of normal and malignant human breast cells based on MPSS and array expression data. Breast Cancer Res. 2006;8:R56. doi: 10.1186/bcr1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agrawal R, Imieliński T, Swami A. Mining association rules between sets of items in large databases. Proc ACM-SIGMOD Management Data. 1993;22:207–216. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.