Abstract
Phospholipids containing sn-2 polyunsaturated fatty acyl residues are primary targets of oxidizing radicals, producing pro-apoptotic and membrane perturbing fragmented phospholipids. The only known phospholipases that specifically select these oxidized and/or short-chained phospholipids as substrates are mammalian group VII phospholipases A2s that were purified and cloned as PAF acetylhydrolases. Platelet-activating factor (PAF) is a short-chained phospholipid, and whether these enzymes actually are PAF hydrolases or evolved as oxidized phospholipid phospholipases is unknown. The fission yeast S. pombe, which does not form or use PAF as a signaling molecule, contains an open reading frame potentially homologous to mammalian group VII phospholipase A2s. We cloned this SPBC106.11c locus and expressed it in distantly related Saccharomyces cerevisiae that lack homologous sequences. The S. pombe locus encoded a functional phospholipase A2, now renamed plg7+, that hydrolyzed PAF and a synthetic oxidized phospholipid. Expression of human type II PAF acetylhydrolase or S. pombe Plg7p enhanced viability of S. cerevisiae subjected to oxidative stress. We conclude a single celled organism with an exceedingly spare genome still expresses an unusually discriminating phospholipase A2, and that selective hydrolysis of phospholipid oxidation products is an early, and critical, way to overcome oxidative membrane damage and oxidant-induced cell death.
Keywords: apoptosis, PAF acetylhydrolase, oxidized phospholipid, phospholipid transport, phospholipase A2
Introduction
The evolutionary relationship of mammals, the fission yeast Schizosaccromyces pombe, and the budding yeast Saccharomyces cerevisiae is approximately equidistant, with S. cerevisiae and S. pombe separating from their common ancestor approximately 420 to 330 million years ago [1]. These yeasts would seem to be protected from the underlying cause of oxidative injury and cell death because they do not contain desaturases that form polyunsaturated fatty acids [2] that are the primary targets of chemical oxidation [3]; but in fact, these free living organisms readily accumulate and incorporate the abundant plant polyunsaturated fatty acids they encounter in crushed grapes and malting barley into their membrane phospholipids [4–6]. These yeasts also accumulate whole phosphatidylcholines from their environment [7]. These exogenous phospholipids include short-chained phospholipids [8, 9] that are components of red wine and its must [10], or that are formed by chemical oxidation or enzymatic attack on plant phospholipids.
Mammalian cells also accumulate exogenous fragmented phospholipids as intact phospholipids [11]. Some of these disrupt membrane lipid packing [12], while others disrupt mitochondrial integrity and activate the intrinsic apoptotic pathway [11]. To deal with these membrane disruptive phospholipids, mammalian genomes encode a small family of phospholipases A2 that selectively degrade phospholipids with oxidatively-fragmented [13] or peroxidized [14, 15] fatty acyl residues. These enzymes, unlike all other phospholipases A2, have no activity against intact, long chained phospholipids, and this specific removal of oxidatively damaged phospholipids protects mammalian cells from oxidative death initiated by H2O2 exposure [16] or UVB irradiation [17]. These enzymes were originally purified and cloned as PAF acetylhydrolases [18, 19], but their role in physiologic and pathologic PAF catabolism is unclear [20] and the presence of a functional ortholog in C. elegans [21] involved in epithelial sheet migration potentially indicates a different original purpose for these enzymes.
The S. pombe genome, but not that of S. cerevisiae, contains a sequence encoding a hypothetical protein that would have 25% identity to human PAF acetylhydrolases [22], and would retain the residues forming the catalytic triad of the mammalian enzyme. Since S. pombe does not use PAF as a signaling molecule, a catalytically active enzyme encoded by this SPBC106.11c locus might instead function as an oxidized phospholipid phospholipase positioned to maintain viability in the face of environmental oxidative stress. Here, we show the S. pombe genome does encode for a functional PAF acetylhydrolase, and that this enzyme will reduce oxidative cell death. Expression of a functional member of the PAF acetylhydrolase family by a unicellular organism shows these enzymes are ancient responses to environmental stresses.
Material and Methods
Strains and Media
S. pombe strain CHP428 (h+ ura4-D18 leu1-32 ade6-M210 his7-366) was purchased from ATCC (201399; Manassas, VA), S. cerevisiae wild type diploid expression strain INVSc1 (MATa his3D1 leu2 trp1-289 ura3-52 MATá his3D1 leu2 trp1-289 ura3-52) was purchased from Invitrogen (Carlsbad, CA), DY1838 (MAT α pep4-3 prb1-1122 HISΔ:pGAL10:GAL4 leu2 trp1 ura3-52) was kindly provided by D. J. Stillman (University of Utah), BY4741fet3Δ (MATa, ura3-52, leu2-3, 2-112, trp1-1, his3-11, 3–15, ade2-1, can1-100, fet3::HIS3) was a gift from D. R. Winge (University of Utah). S. cerevisiae were grown in synthetic complete (SC) minimal media without uracil (2% glucose or 2% galactose and 1% raffinose for expression studies) or YEPD (1% yeast extract, 2% peptone, 2% glucose). S. pombe were grown in SC minimal media without histidine or YES media (0.5% yeast extract, 3% glucose, 0.2% ade, his, leu, ura, lys).
Plasmids
pENTR™/D-TOPO® and pYES-DEST52 were purchased from Invitrogen. Cosmid 106 was the kind gift of the Wellcome Trust Sanger Institute (Cambridge, UK). This cosmid was used to clone SPBC106.11c into pYES-DEST52 using the pENTR™/D-TOPO® cloning kit. PCR of SPBC106.11c (plg7+) with Platinum® Taq (Invitrogen) to incorporate into pYES-DEST52 with a V5/6xHis C-terminus tag was generated using forward primer 5’-CAC CGA AAT GGG ATT GGG ATT TTC TTC G and reverse primer 5’-GTA CAT AA T TCT TTC CCA CCC AGG. Mutation of SPBC106.11c serine257 to alanine was made with the QuikChange® II site-directed mutagenesis kit (Stratagene, Cedar Creek, TX) using the primers: 5’-AAT TGA TTG TTG CTG GTC ATG CAT TTG GTG CCG CTA CTT GC and 5’-GCA AGT AGC GGC ACC AAA TGC ATG ACC AGC AAC AAT CAA TT. The V5/6xHis tag of plg7+ and plg7-S257A was removed using the primers 5’-TCC CTC GCA TGG TAC TAA GG and 5’-GGT CGG CGC GCC CAC CCT TTC ACA TAA TTC TTT CCC AC. The PCR product was cleaved with HindIII and BssHII and inserted into plg7+-pDEST52 or plg7-S257A-pDEST52 cut with the same enzymes. Empty vector pDEST52 was made by cleaving plg7+-pDEST52 with BssHII and SacII, blunted using Klenow fragment polymerase (Promega, Madison, WI), and the blunt ends were ligated with T4 Ligase (Promega). A human PAF acetylhydrolase type 2 (HPAFAH2) cDNA clone was purchased from Origene (Rockville, MD, NM_000437, Clone AB3241_B06) and shuttled into pYES-DEST52 using the pENTR™/D-TOPO® cloning kit with the primers 5’-CAC CCT GGG TCG TTT CTC ATT TCC and 5’-GGA AAT GGC CAG TTG TGC GTA C. The resulting plasmid (HPAFAH2-pDEST52) did not have a V5/6xHis tag. The V5/6xHis tag was added onto HPAFAH2-pDEST52 using the primers: 5’-GGA ATG GAT CCC TTT CCG TC and 5’-GGT CGG CGC GCC CAC CCT TGG AAA TGG CCA GTT GTG CCC GCA GGC TGG AC. The PCR product was then cleaved with BamHI and BssHII and inserted into HPAFAH2-pDEST52 cut with the same enzymes. The Δplg7 construct was made using the his7+ gene from pEA2 (purchased from ATCC) cut with XbaI and EcoRI and inserted into pCI-neo (Promega). The PCR product of the N-terminus of plg7+ using the primers 5’-CCT AGC TAG CGG GAT TGG GAT TTT CTT CG and 5’-CGG AAT TCC GAA AAC CTT TCG CAA CTT C was cut with NheI and EcoRI and inserted into pCI-neo-his7. The PCR product of the C-terminus of plg7+ using the primers 5’-TGC TCT AGA TTC CCA CGT GTT TGT TTA TGA and 5’-ATA AGA ATG CGG CCG CAT TCT TTC CCA CCC AGG AAT was cut with XbaI and NotI and inserted into pCI-neo-his7-N-terminus plg7+ to make the Δplg7 construct. S. cerevisiae strains were transformed using the S.c. EasyComp™ Transformation Kit (Invitrogen); S. pombe was transformed with the YEASTMAKER™ yeast transformation system 2 (BD Clontech, San Jose CA).
Sequence analysis
BLAST analysis comparing SPBC106.11c to human plasma (PLA2g7) and type 2 PAF acetylhydrolase (PAFAH2) used the NCBI website http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi. ClustalW alignment of SPBC106.11c, PLA2g7 and PAFAH2 was done using the default settings at website http://align.genome.jp (CLUSTAL output, BLOSUM weight matrix).
Expression studies
A single colony from transformed S. cerevisiae was grown in 5 ml SC-ura (glucose) at 30°C overnight, centrifuged at 400xg for 5 min, washed once with sterile water and resuspended in 25 ml SC-ura (galactose and raffinose). Cells were grown overnight at 30°C, centrifuged and resuspended to an optical density (OD) at 550 nm of 100 (GENESYS™ 5, Spectronic) in SDS buffer (1% SDS, 5% glycerol, 125 mM Tris pH 6.8) + Halt™ protease inhibitor cocktail (Pierce, Rockford, IL). Cells were lysed by vortexing with glass beads, boiling for 5 min, then clarified by centrifugation for 2 min at 20,000xg in a microcentrifuge. Twenty µl of clarified lysates were loaded onto a 12% SDS-PAGE gel, transferred to a PVDF membrane (Immobilon™, Millipore, Billerica, MA) and blotted for the V5 epitope (R960, Invitrogen) at 1:2500 and actin (691001, MP Biomedicals, Solon, OH) at 1:1000 dilutions. For activity assays, cells were lysed in 50 mM Tris buffer pH 7.8 with 20 uM E-64, pepstatin A, and 2 mM benzamidine HCl. PAF acetylhydrolase activity assays used 25 µl cell lysates and incubating at 37°C for 15 min with 40 µl 1 nmol [3H]PAF, 5 µl 400 mM DTT and 0.5 µl 0.5 M EDTA. The PAF acetylhydrolase inhibitors Pefabloc SC (Boehringer Mannheim 1429 868, Ingelheim, GE) and methyl arachidonyl fluorophosphonate (MAFP, BIOMOL, Plymouth Meeting, PA, ST 360) were added to lysates at a final concentration of 1 mM and incubated for 30 min at 37°C prior to incubation with 3H-PAF. The reactions were quenched with 50 µl of 10 M glacial acetic acid and excess 0.1 M sodium acetate. Cleaved [3H]acetate was isolated using BAKERBOND™ spe Octadecyl (C18) extraction columns (J.T. Baker, Phillipsburg, NJ). Lysates were normalized by Bradford assay using the Coomassie Protein Assay Kit (Pierce).
Δplg7 generation
S. pombe strain CHP428 was transformed with the Δplg7 construct and grown on SC-his media (3% agar) at 30°C. Colonies were replica plated 3 times on SC complete media, then transferred back to SC-his plates to select for stable transformants. Colonies were screened for correct insertion of construct by colony PCR as described previously [23, 24]. Briefly, a colony was resuspended in 10 uL zymolyase mixture (2.5 mg/ml zymolyase (Sigma, St. Louis, MO), 1.2 M sorbitol, 0.1 M sodium phosphate, pH 7.4) and incubated for 15 min at 37°C. Two µl of the spheroplasts were used in a 25 µL PCR reaction with Taq DNA polymerase (Promega) using primers: d1 5’ CCT TAA TCA TCG CGG TCC TA with d2 5’ AGG CTT TTT CCA TCT CCT GA, or d3 5’ TG CAA ACG AAA GAT TCA CA with d4 5’ AAA ACG AAC CGG CTA AAA GG to detect successful integration. The d4 primer with the Y5 primer (5’ TCT CGC GAT ACT GAA CAA CG) was used to detect the presence of wild type plg7+.
Supplementation and oxidation assays
S. cerevisiae were grown for 2 days in 25 ml SC-ura (galactose and raffinose) + 1% Igepal at 25°C with shaking. Cells were recovered by centrifugation at 400xg, washed once with sterile water and a portion of the cells resuspended to an OD (550 nm) of approximately 0.2 in 19 ml of SC-ura (galactose and raffinose) + 1 ml filter sterilized 5% Igepal or 1 ml of 20 mM linolenic acid (Sigma) in 5% Igepal. The cells were grown overnight at 25°C shaking, and 1 ml was removed for oxidation detection by fluorescence. Five µl of 2 mM Bodipy® 581/591 C11 (Invitrogen) was added to 1 ml of cells, and incubated rocking at room temperature for 30 min. Cells were collected by centrifugation at 400xg for 5 min and resuspended in 2-(4-morpholino)-ethane sulfonic acid (MES) buffer pH 5.5, 1% glucose to obtain an OD (550 nm) of 1. Two hundred fifty µl were removed as nontreated samples, and the remaining amount treated with a final concentration of 50 µM CuSO4. One hundred twenty five µl of cells were aliquoted in duplicate into a flat-bottomed black 96-well plate and fluorescence was measured in 5 min increments with a 485 nm, 20 nm bandwidth excitation and a 528 nm, 20 nm bandwidth emission filter at a sensitivity of 50 in a Synergy HT fluorimeter (BIO-TEK®). For microscopy, cells were allowed to adhere to 8-well chamber coverslips coated with 1 mg/ml poly-L-lysine (Sigma). Thirty µl of cells were incubated on coverslips for 1–2 min, and washed once with MES buffer. Cells were treated, or not, with a final concentration of 50 µM CuSO4 and visualized after 60 min by confocal microscopy using a 488 nm Argon laser, 520/10 nm emission filter (60X 1.42NA oil objective on an FV300 Olympus IX81 microscope).
Phospholipid mass spectrometry
Hexadecyl azelaoyl choline phosphoglyceride (AzPC; Cayman Chemical Co., Ann Arbor, MI) was quantified in S. cerevisiae expressing Plg7p or its S257 mutant by supplementing the cells with linolenate and then exposing the cells to Cu+ for 0, 30 or 60 min as above. The cell wall was digested with zymolase, the spheroplasts recovered by centrifugation, lysed by dounce homogenizer and their lipids extracted [25]. Polar phospholipids were resolved from free fatty acids with aminopropyl extraction columns and the recovered PAF-like lipids were reconstituted in methanol for analysis by LC/ESI/MS/MS [26] in comparison to [2H]PAF as described [11].
Viability assays
S. cerevisiae were grown in mock or linolenic acid supplemented media as with the oxidation detection assay and collected the next day at 400xg for 5 min. Pellets were washed twice with sterile water before the cell pellets were resuspended in water. Suspensions were added to 20 ml of sterile MES buffer to an OD (550 nm) of 0.6–0.8 (culture ODs were matched within 1%). After 10 min equilibration, a portion was removed for nontreated control, and the rest of the cells were treated with 50 µM CuSO4. Aliquots were removed after 15 and 60 min, diluted 10-fold serially and 7 µl of each dilution was spot plated on YEPD (3% agar). Cells were grown for 4 days at room temperature. For cell counting, 100 µl of 10−3 and 10−4 cell dilutions were plated on YEPD media and grown for 4 days at room temperature. Viability was determined as the fraction of cells that formed colonies after treatment with CuSO4 compared to untreated cells.
RESULTS
S. pombe expresses an enzymatically active group VII PAF acetylhydrolase homolog
BLAST analysis against the human plasma PAF acetylhydrolase (PLA2g7) revealed a putative PAF acetylhydrolase open reading frame in the S. pombe genome. This open reading frame, SPBC106.11c, has 25% identity and 44% similarity to PLA2g7 (Fig. 1) and 24% identity and 41% similarity to human type 2 PAF acetylhydrolase (gene name PAFAH2). SPBC106.11c does not share homology with the N-terminus of PLA2g7, including the 17-amino acid sequence predicted to be a secretion signal [27]. ClustalW analysis reveals the consensus lipase sequence GXSXG is conserved in the yeast and human isoforms (Fig. 1). Also conserved are the amino acids Ser257, Asp291, and His368, which were previously shown to be critical for enzyme activity (Ser273/236, Asp296/259, His351/315 for PLA2g7 and PAFAH2 respectively) [28, 29]. These residues form a catalytic triad characteristic of esterases and lipases, and retention of all the essential residues suggest SPBC106.11c might encode a functional enzyme. Based on the information below, we have renamed the open reading frame SPBC106.11c as plg7, and hence the encoded protein as Plg7p, to emphasize the similarity of this gene to mammalian group VII phospholipases A2 [30].
Figure 1. The S. pombe genome contains a sequence homologous to the human plasma and type 2 PAF-acetylhydrolases.
Sequence alignment of the S. pombe locus SPBC106.11c, human plasma PAF acetylhydrolase (PLA2g7) and type 2 PAF-acetylhydrolase (HPAFAH2) using ClustalW, BLOSUM program. Characters highlighted in black are exact matches, characters highlighted in gray are similar in identity. Amino acids marked (*) are essential for enzyme activity.
Δplg7 S. pombe have decreased, but residual, PAF acetylhydrolase activity
We targeted the single homolog of PAF acetylhydrolase in S. pombe by sequence analysis to generate a knockout of plg7+. A plasmid construct was generated with approximately 200 bp of the 5’ and 3’ ends of plg7+ flanking the his7+ gene from pEA2 (Fig. 2A). Transformation of strain CHP428 (ATCC# 201399) with the knockout construct yielded colonies that were selected for stable incorporation of the plasmid. Successful integration was analyzed in 85 colonies using PCR primers internal to the his7+ gene and external to plg7+ for detection of Δplg7 (Fig. 2A). Generation of Δplg7 was successful in 5 out of 85 colonies, illustrated in the PCR results from one of the colonies (Fig. 2B). Appropriate sized bands were present in Δplg7 but not in wild type cells; the opposite was true for detection of intact plg7+ (Fig. 2B) and confirmed by sequence analysis (data not shown). These results show the previous inability to recover an insertional mutant at this locus by the S. pombe genome project was not due to a lethal defect caused by an absence of Plg7p.
Figure 2. Δplg7 cells have decreased but residual PAF-AH activity.
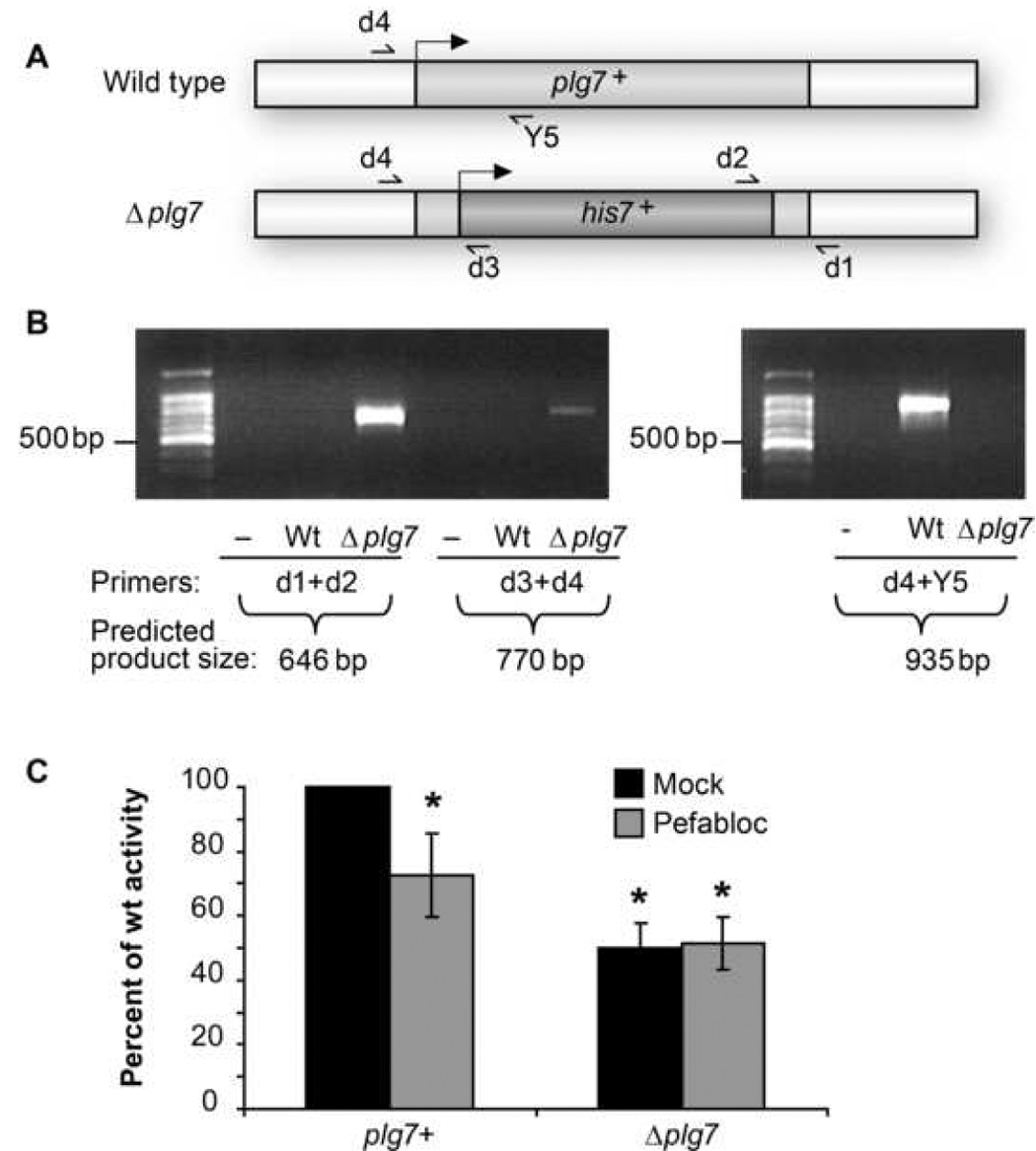
(A) Gene deletion scheme to replace plg7+ with his7+ in S. pombe strain CHP428. Approximately 200 base pairs of both ends of plg7+ were added to a construct with the his7+ gene. 5’ ends of the genes are indicated with black arrows. (B) PCR analysis of plg7+::his7+ gene replacement in wild type (Wt) and Δplg7 cells using genomic and his7+ specific primers; (−) = negative PCR reaction. PCR detecting intact genomic plg7+ in wild type and Δplg7 cells. (C) PAF-AH activity assay of OD (550 nm) normalized wild type and Δplg7 cells, lysed and pre-treated with mock or 100 µM Pefabloc SC. Values are expressed relative to mock treated wild type lysates, n=2 in duplicate * = P<0.001 vs plg7+ (One Way Anova, Student t-test).
We found that Δplg7 displayed reduced PAF hydrolytic activity compared to wild type S. pombe, but that half of this activity remained after plg7+ deletion (Fig. 2C). Accordingly, the selective PAF acetylhydrolase inhibitor Pefabloc SC [31] reduced wild type PAF hydrolysis by 50%, and this residual hydrolytic activity in Δplg7 was not sensitive to Pefabloc inhibition. Similar results were obtained in experiments using the PAF acetylhydrolase inhibitor [32] methyl arachidonoyl fluorophosphonate (MAFP; data not shown). A significant change in phenotype of Δplg7 versus wild type was not observed using various challenges (heavy metal, oxidative stress, and temperature). S. pombe therefore constitutively express at least two enzymes with phospholipase activity against short chain phospholipids, with only Plg7p containing a catalytically essential serine residue.
plg7+ expressed in Saccharomyces cerevisiae encodes a functional PAF acetylhydrolase
S. cerevisiae are as distantly related to S. pombe as humans are to either yeast, and a search of the S. cerevisiae genome revealed no homologous sequence to plg7 or mammalian PAF acetylhydrolase genes (not shown). Accordingly, the lysate of wild type S. cerevisiae INVSc1 hydrolyzed PAF significantly less well (16±6 vs. 196±54 dpm/h/µg) than a lysate of a wild type S. pombe (CHP 428), suggesting this lower background would reveal whether plg7p is active and has a role in protecting cells against oxidative stress. We cloned plg7 and a mutant (Ser257Ala), which replaces what is predicted to be the active site serine, into the S. cerevisiae pYES-DEST52 vector that introduced V5 and 6xHis tags at the C-terminus. We transformed these clones into S. cerevisiae strain DY 1838 (Fig. 3A) and found both clones expressed proteins at the predicted size of 55 kDa. The locus SPBC106.11c therefore is a functional gene, now plg7+, encoding the protein Plg7p.
Figure 3. plg7+ encodes a protein with Ser257 dependent PLA2 activity.
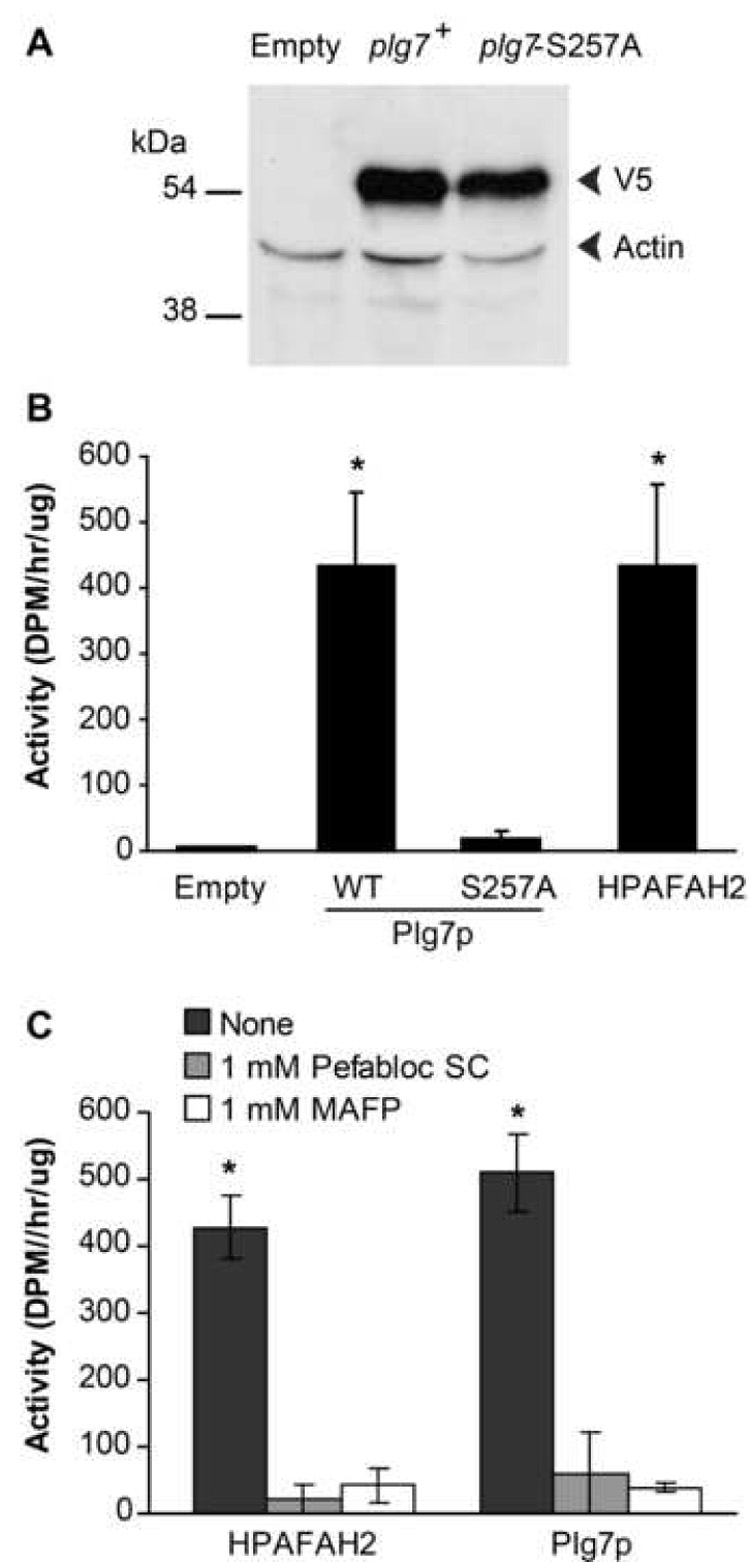
(A) Western blot of lysates from empty vector, plg7+, or plg7-S257A-pDEST52 transformed DY 1838 cells using a combination of antibodies against the V5 epitope tag (upper band) and a diluted antibody to actin (lower band). (B) PAF-AH activity assay of cell lysates from empty vector, non-tagged plg7+, plg7-S257A, or human PAFAH2-pDEST52 transformed INVSc1 cells, normalized by total protein. (C) PAF-AH activity assay of cell lysates from non-tagged plg7+ or human PAFAH2-pDEST52 DY 1838 transformed cells, treated with 1 mM Pefabloc SC or MAFP. Results for B: n=4, in duplicate * = P<0.05 vs empty, Plg7p-S257A; results for C: n=3, in duplicate * = P<0.05 vs Pefabloc SC, MAFP (One Way ANOVA, Tukey Test).
We determined whether Plg7p is a catalytically active enzyme, but found that the chimera Plg7p/V5/6xHis in crude lysates or after purification on a chelation affinity column did not hydrolyze PAF (data not shown). Similarly, expression constructs with the V5 and 6xHis tags moved to the amino terminus were without activity, as were constructs containing Protein A tags. However, expression of native Plg7p without modified termini in the S. cerevisiae strain INVSc resulted in increased PAF hydrolytic activity compared to empty vector (Fig. 3B). The activity levels of Plg7p were similar to the human intracellular type 2 PAF acetylhydrolase expressed under the same conditions. We also found that the mutant Plg7p-S257A had little or no activity suggesting the conserved serine residue is essential for catalysis by Plg7p. We tested whether targeted covalent modification of the active site serine by Pefabloc SC [31], a modified sulfonyl fluoride that inhibits PLA2g7 and PAFAH2 encoded enzymes, or methyl arachidonyl fluorophosphonate [32] would inhibit Plg7p. We found that each reagent greatly reduced, but did not abolish, Plg7p and HPAFAH2 activity in crude lysates (Fig. 2C). We attribute the residual activity to endogenous serine- and Ca++- independent esterolytic activity of unknown origin.
We tested the ability of Plg7p to hydrolyze the synthetic oxidized phospholipid azelaoyl phosphatidylcholine. This cytotoxic phospholipid arises from oxidative fragmentation of the 9,10 bond of sn-2 oleoyl, linoleoyl and linolenoyl residues, and is the prominent phospholipid fragmentation product of phospholipids containing these residues [33]. We found that a crude S. cerevisiae Plg7p lysate hydrolyzed azelaoyl phosphatidylcholine (0.6 ± 0.1 nmol/mg/h), while the S257 mutant had little activity (0.03 ± 0.02 nmol/mg/h) in this assay.
S. cerevisiae expressing Plg7p are protected against oxidative death
Over-expression of type II PAF acetylhydrolase protects mammalian cells against oxidative stress and death [16, 17], potentially because its short-chained phospholipid substrates [13, 34, 35] either initiate apoptosis via the mitochondria-dependent apoptotic caspase cascade [11] or from Ca++ overload subsequent to PAF receptor activation [36]. To avoid the complication of receptor signaling in mammalian cells, we used the yeast transition metal-dependent oxidation model developed by Avery et al. [37] to test whether Plg7p expressed in S. cerevisiae conferred protection from oxidant injury. We found (Fig. 4A) that supplementation of the wild type S. cerevisiae strain INVSc1 with linolenic acid (C18:3) caused a time dependent increase in fluorescence from Bodipy® 581/591 C11, a membrane-intercalated indicator lipid that shifts its fluorescence in response to oxidation. The addition of the transition metal Cu+ alone to INVSc1 did not initiate membrane oxidation, but a combination of C18:3 supplementation followed by exposure to Cu+ increased the fluorescence over that produced by C18:3 supplementation alone.
Figure 4. S. cerevisiae supplemented with linolenic acid is sensitive to oxidation.
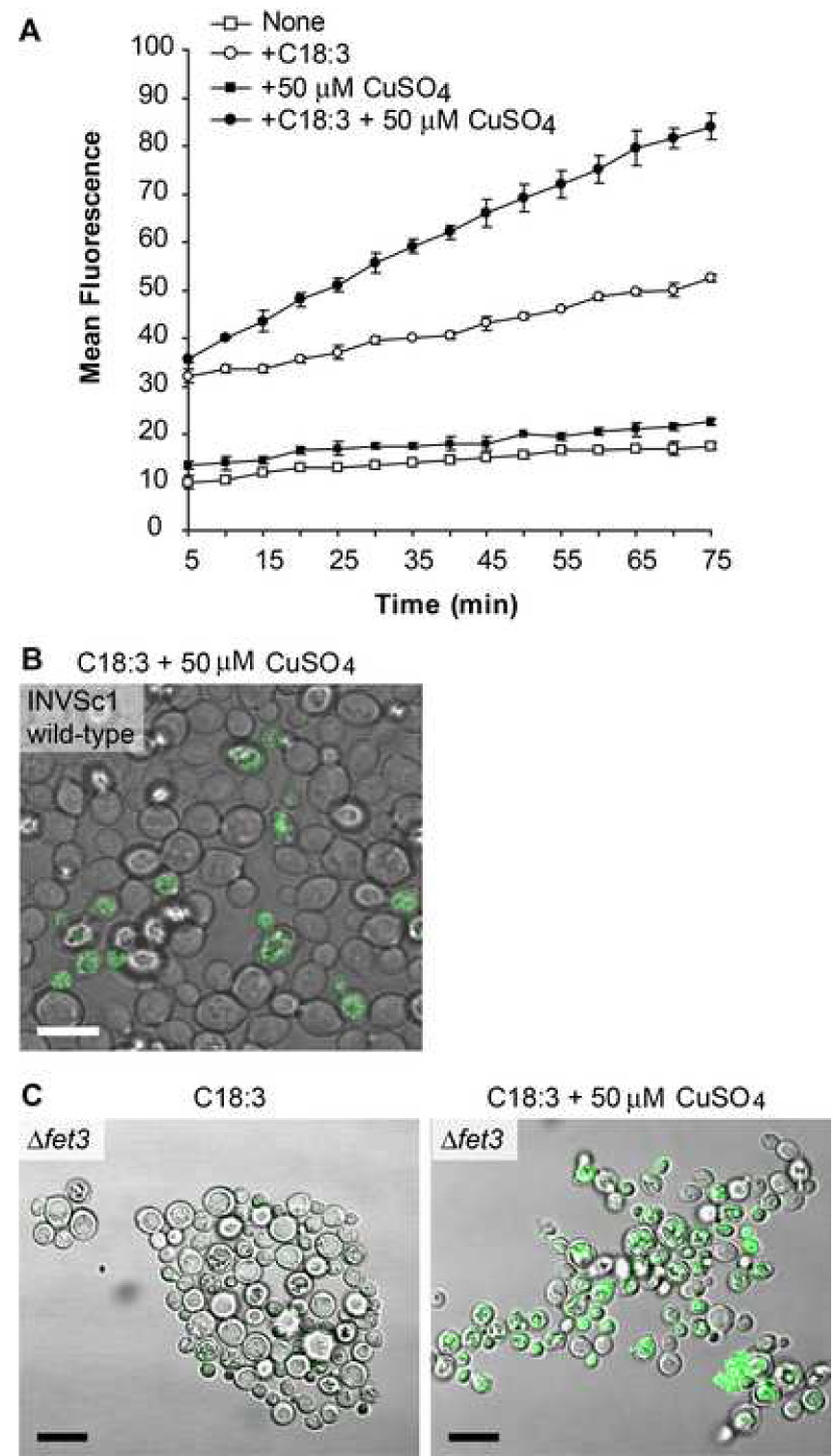
(A) INVSc1 wildtype cells loaded with the lipophilic dye Bodipy® 581/591 C11 that fluoresces after oxidation were examined by fluorimetry using 488/20 nm bandwidth excitation, 528/20 nm bandwidth emission in a representative experiment done in duplicate. Key: Non treated (open square), copper treated (filled square), C18:3 supplemented (open circle), and C18:3 supplemented and copper treated (filled circle). (B) INVSc1 strain was supplemented with C18:3, loaded with Bodipy® 581/591 C11, and treated with 50 µM CuSO4 for 60 min. Cells were visualized by confocal microscopy using a 488 nm excitation laser. (C) fet3Δ cells supplemented with C18:3 and loaded with Bodipy® 581/591 C11 were visualized after 60 min of mock or 50 µM CuSO4 treatment. Scale bar = 10 µm.
We used confocal microscopy to image the oxidative environment detected by the Bodipy® 581/591 C11 dye, and found punctate intracellular staining with a diffuse halo closely associated with the plasma membrane (Fig. 4B). Remarkably, there was great variation among these genetically identical cells in the level of membrane oxidative stress that individual cells experienced, with some cells apparently being completely unaffected by Cu+ exposure. S. cerevisiae peroxiredoxins metabolize hydroperoxides [38] and lipid hydroperoxides [39] to reduce oxidative stress that can vary with the age of the culture [40], and previous work [41] shows there is significant cell cycle variation in oxidant sensitivity in this system with a 5-fold variation in a critical Cu+ resistance protein with cell cycle and cell replicative age [42, 43]. Alternatively, we considered that intracellular Cu+ might be limiting, and hence limit oxidant stress in some of the cells, because Cu+ limits its own uptake [44]. We therefore used a fet3Δ strain that displays extreme sensitivity to Cu+ due to abnormalities in Fe/Cu homeostasis [41, 45]. The fet3Δ cells displayed increased levels of Bodipy® 581/591 C11 fluorescence in an oxidized lipid environment when supplemented with C18:3 and treated with Cu+ (Fig. 4C), but still these cells were heterogeneous in their response to oxidative stress.
We reduced the effect of the variation of oxidative stress among individual cells [41, 43] by plating and immobilizing INVSc1 to assess colony number, an approach where the background arising from cells not subjected to oxidative stress is quantifiable. We treated INVSc1 with Cu+ for 0, 10 or 60 min in culture and then plated serial dilutions to immobilize the cells. Exposure to Cu+ reduced the number of viable cells in a time-dependent way (Fig. 5A), and cells supplemented with C18:3 prior to this exposure displayed enhanced sensitivity to the transition metal. Introduction of a plg7+ expression plasmid into copper challenged, C18:3-supplemented INVSc1 suppressed Cu+ toxicity (Fig. 5B). Quantitation showed the presence of plg7+ resulted in an average 6-fold increase in viability compared to no plasmid (n=3; p < 0.05).
Figure 5. plg7+ protects S. cerevisiae against oxidative stress.
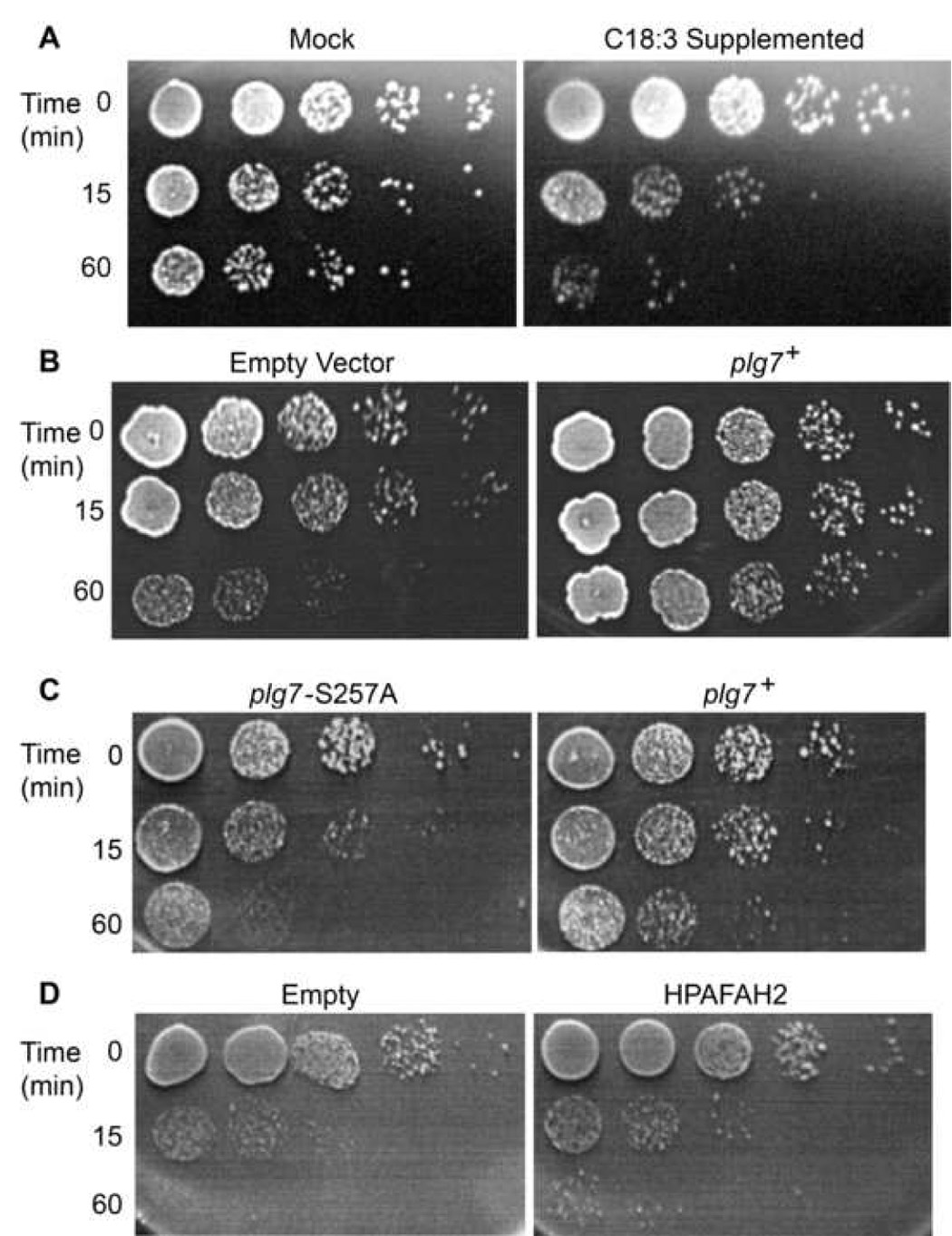
(A) Viability assay of INVSc1 cells −/+ C18:3 supplementation, treated with 50 µM CuSO4 for 0, 10 or 60 min, diluted 10-fold serially and plated on YEPD plates. (B) C18:3 supplemented INVSc1 cells were transformed with empty vector or plg7+pDEST52 and challenged with 50 µM CuSO4 for the stated times, then serially diluted and plated to test viability. (C) Viability assay using fet3Δ cells transformed with plg7+, plg7-S257A. (D) HPAFAH2-pDEST52, or empty vector, supplemented with 1 mM C18:3, treated with 50 µM CuSO4, serially diluted and grown on YEPD plates for 4 days at room temperature.
We challenged C18:3-supplemented fet3Δ cells expressing Plg7p with Cu+ and found that cells also had increased viability compared to cells expressing the inactive Plg7p-S257A (Fig. 5C). Similar levels of oxidation were detected by Bodipy® 581/591 C11 fluorimetry in both Plg7p and Plg7p-S257A expressing cells (not shown), so each strain encountered the same level of oxidative stress. We observed similar results in fet3Δ expressing HPAFAH2 compared to cells with empty vector (Fig. 5D). These results show that S. cerevisiae cells are sensitive to oxidative death when supplemented with a polyunsaturated fatty acid, and that viability can be enhanced by over-expression of active Plg7p.
Discussion
The genome of the yeast S. pombe, a fission yeast distantly related to both animals and the budding yeast S. cerevisiae, has been fully sequenced. This revealed it to be the smallest sequenced eukaryotic genome, yet it contains genes with introns, genes homologous to human disease genes, and clusters of genes regulating cell function related to those found in higher eukaryotes [22]. The Gene Ontology Project (http:www.geneontology.org) infers that SPBC106.11c should be a member of the phospholipase family, but the function of this locus remained undefined because it is among the ~10% of the potential genes that could not be ablated in a high throughput insertional mutagenesis screen [46]. Expression tagging shows the SPBC106.11c locus is among the several hundred genes induced in response to H2O2 and Cd++ exposure [47], and so may play a cytoprotective role.
We cloned the S. pombe open reading frame SPBC106.11c and expressed it in S. cerevisiae, which lacks confounding homologous sequences, to find that this locus is a functional gene encoding an enzymatically active member of the group VII phospholipase A2 family [30]. We found that, like the mammalian group VII enzymes, the S. pombe member Plg7p was Ca++ -independent and it required the Ser residue in the midst of the GXSXG lipase motif for catalysis. Additional evidence that Plg7p contains essential features of the mammalian homolog is that the selective serine-directed inhibitors Pefabloc SC and methyl arachidonyl fluorophosphonate that irreversibly inhibit human plasma PAF acetylhydrolase also inhibited Plg7p.
We ectopically expressed Plg7p in S. cerevisiae to test its function in a system where lipid peroxidation is lethal, but yet also does not employ PAF as a signaling entity. Polyunsaturated fatty acids can be incorporated into S. cerevisiae lipid by supplementing their growth media with linoleate, rendering the cells more susceptible to Cu+-induced lipid peroxidation and cell death [48]. We found by serial plating that S. cerevisiae expressing Plg7p were protected against this oxidative stress, and that this protection required the deduced active site serine. We found a similar level of protection when we expressed the mammalian phospholipase A2 group VII enzyme PAFAH2. The results from this reduced system indicate that membrane lipid peroxidation and death can be suppressed after an oxidative attack has been initiated, and that both the human and S. pombe enzyme act in the same way to hydrolyze structurally damaged phospholipids and maintain viability.
The group VII family of phospholipases A2 was originally described in studies of PAF catabolism [49], but their role in inflammation or homeostasis in an oxidizing environment has not yet been fully defined. One family member, the liver type II gene (PAFAH2), recently has been genetically ablated to discover that the mice develop and behave normally, but that repair of liver after CCl4 damage is delayed [50]. In contrast, genetic ablation of a C. elegans group VII phospholipase A2 ortholog interfered with normal epithelial morphogenesis [21]. Over-expression studies show the mammalian enzymes protect against complex disease and apoptosis [51–56], protect skin epidermis from UVB irradiation [17], and protect transfected CHO cells from exogenous peroxides [16]. However, whether the process(es) affected by the PAF acetylhydrolases in these studies derive from the signaling role of PAF or from the deleterious effect of oxidized phospholipids, and hence whether the relevant substrate(s) of the enzymes are PAF or oxidized phospholipids, cannot be easily be distinguished in mammalian systems that use PAF as a signaling molecule.
The natural habitat of the budding and fission yeasts S. cerevisiae and S. pombe, respectively, are largely undefined. S. cerevisiae inhabits the skin of grapes, likely a consequence of cultivation, but also exists as a free-living organism in soil beneath broad-leafed trees [57]. These free-living cells encounter unknown and uncontrollable environments far different from that established in laboratories. The lipid composition of S. cerevisiae grown in culture is simple because these cells lack a dehydrogenase other than the Δ9 desaturase that generates pamitoleate and oleate [2]. Defined culture media contains no polyunsaturated fatty acids, and so neither do laboratory-grown yeast [58]. In contrast, S. cerevisiae [4, 5] and S. pombe [6] grown outside the laboratory do contain polyunsaturated fatty acids because polyunsaturated fatty acids are abundant lipids in plants and their seeds [59]. The must of crushed grapes contains micromolar levels of free linoleate (C18:2)—the primary polyunsaturated fatty acid of plants—and an equivalent amount esterified as linoleoyl-phosphatidylcholine [60]. Depending on the variety [61], crushed grapes contain a third to half as much linolenate (C18:3) as linoleate. Ethanol generated by the fermentation of grape must, malt wort, and cereal flour reduces yeast viability [62]. This ethanol-induced loss of viability is ameliorated by increasing membrane fluidity by capturing environmental polyunsaturated fatty acids [62], so free living yeast will contain oxidizable polyunsaturated fatty acids.
Yeast also encounter oxidized polyunsaturated fatty acids, oxidized polyunsaturated phospholipids, and phospholipid cleavage products with short and functionalized sn-2 residues in their environment. Plants contain 9-lipoxygenase (Lox1) and 13-lipoxygenase (Lox2) that form free hydroperoxy fatty acids, but the preferred substrates of both enzymes are esterified fatty acyl esters, neutral lipids and phosphatidylcholine [63]. Accordingly, the vast majority of these enzymatic lipid hydroperoxide products in plants are esterified [64], and esterified fatty acyl hydroperoxides can account for up to 6% of the total polyunsaturated fatty acyl pool [65, 66].
Plants also contain a fatty acid hydroperoxide lyase that cleaves the carbon backbone of free and esterified [67] hydroperoxy lipids adjacent to the hydroperoxy function to form two aldehyde-bearing fragments [65]. Cleavage of free or esterified 9-hydroperoxy fatty acids by this enzyme generates precursors, derived from the ω-end of the fatty acid, used in the synthesis of oxilipins and volatile organic molecules that signal the plant, its neighbors, and beneficial insects [68–70]. This cleavage simultaneously generates phospholipids with a 9-carbon aldehyde esterified at the sn-2 position that can then be oxidized to the acid, which are the azelaoyl phosphatidylcholines. S. pombe and S. cerevisiae internalize the oxidatively- and enzymatically-generated short-chained phospholipids they encounter in their environment in a process requiring ATP cassette proteins and the interacting gene product lem3p/ROS3p [9, 71]. This provides a ready-made source of ethanolamine and choline headgroups [72, 73], but also means they must be prepared to appropriately metabolize cytotoxic short-chained phospholipids. This, we now show, is accomplished by Plg7p.
Even though S. pombe has one of the smallest eukaryote genomes—encoding just 4,824 proteins [22]—among this handful of genes is one that codes for an enzyme that specifically hydrolyzed oxidized phospholipids and protects against oxidative cell death. Actually, our data suggest that at least one other activity is also present that degrades or remodels incoming phospholipid oxidation products. This metabolic function is retained by the homologous group VII mammalian phospholipases A2, suggesting the original, and potentially remaining, purpose of PAF acetylhydrolases is catabolism of phospholipid oxidation products.
Acknowledgements
We are extremely grateful for the insightful discussions and materials from Drs. David Stillman, Dennis Winge, Jerry Kaplan and John Weis. We appreciate many fruitful interactions with Gopal Marathe, and we thank Diana Lim for preparation of the figures. We appreciate the aid of Jiawei Chen and Vincient R. Barnes, Sr. in the hydrolysis experiments, and Rui Chen and Lili Yang in the mass spectrometry experiments along with the expert advice and services of Renliang Zhang of the Cleveland Clinic mass spectrometry core II. This work was supported by NIH 1 R01 HL44513 and 1 P50HL087018 (T.M.M.).
Abbreviations
- HAzPC
hexadecyl azelaoyl phosphatidylcholine
- MAFP
methyl arachidonoyl fluorophosphonate
- PAF
Platelet-activating Factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sipiczki M. Where does fission yeast sit on the tree of life? Genome biology. 2000;1 doi: 10.1186/gb-2000-1-2-reviews1011. 1011.1011-1011-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stukey JE, et al. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J Biol Chem. 1989;264:16537–16544. [PubMed] [Google Scholar]
- 3.Niki E, et al. Lipid peroxidation: mechanisms, inhibition, and biological effects. Biochem Biophys Res Commun. 2005;338:668–676. doi: 10.1016/j.bbrc.2005.08.072. [DOI] [PubMed] [Google Scholar]
- 4.Valero E, et al. Influence of pre-fermentative treatment on the fatty acid content of Saccharomyces cerevisiae (M(3)30-9) during alcoholic fermentation of grape must. J Biosci Bioeng. 2001;91:117–122. doi: 10.1263/jbb.91.117. [DOI] [PubMed] [Google Scholar]
- 5.Blagovic B, et al. Lipid analysis of the plasma membrane and mitochondria of brewer's yeast. Folia Microbiol (Praha) 2005;50:24–30. doi: 10.1007/BF02931290. [DOI] [PubMed] [Google Scholar]
- 6.McDonough VM, Roth TM. Growth temperature affects accumulation of exogenous fatty acids and fatty acid composition in Schizosaccharomyces pombe. Antonie Van Leeuwenhoek. 2004;86:349–354. doi: 10.1007/s10482-004-0515-0. [DOI] [PubMed] [Google Scholar]
- 7.Elvington SM, et al. Fluorescent, acyl chain-labeled phosphatidylcholine analogs reveal novel transport pathways across the plasma membrane of yeast. J Biol Chem. 2005;280:40957–40964. doi: 10.1074/jbc.M507926200. [DOI] [PubMed] [Google Scholar]
- 8.Zaremberg V, McMaster CR. Differential partitioning of lipids metabolized by separate yeast glycerol-3-phosphate acyltransferases reveals that phospholipase D generation of phosphatidic acid mediates sensitivity to choline-containing lysolipids and drugs. J Biol Chem. 2002;277:39035–39044. doi: 10.1074/jbc.M207753200. [DOI] [PubMed] [Google Scholar]
- 9.Hanson PK, et al. Lem3p is essential for the uptake and potency of alkylphosphocholine drugs, edelfosine and miltefosine. J Biol Chem. 2003;278:36041–36050. doi: 10.1074/jbc.M305263200. [DOI] [PubMed] [Google Scholar]
- 10.Fragopoulou E, et al. Biological activity of total lipids from red and white wine/must. J Agric Food Chem. 2001;49:5186–5193. doi: 10.1021/jf0106392. [DOI] [PubMed] [Google Scholar]
- 11.Chen R, et al. Cytotoxic phospholipid oxidation products: Cell death from mitochondrial damage and the intrinsic caspase cascade. J Biol Chem. 2007;282:24842–24850. doi: 10.1074/jbc.M702865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg ME, et al. The lipid whisker model of the structure of oxidized cell membranes. J Biol Chem. 2008;283:2385–2396. doi: 10.1074/jbc.M707348200. [DOI] [PubMed] [Google Scholar]
- 13.Stremler KE, et al. Human plasma platelet-activating factor acetylhydrolase: Oxidatively-fragmented phospholipids as substrates. J. Biol. Chem. 1991;266:11095–11103. [PubMed] [Google Scholar]
- 14.Kriska T, et al. Phospholipase action of platelet-activating factor acetylhydrolase, but not paraoxonase-1, on long fatty acyl chain phospholipid hydroperoxides. J Biol Chem. 2007;282:100–108. doi: 10.1074/jbc.M608135200. [DOI] [PubMed] [Google Scholar]
- 15.Stafforini DM, et al. Release of free F2-isoprostanes from esterified phospholipids is catalyzed by intracellular and plasma platelet-activating factor acetylhydrolases. J Biol Chem. 2006;281:4616–4623. doi: 10.1074/jbc.M507340200. [DOI] [PubMed] [Google Scholar]
- 16.Matsuzawa A, et al. Protection against oxidative stress-induced cell death by intracellular platelet-activating factor-acetylhydrolase II. J Biol Chem. 1997;272:32315–32320. doi: 10.1074/jbc.272.51.32315. [DOI] [PubMed] [Google Scholar]
- 17.Marques M, et al. Identification of platelet-activating factor acetylhydrolase II in human skin. J Invest Dermatol. 2002;119:913–919. doi: 10.1046/j.1523-1747.2002.01859.x. [DOI] [PubMed] [Google Scholar]
- 18.Tjoelker LW, et al. Plasma platelet-activating factor acetylhydrolase is a secreted phospholipase A2 with a catalytic triad. J Biol Chem. 1995;270:25481–25487. doi: 10.1074/jbc.270.43.25481. [DOI] [PubMed] [Google Scholar]
- 19.Hattori K, et al. cDNA cloning and expression of intracellular platelet-activating factor (PAF) acetylhydrolase II. Its homology with plasma PAF acetylhydrolase. J Biol Chem. 1996;271:33032–33038. doi: 10.1074/jbc.271.51.33032. [DOI] [PubMed] [Google Scholar]
- 20.Karabina SA, Ninio E. Plasma PAF-acetylhydrolase: an unfulfilled promise? Biochimica et biophysica acta. 2006;1761:1351–1358. doi: 10.1016/j.bbalip.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Inoue T, et al. Type II platelet-activating factor-acetylhydrolase is essential for epithelial morphogenesis in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2004;101:13233–13238. doi: 10.1073/pnas.0405507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 23.Ling M, et al. A rapid and reliable DNA preparation method for screening a large number of yeast clones by polymerase chain reaction. Nucleic Acids Res. 1995;23:4924–4925. doi: 10.1093/nar/23.23.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen HR, et al. Spheroplast preparation facilitates PCR screening of yeast sequence. Bio Techniques. 1995;19:744–746. 748. [PubMed] [Google Scholar]
- 25.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 26.Podrez EA, et al. Identification of a novel family of oxidized phospholipids that serve as ligands for the macrophage scavenger receptor CD36. J Biol Chem. 2002;277:38503–38516. doi: 10.1074/jbc.M203318200. [DOI] [PubMed] [Google Scholar]
- 27.Tjoelker LW, et al. Anti-inflammatory properties of a platelet-activating factor acetylhydrolase. Nature. 1995;374:549–553. doi: 10.1038/374549a0. [DOI] [PubMed] [Google Scholar]
- 28.Arai H, et al. Platelet-activating factor acetylhydrolase (PAF-AH) J Biochem (Tokyo) 2002;131:635–640. doi: 10.1093/oxfordjournals.jbchem.a003145. [DOI] [PubMed] [Google Scholar]
- 29.Wei Y, et al. Structure of a microbial homologue of mammalian platelet-activating factor acetylhydrolases: Streptomyces exfoliatuslipase at 1.9 A resolution. Structure. 1998;6:511–519. doi: 10.1016/s0969-2126(98)00052-5. [DOI] [PubMed] [Google Scholar]
- 30.Six DA, Dennis EA. The expanding superfamily of phospholipase A(2) enzymes: classification and characterization. Biochim biophys acta. 2000;1488:1–19. doi: 10.1016/s1388-1981(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 31.Dentan C, et al. Pefabloc, 4-[2-aminoethyl]benzenesulfonyl fluoride, is a new, potent nontoxic and irreversible inhibitor of PAF-degrading acetylhydrolase. Biochim biophys acta. 1996;1299:353–357. doi: 10.1016/0005-2760(95)00226-x. [DOI] [PubMed] [Google Scholar]
- 32.Kell PJ, et al. Inhibition of platelet-activating factor (PAF) acetylhydrolase by methyl arachidonyl fluorophosphonate potentiates PAF synthesis in thrombin-stimulated human coronary artery endothelial cells. J Pharmacol Exp Ther. 2003;307:1163–1170. doi: 10.1124/jpet.103.055392. [DOI] [PubMed] [Google Scholar]
- 33.Itabe H, et al. Identification of a 2-azelaoylphosphatidylcholine as one of the cytotoxic products generated during oxyhemoglobin-induced peroxidation of phosphatidylcholine. Biochim. Biophys. Acta. 1988;962:8–15. [PubMed] [Google Scholar]
- 34.Stremler KE, et al. An oxidized derivative of phosphatidylcholine is a substrate for the platelet-activating factor acetylhydrolase from human plasma. J. Biol. Chem. 1989;264:5331–5334. [PubMed] [Google Scholar]
- 35.Hattori K, et al. Purification and characterization of platelet-activating factor acetylhydrolase II from bovine liver cytosol. J Biol Chem. 1995;270:22308–22313. doi: 10.1074/jbc.270.38.22308. [DOI] [PubMed] [Google Scholar]
- 36.Silomon M, et al. Role of platelet-activating factor in hepatocellular Ca2+ alterations during hemorrhagic shock. J surg res. 1997;72:101–106. doi: 10.1006/jsre.1997.5171. [DOI] [PubMed] [Google Scholar]
- 37.Avery SV, et al. Copper toxicity towards Saccharomyces cerevisiae: dependence on plasma membrane fatty acid composition. Appl Environ Microbiol. 1996;62:3960–3966. doi: 10.1128/aem.62.11.3960-3966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogusucu R, et al. Reactions of yeast thioredoxin peroxidases I and II with hydrogen peroxide and peroxynitrite: rate constants by competitive kinetics. Free radic biol med. 2007;42:326–334. doi: 10.1016/j.freeradbiomed.2006.10.042. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka T, et al. GPX2, encoding a phospholipid hydroperoxide glutathione peroxidase homologue, codes for an atypical 2-Cys peroxiredoxin in I. J Biol Chem. 2005;280:42078–42087. doi: 10.1074/jbc.M508622200. [DOI] [PubMed] [Google Scholar]
- 40.Lee JH, Park JW. Role of thioredoxin peroxidase in aging of stationary cultures of I. Free radic res. 2004;38:225–231. doi: 10.1080/10715760310001649009. [DOI] [PubMed] [Google Scholar]
- 41.Howlett NG, Avery SV. Flow cytometric investigation of heterogeneous copper-sensitivity in asynchronously grown I. FEMS Microbiol Lett. 1999;176:379–386. doi: 10.1111/j.1574-6968.1999.tb13687.x. [DOI] [PubMed] [Google Scholar]
- 42.Sumner ER, et al. Cell cycle- and age-dependent activation of Sod1p drives the formation of stress resistant cell subpopulations within clonal yeast cultures. Mol Microbiol. 2003;50:857–870. doi: 10.1046/j.1365-2958.2003.03715.x. [DOI] [PubMed] [Google Scholar]
- 43.Bishop AL, et al. Phenotypic heterogeneity can enhance rare-cell survival in 'stress-sensitive' yeast populations. Mol Microbiol. 2007;63:507–520. doi: 10.1111/j.1365-2958.2006.05504.x. [DOI] [PubMed] [Google Scholar]
- 44.Eide DJ. The molecular biology of metal ion transport in Saccharomyces cerevisiae. Annu Rev Nutr. 1998;18:441–469. doi: 10.1146/annurev.nutr.18.1.441. [DOI] [PubMed] [Google Scholar]
- 45.Shi X, et al. Fre1p Cu2+ reduction and Fet3p Cu1+ oxidation modulate copper toxicity in I. J Biol Chem. 2003;278:50309–50315. doi: 10.1074/jbc.M307019200. [DOI] [PubMed] [Google Scholar]
- 46.Decottignies A, et al. I essential genes: a pilot study. Genome Res. 2003;13:399–406. doi: 10.1101/gr.636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen D, et al. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howlett NG, Avery SV. Induction of lipid peroxidation during heavy metal stress in Saccharomyces cerevisiae and influence of plasma membrane fatty acid unsaturation. Appl Environ Microbiol. 1997;63:2971–2976. doi: 10.1128/aem.63.8.2971-2976.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farr RS, et al. Preliminary studies of an acid-labile factor (ALF) in human sera that inactivates platelet-activating factor (PAF) Clin. Immunol. Immnopathol. 1980;15:318–330. doi: 10.1016/0090-1229(80)90044-6. [DOI] [PubMed] [Google Scholar]
- 50.Kono N, et al. Protection against oxidative stress-induced hepatic injury by intracellular type II platelet-activating factor acetylhydrolase by metabolism of oxidized phospholipids in vivo. J Biol Chem. 2008;283:1628–1636. doi: 10.1074/jbc.M708622200. [DOI] [PubMed] [Google Scholar]
- 51.Quarck R, et al. Adenovirus-mediated gene transfer of human platelet-activating factor- acetylhydrolase prevents injury-induced neointima formation and reduces spontaneous atherosclerosis in apolipoprotein E-deficient mice. Circulation. 2001;103:2495–2500. doi: 10.1161/01.cir.103.20.2495. [DOI] [PubMed] [Google Scholar]
- 52.Theilmeier G, et al. HDL-associated PAF-AH reduces endothelial adhesiveness in apoE−/− mice. Faseb J. 2000;14:2032–2039. doi: 10.1096/fj.99-1029com. [DOI] [PubMed] [Google Scholar]
- 53.Biancone L, et al. Platelet-activating factor inactivation by local expression of platelet-activating factor acetyl-hydrolase modifies tumor vascularization and growth. Clin Cancer Res. 2003;9:4214–4220. [PubMed] [Google Scholar]
- 54.Iso ON, et al. Adenovirus-mediated gene transfer and lipoprotein-mediated protein delivery of plasma PAF-AH ameliorates proteinuria in rat model of glomerulosclerosis. Mol Ther. 2006;13:118–126. doi: 10.1016/j.ymthe.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 55.Gomes RN, et al. Exogenous platelet-activating factor acetylhydrolase reduces mortality in mice with systemic inflammatory response syndrome and sepsis. Shock. 2006;26:41–49. doi: 10.1097/01.shk.0000209562.00070.1a. [DOI] [PubMed] [Google Scholar]
- 56.Umemura K, et al. Neuroprotective role of transgenic PAF-acetylhydrolase II in mouse models of focal cerebral ischemia. Stroke. 2007;38:1063–1068. doi: 10.1161/01.STR.0000257981.09329.d2. [DOI] [PubMed] [Google Scholar]
- 57.Sniegowski PD, et al. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 58.Schneiter R, et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dorne AJ, et al. Polar Lipid Composition of a Plastid Ribosome-Deficient Barley Mutant. Plant Physiol. 1982;69:1467–1470. doi: 10.1104/pp.69.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beltran G, et al. Effect of fermentation temperature and culture media on the yeast lipid composition and wine volatile compounds. Int J Food Microbiol. 2008;121:169–177. doi: 10.1016/j.ijfoodmicro.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 61.Yunoki K, et al. Fatty acids in must prepared from 11 grapes grown in Japan: comparison with wine and effect on fatty acid ethyl ester formation. Lipids. 2005;40:361–367. doi: 10.1007/s11745-006-1395-z. [DOI] [PubMed] [Google Scholar]
- 62.Alexandre H, et al. Ethanol adaptation mechanisms in Saccharomyces cerevisiae. Biotechnol Appl Biochem. 1994;20(Pt 2):173–183. [PubMed] [Google Scholar]
- 63.Garbe LA, et al. Dual positional and stereospecificity of lipoxygenase isoenzymes from germinating barley (green malt): biotransformation of free and esterified linoleic acid. J Agric Food Chem. 2006;54:946–955. doi: 10.1021/jf051993t. [DOI] [PubMed] [Google Scholar]
- 64.Hubke H, et al. Characterization and quantification of free and esterified 9- and 13-hydroxyoctadecadienoic acids (HODE) in barley, germinating barley, and finished malt. J Agric Food Chem. 2005;53:1556–1562. doi: 10.1021/jf048490s. [DOI] [PubMed] [Google Scholar]
- 65.Weichert H, et al. Metabolic profiling of oxylipins in germinating cucumber seedlings--lipoxygenase-dependent degradation of triacylglycerols and biosynthesis of volatile aldehydes. Planta. 2002;215:612–619. doi: 10.1007/s00425-002-0779-4. [DOI] [PubMed] [Google Scholar]
- 66.Griffiths G, et al. Lipid hydroperoxide levels in plant tissues. J Exp Bot. 2000;51:1363–1370. [PubMed] [Google Scholar]
- 67.Kandzia R, et al. On the specificity of lipid hydroperoxide fragmentation by fatty acid hydroperoxide lyase from Arabidopsis thaliana. J Plant Physiol. 2003;160:803–809. doi: 10.1078/0176-1617-01026. [DOI] [PubMed] [Google Scholar]
- 68.Arimura G, et al. Herbivore-induced, indirect plant defences. Biochimica et biophysica acta. 2005;1734:91–111. doi: 10.1016/j.bbalip.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 69.Matsui K. Green leaf volatiles: hydroperoxide lyase pathway of oxylipin metabolism. Curr Opin Plant Biol. 2006;9:274–280. doi: 10.1016/j.pbi.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 70.Wei J, et al. Plants attract parasitic wasps to defend themselves against insect pests by releasing hexenol. PLoS ONE. 2007;2:e852. doi: 10.1371/journal.pone.0000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kato U, et al. A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J Biol Chem. 2002;277:37855–37862. doi: 10.1074/jbc.M205564200. [DOI] [PubMed] [Google Scholar]
- 72.Riekhof WR, et al. Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: the role of P-type ATPases in transport and a broad specificity acyltransferase in acylation. J Biol Chem. 2007;282:36853–36861. doi: 10.1074/jbc.M706718200. [DOI] [PubMed] [Google Scholar]
- 73.Riekhof WR, Voelker DR. Uptake and utilization of lyso-phosphatidylethanolamine by Saccharomyces cerevisiae. J Biol Chem. 2006;281:36588–36596. doi: 10.1074/jbc.M608851200. [DOI] [PubMed] [Google Scholar]



