What seems to be a ‘mere' cofactor of a core factor to some, might be the main component to others; it is all a question of perspective and, probably quite often, pure semantics. SGT1 (SUPPRESSOR OF G2 ALLELE OF SKP1) was discovered genetically before it was recognized as being a cofactor of the molecular chaperone HSP90 (HEAT-SHOCK PROTEIN 90). The molecular details of this interaction and its physiological relevance are the subject of a paper from the Shirasu and Guerois groups, in which they combine structural biology, biochemistry and genetics (Kadota et al, 2008, this issue). The interaction surfaces were analysed by nuclear magnetic resonance (NMR) spectroscopy and by testing point mutants with pull-down experiments for interaction in vitro and by testing them for function in the resistance of plants against pathogens in vivo. If these approaches left any doubt that the protein–protein interactions are real, then the in vivo complementation by mutants with compensatory swaps of amino acids in the interface between the two partner proteins leaves no room for appeal. The purpose of this protein complex is to support crucial pathogen-sensor proteins of the plant immune system—the NB-LRR (NUCLEOTIDE BINDING AND LEUCINE-RICH REPEAT) proteins—of which Arabidopsis encodes approximately 125 (Jones & Dangl, 2006). Remarkably, both the SGT1–HSP90 molecular chaperone machine and its targets, the NB-LRR proteins, also have a function in the innate immune responses of animals (da Silva Correia et al, 2007; Mayor et al, 2007).
HSP90 is usually considered to be the central subunit of a molecular machine. It owes this prominent role to the fact that it is an abundant cytosolic protein, perhaps even the most abundant protein in unstressed cells (Lai et al, 1984), and that it has ATPase activity, suggesting that it might ‘burn' ATP to function as a molecular chaperone (Picard, 2002; Pearl & Prodromou, 2006; Neckers, 2007; Wandinger et al, 2008). Proteins that depend on HSP90 for folding, stability, assembly and/or function are known as substrates or ‘clients', and their number continues to increase (for an updated list, see http://www.picard.ch/downloads/Hsp90interactors.pdf). A characteristic of HSP90 substrates is that they are degraded by the proteasome when HSP90 function is inhibited pharmacologically or genetically (Whitesell & Lindquist, 2005; Pearl et al, 2008). However, exactly what HSP90 does to its substrates and how remains poorly understood and is the subject of intense research.
Over the past two decades, it has been recognized that HSP90 does not act alone. Cofactors—known as co-chaperones in this context—bind to HSP90 in a multitude of assortments to regulate its interactions with and effects on substrates, or its ATPase activity. So far, there are approximately 30 proteins in this category, which is far too many to have a clear picture of how they work together. Even the term ‘co-chaperone' is not as clear as it might seem. The distinction between a substrate and a co-chaperone can be difficult to make; for example, the ‘established' co-chaperones AHA1 (ACTIVATOR OF HSP90 ATPASE 1) and CPR6 (CYCLOSPORIN-SENSITIVE PROLINE ROTAMASE 6) stimulate the ATPase activity of HSP90 (Panaretou et al, 2002), but it seems that some substrates might also stimulate its activity (McLaughlin et al, 2002). Substrates, but not co-chaperones, are expected to be degraded on inhibition of HSP90 with a drug, but some are not degraded or even stabilized (see, for example, McClellan et al, 2005; Chen & Balch, 2006; He et al, 2007). The histone deacetylase HDAC6 is a regulator of HSP90 because it keeps it in the deacetylated active form and also has features of an HSP90 substrate (Rao et al, 2008).
The pejorative connotation of the ‘co' in co-chaperones is unfair and inappropriate for a number of other reasons; several co-chaperones display HSP90-independent chaperone activities themselves (Bose et al, 1996; Freeman et al, 1996; Kimura et al, 1997). Thus, given the large number of HSP90 co-chaperones—and despite the abundance of HSP90—it is likely that some co-chaperones have independent functions. These might be completely HSP90-independent or inverted with HSP90 playing the role of the ATP-churning servant, the ATPase-driven conformational gymnastics of which (Bron et al, 2008; Krukenberg et al, 2008; Richter et al, 2008) serve the purposes of the ‘co-'chaperone. Therefore, now that I turn to discuss SGT1, the reader will understand that I call it an HSP90 co-chaperone for lack of a better term... and out of ignorance of the breadth of SGT1 functions.
The gene encoding SGT1 was discovered in the budding yeast as a high-copy suppressor of a mutation in the gene for SKP1—a component of the SCF (Skp1–Cullin–F-box) ubiquitin ligase complex required for kinetochore function (Kitagawa et al, 1999). It was found to be linked to plant pathogen resistance when its plant orthologue was identified as an interactor of RAR1 (REQUIRED FOR MLA12 RESISTANCE) in a yeast two-hybrid screen (Azevedo et al, 2002), as RAR1 was known to be an essential factor for resistance mediated by NB-LRR genes (Shirasu et al, 1999). Shortly thereafter, it was recognized that HSP90 is also involved, both for kinetochore function (Stemmann et al, 2002) and for plant resistance (Hubert et al, 2003; Takahashi et al, 2003).
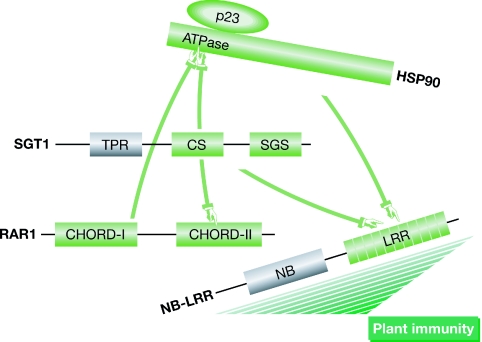
To bind to HSP90, RAR1 and SGT1 use a CHORD (cysteine- and histidine-rich domain) and a CS motif (present in metazoan CHORD and SGT1 proteins), respectively (Fig 1). The two domains structurally resemble those of the co-chaperone p23 (Dubacq et al, 2002; Garcia-Ranea et al, 2002), a well-characterized regulator of the HSP90 ATPase domain (Ali et al, 2006). In the case of SGT1, it came as a surprise that its TPR (tetratricopeptide repeat) domain—a protein–protein interaction domain used by many other co-chaperones to dock to HSP90—is not involved in this interaction (Lee et al, 2004; Catlett & Kaplan, 2006; Botër et al, 2007; Zhang et al, 2008). Instead, the TPR domain—which is not essential for SGT1 function in plant immunity—might be important in regulating the relative protein levels of the isoforms SGT1a and SGT1b in Arabidopsis (Azevedo et al, 2006), possibly by allowing the HSP90–SGT1 complex to connect to SKP1 and its partners in the SCF ubiquitin ligase complex (Catlett & Kaplan, 2006). SGT1 also uses the CS domain to bind to RAR1 through a second CHORD domain (Azevedo et al, 2002). Previous work from the Shirasu and Guerois groups has shown that opposite sides of the CS domain are used to bind to HSP90 and RAR1 (Botër et al, 2007). The plot thickens with the new report (Kadota et al, 2008); here, the in-depth analysis of the interaction surfaces by NMR and mutagenesis reveals that SGT1 and RAR1, with the CS and CHORD-I domains, respectively, bind to overlapping surfaces on the ATPase domain of HSP90. The CHORD-I domain of RAR1 competes with SGT1 for HSP90 binding, but ternary complexes can nevertheless form (Botër et al, 2007). In any case, this is not a quiet threesome, as there are several other co-chaperones that bind to HSP90 nearby, notably p23. On the basis of their structural analysis of the CS–HSP90 complex, the authors had previously suggested that p23 binds to HSP90 differently, owing, in part, to an additional strand missing in the CS domain of SGT1 (Botër et al, 2007); they have now confirmed this experimentally. Despite the structural similarities between p23 and the CS domain, the interfaces on HSP90 do not overlap. The crystal structure of the HSP90–SGT1 CS complex, recently reported in a study to which the Shirasu group also contributed (Zhang et al, 2008), confirms the structure modelled on the NMR data. The fact that full-length SGT1 competes with p23 for HSP90 binding (Kadota et al, 2008) might indicate an indirect conformational effect or steric hindrance. The various binding modes of the co-chaperones correlate with different functional consequences for HSP90: p23 preferentially binds to the ATP-bound form of HSP90 and inhibits its ATPase activity, whereas SGT1 binds to the ADP-bound form and does not influence the ATPase (Catlett & Kaplan, 2006). In fact, the functional consequences of SGT1 binding to HSP90, other than preventing the access of some other co-chaperones, remain unknown.
Figure 1.
Schematic representation of HSP90, SGT1, RAR1 and NB-LRR interactions. For clarity, the protein–protein interfaces are only indicated by arrows, as a dynamic three-dimensional representation would be necessary to do justice to all known interactions. Note that the lengths of proteins and structural motifs are not to scale. The signalling pathways downstream from the NB-LRR proteins that lead to immunity in plants are largely unknown. HSP90, HEAT-SHOCK PROTEIN 90; NB-LRR, NUCLEOTIDE BINDING AND LEUCINE-RICH REPEAT; RAR1, REQUIRED FOR MLA12 RESISTANCE; SGT1, SUPPRESSOR OF G2 ALLELE OF SKP1.
What about the substrates of all this? Although there is no doubt that SGT1 and HSP90 are required for maintaining NB-LRR proteins in a state that allows them to respond to an infection, the details of the interactions of the NB-LRR proteins with SGT1 and/or HSP90 are far less well understood. The structural consequences of these interactions for the NB-LRR proteins are completely unknown. SGT1 uses the carboxy-terminal SGS (SGT1-specific) domain to interact directly or indirectly with the LRR domain of plant R (RESISTANCE) proteins (Dubacq et al, 2002; Bieri et al, 2004), whereas SGT1 apparently needs both the CS and SGS domains to bind to the LRR domain of animal NOD-like receptors; Mayor et al, 2007). HSP90 was shown to interact directly with part of the LRR domain of the I-2 protein of tomato (de la Fuente van Bentem et al, 2005), but a more extensive analysis in the animal system, albeit not with purified proteins, indicated an interaction with the nucleotide-binding and oligomerization domain (the NACHT domain) of the NOD-like receptors (Mayor et al, 2007). In this system, the interaction of HSP90 with the LRR domain of the NOD-like receptor NALP3 was found to be stimulated by SGT1, suggesting a far more complex interplay between substrate, HSP90 and SGT1.
I have alluded several times to other interactions in which SGT1 (and HSP90) can engage and there are others that could be discussed. This bewildering network calls for a more careful and detailed analysis, for which the paper by Kadota et al (2008) provides an excellent roadmap. Obtaining a more detailed view of the protein–protein interfaces will be the first step towards drawing a clearer picture of the higher order protein complexes and their dynamics. Whether other interfaces rely on salt bridges—as does the one between the CS domain of SGT1 and HSP90—remains to be seen. With such an understanding of the molecular contacts, it might also be possible to generate compensatory amino-acid changes across other types of interface. If this approach were applicable to the in vivo interactions between substrates such as an NB-LRR protein and the HSP90–SGT1 tandem, it would be an exciting quantum leap for the field.

Acknowledgments
I thank Pablo Echeverría for critical comments on the manuscript. Work in the laboratory is supported by the Swiss National Research Foundation and the Canton de Genève.
References
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH (2006) Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440: 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noël L, Sadanandom A, Casais C, Parker J, Shirasu K (2006) Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J 25: 2007–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri S et al. (2004) RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16: 3480–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Weikl T, Bügl H, Buchner J (1996) Chaperone function of Hsp90-associated proteins. Science 274: 1715–1717 [DOI] [PubMed] [Google Scholar]
- Botër M et al. (2007) Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell 19: 3791–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron P, Giudice E, Rolland JP, Buey RM, Barbier P, Diaz JF, Peyrot V, Thomas D, Garnier C (2008) Apo–Hsp90 coexists in two open conformational states in solution. Biol Cell 100: 413–425 [DOI] [PubMed] [Google Scholar]
- Catlett MG, Kaplan KB (2006) Sgt1p is a unique co-chaperone that acts as a client adaptor to link Hsp90 to Skp1p. J Biol Chem 281: 33739–33748 [DOI] [PubMed] [Google Scholar]
- Chen CY, Balch WE (2006) The Hsp90 chaperone complex regulates GDI-dependent Rab recycling. Mol Biol Cell 17: 3494–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Correia J, Miranda Y, Leonard N, Ulevitch R (2007) SGT1 is essential for Nod1 activation. Proc Natl Acad Sci USA 104: 6764–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Vossen JH, Vries KJ, Wees S, Tameling WI, Dekker HL, Koster CG, Haring MA, Takken FL, Cornelissen BJ (2005) Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I-2 disease resistance protein. Plant J 43: 284–298 [DOI] [PubMed] [Google Scholar]
- Dubacq C, Guerois R, Courbeyrette R, Kitagawa K, Mann C (2002) Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot Cell 1: 568–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Toft DO, Morimoto RI (1996) Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science 274: 1718–1720 [DOI] [PubMed] [Google Scholar]
- Garcia-Ranea J, Mirey G, Camonis J, Valencia A (2002) p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett 529: 162–167 [DOI] [PubMed] [Google Scholar]
- He F, Qiao ZH, Cai J, Pierce W, He DC, Song ZH (2007) Involvement of the 90-kDa heat shock protein (Hsp-90) in CB2 cannabinoid receptor-mediated cell migration: a new role of Hsp-90 in migration signaling of a G protein-coupled receptor. Mol Pharmacol 72: 1289–1300 [DOI] [PubMed] [Google Scholar]
- Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J 22: 5679–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Amigues B, Ducassou L, Madaoui H, Ochsenbein F, Guerois R, Shirasu K (2008) Structural and functional analysis of SGT1–HSP90 core complex required for innate immunity in plants. EMBO Rep 9: 1209–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Rutherford SL, Miyata Y, Yahara I, Freeman BC, Yue L, Morimoto RI, Lindquist S (1997) Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes Dev 11: 1775–1785 [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P (1999) SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell 4: 21–33 [DOI] [PubMed] [Google Scholar]
- Krukenberg KA, Forster F, Rice LM, Sali A, Agard DA (2008) Multiple conformations of E. coli Hsp90 in solution: insights into the conformational dynamics of Hsp90. Structure 16: 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B-T, Chin NW, Stanek AE, Keh W, Lanks KW (1984) Quantitation and intracellular localization of the 85K heat shock protein by using monoclonal and polyclonal antibodies. Mol Cell Biol 4: 2802–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YT, Jacob J, Michowski W, Nowotny M, Kuznicki J, Chazin WJ (2004) Human Sgt1 binds HSP90 through the CHORD–Sgt1 domain and not the tetratricopeptide repeat domain. J Biol Chem 279: 16511–16517 [DOI] [PubMed] [Google Scholar]
- Mayor A, Martinon F, De Smedt T, Petrilli V, Tschopp J (2007) A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat Immunol 8: 497–503 [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Scott MD, Frydman J (2005) Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121: 739–748 [DOI] [PubMed] [Google Scholar]
- McLaughlin SH, Smith HW, Jackson SE (2002) Stimulation of the weak ATPase activity of human Hsp90 by a client protein. J Mol Biol 315: 787–798 [DOI] [PubMed] [Google Scholar]
- Neckers L (2007) Heat shock protein 90: the cancer chaperone. J Biosci 32: 517–530 [DOI] [PubMed] [Google Scholar]
- Panaretou B et al. (2002) Activation of the ATPase activity of Hsp90 by the stress-regulated cochaperone Aha1. Mol Cell 10: 1307–1318 [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C (2006) Structure and mechanism of the hsp90 molecular chaperone machinery. Annu Rev Biochem 75: 271–294 [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C, Workman P (2008) The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem J 410: 439–453 [DOI] [PubMed] [Google Scholar]
- Picard D (2002) Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci 59: 1640–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R, Fiskus W, Yang Y, Lee P, Joshi R, Fernandez P, Mandawat A, Atadja P, Bradner JE, Bhalla K (2008) HDAC6 inhibition enhances 17-AAG–mediated abrogation of hsp90 chaperone function in human leukemia cells. Blood 112: 1886–1893 [DOI] [PubMed] [Google Scholar]
- Richter K, Soroka J, Skalniak L, Leskovar A, Hessling M, Reinstein J, Buchner J (2008) Conserved conformational changes in the ATPase cycle of human Hsp90. J Biol Chem 283: 17757–17765 [DOI] [PubMed] [Google Scholar]
- Shirasu K, Lahaye T, Tan MW, Zhou F, Azevedo C, Schulze-Lefert P (1999) A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99: 355–366 [DOI] [PubMed] [Google Scholar]
- Stemmann O, Neidig A, Köcher T, Wilm M, Lechner J (2002) Hsp90 enables Ctf13p/Skp1p to nucleate the budding yeast kinetochore. Proc Natl Acad Sci USA 99: 8585–8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Casais C, Ichimura K, Shirasu K (2003) HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci USA 100: 11777–11782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger SK, Richter K, Buchner J (2008) The Hsp90 chaperone machinery. J Biol Chem 283: 18473–18477 [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL (2005) HSP90 and the chaperoning of cancer. Nat Rev Cancer 5: 761–772 [DOI] [PubMed] [Google Scholar]
- Zhang M, Boter M, Li K, Kadota Y, Panaretou B, Prodromou C, Shirasu K, Pearl LH (2008) Structural and functional coupling of Hsp90- and Sgt1-centred multi-protein complexes. EMBO J 27: 2789–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]