Abstract
Sarcopenia, skeletal muscle loss during aging, is associated with increased falls, fractures, morbidity, and loss of independence. MicroRNAs (miRNAs) are novel posttranscriptional regulators. The role of miRNAs in cell size regulation after an anabolic stimulus in human skeletal muscle is unknown. We hypothesized that aging would be associated with a differential expression of skeletal muscle primary miRNA (pri-miRNA) and mature miRNA (miR). To test this hypothesis, we used real-time PCR and immunoblotting before and after an anabolic stimulus (resistance exercise + ingestion of a 20-g leucine-enriched essential amino acid solution) to measure the expression of muscle-specific miRNAs (miR-1, miR-133a, and miR-206), upstream regulators (MyoD and myogenin), and downstream targets [insulin-like growth factor-I, histone deacetylase-4, myocyte enhancing factor-2, and Ras homolog enriched in brain (Rheb)] in skeletal muscle of young and older men. Muscle biopsies were obtained at baseline and 3 and 6 h after exercise. At baseline, we found pri-miRNA-1-1, -1-2, -133a-1, and -133a-2 expression elevated in older compared with young men (P < 0.05). Pri-miRNA-1-2, -133a-1, and -133a-2 were reduced at 6 h after exercise only in the young men compared with baseline, whereas pri-miRNA-206 was elevated at different postexercise time points in older and young men (P < 0.05). Compared with baseline, miR-1 was reduced only in the young men, whereas Rheb protein was increased in both age groups after the anabolic stimulus (P < 0.05). We conclude that skeletal muscle primary and mature miRNA expression in young men is readily altered by an anabolic stimulus of resistance exercise + essential amino acid ingestion. However, aging is associated with higher basal skeletal muscle primary miRNA expression and a dysregulated miRNA response after the anabolic stimulus.
Keywords: muscle growth, sarcopenia, posttranscriptional regulators
recent discoveries have shown that small, ∼20-nucleotide, RNA molecules [microRNAs (miRNAs)] have the ability to repress protein synthesis in many cell types, including skeletal muscle (5, 45). miRNAs appear to be important during various cellular processes, such as skeletal muscle proliferation and differentiation (5), fat and amino acid metabolism (13, 23, 34), apoptosis (4, 53), and aging (3, 17, 51). It is thought that one-third of the mammalian genome is regulated by miRNAs (26). Unfortunately, little is known about how miRNAs operate in human skeletal muscle.
miRNAs are initially transcribed as primary miRNAs (pri-miRNAs) and then processed into a ∼70-bp stem-loop precursor miRNA by the enzyme Drosha. The pri-miRNA is transported out of the nucleus by Exportin-5 and then processed by another enzyme (Dicer) into the mature ∼20-bp miRNA (miR). The mature miR is thought to guide the RNA-induced silencing complex to target specific transcripts (7, 25). miRNAs repress gene expression posttranscriptionally by binding to the 3′-untranslated region of a target gene and preventing translation and/or by promoting transcript degradation (36, 46). Several reports have indicated that inhibition occurs rather rapidly at the level of translation initiation (16, 20, 30, 44). Interestingly, enhanced translation initiation is considered one of the primary mechanisms by which protein synthesis is increased in humans during the recovery period after an acute bout of resistance exercise and/or immediately after essential amino acid (EAA) ingestion (9–11, 14).
miRNA-1, -133, and -206 have been identified to be highly expressed in skeletal muscle (1, 27, 42), thereby indicating a possible regulatory role in skeletal muscle adaptation. Recent reports have documented that these miRNAs are altered in response to skeletal muscle wasting (12, 33) and muscle overload (32). McCarthy et al. (33) reported that miR-206 was elevated in dystrophin-deficient (mdx) mouse diaphragm. McCarthy and Esser (32) indicated that miR-1 and -133a were downregulated after 7 days of functional overload in mice and that this downregulation was accompanied by a 45% increase in plantaris muscle weight. Indeed, with validated [e.g., follistatin-like-1 (38), serum response factor (5), DNA polymerase-α (18), histone deacetylase 4 (HDAC4) (5), myocyte enhancing factor-2 (MEF2) (43), Delta (24), and Ras homolog enriched in brain (Rheb) (41)] and predicted [e.g., insulin-like growth factor I (IGF-I), hepatic growth factor, Pax3, and c-Met (http://targetscan.org)] transcriptional targets that influence cell growth and satellite cell function, it is not surprising that miR-1, -133a, and -206 may have a role in regulating adult skeletal muscle size.
The role of miRNAs in various tissue types has been explored using animal models, cell lines, and other organisms. However, only one study has translated the findings of these novel regulators to human skeletal muscle (12), and none has explored miRNA expression or downstream targets in aging or after stimulation of muscle protein synthesis in human skeletal muscle. Our laboratory has reported that heavy resistance exercise (10) and EAAs (14) can increase muscle protein synthesis individually, but the combination further augments this response (9). Therefore, to detect changes in miRNA expression, we sought to use this model to maximally stimulate protein synthesis. We hypothesized that aging would be associated with differential expression of miRNAs (as well as their downstream targets) at rest and after an acute stimulation of skeletal muscle protein synthesis.
MATERIALS AND METHODS
Subjects
We studied six young (29 ± 2 yr old) and six older (70 ± 2 yr old) men. Subject characteristics are described elsewhere (11). Subjects were healthy, as evaluated with a clinical history, physical examination, and laboratory blood and urine tests during the screening process, and were not engaged in any regular exercise training at the time of enrollment. On two separate occasions, subjects performed a one-repetition maximum (1 RM) exercise bout on a leg extension machine (model VR2, Cybex, Medway, MA) in a test for maximal strength. All subjects gave informed written consent before participating in the study, which was approved by the Institutional Review Board of the University of Texas Medical Branch (which is in compliance with the Declaration of Helsinki).
Experimental Design
All subjects were admitted to the General Clinical Research Center of the University of Texas Medical Branch on the day before the exercise study, and a dual-energy X-ray absorptiometry scan (model QDR 4500W, Hologic, Bedford, MA) was performed to measure body composition and lean mass. The subjects were then fed a standard dinner and a snack at 2200. All subjects refrained from exercise for 24 h before study participation and were studied after an overnight fast under basal conditions and at the same time during the procedure (i.e., between 0500 and 1500).
A baseline muscle biopsy was obtained from the lateral portion of the vastus lateralis using a 5-mm Bergström biopsy needle under sterile procedure and local anesthesia (1% lidocaine). The subject was escorted to the leg extension machine and then performed 8 sets of 10 repetitions of two-legged extension exercises set to 70% of their 1 RM. The rest period between sets was 3 min. After the last exercise set, the subject was returned to his hospital bed for the remainder of the study. At 1 h after the completion of exercise, another muscle biopsy was taken. Immediately thereafter, the subject ingested 20 g of a leucine-enriched EAA solution. The composition of the EAA solution has been published previously (11). Subsequent muscle biopsies were sampled at 3 and 6 h after exercise. The baseline and 1-h muscle biopsy and the 3- and 6-h muscle biopsy were sampled from incision sites 1 and 2, respectively, which were ∼7 cm apart. The needle for each muscle biopsy was angled in a different direction to prevent needle biopsy effects from repeated sampling. Muscle tissue was immediately blotted and frozen in liquid nitrogen and stored at −80°C until analysis.
To test our hypothesis, we used only the baseline and 3- and 6-h postexercise muscle biopsies to assess changes in the variables of interest. These two postexercise time points (3 and 6 h) were sampled after the potent anabolic combination of resistance exercise and EAA ingestion.
RNA Isolation
Total RNA was isolated by homogenization of 30–40 mg of tissue with a homogenizing dispenser (T10 Basic Ultra Turrax, IKA, Wilmington, NC) in a solution containing 1.0 ml of TriReagent and 4 μl of polyacryl carrier (Molecular Research Center, Cincinnati, OH). Separation of RNA into an aqueous phase was accomplished using 0.2 ml of chloroform, and 0.50 ml of isopropanol was used for precipitation from the aqueous phase. Extracted RNA was washed with 1 ml of 75% ethanol, dried, and then suspended in a known amount (1.5 μl/mg tissue) of nuclease-free water. RNA was quantified spectrophotometrically (SmartSpec, Bio-Rad, Hercules, CA) at a wavelength of 260 nm. RNA concentration was calculated on the basis of total RNA yield. RNA quality was assessed by RNA agarose gel electrophoresis followed by visualization of the 18S and 28S rRNA bands under ultraviolet light. RNA was treated with DNase using a commercially available kit (DNA-free, Ambion, Austin, TX).
Primary miRNA Expression Analysis
First-strand cDNA synthesis from total RNA (1 μg) was performed using SuperScript III First-Strand Synthesis SuperMix for quantitative RT-PCR (Invitrogen, Carlsbad, CA) according to the manufacturer's directions. Real-time PCR was used to detect primary miRNA transcript levels for human pri-miRNA-1-1, -1-2, -133a-1, -133a-2, and -206, as well as Drosha, Exportin-5, and Dicer genes. Primer sets were designed using the Biology Workbench 3.2 PrimerTm program (http://seqtool.sdsc.edu/CGI/BW.cgi) with a melting temperature of ∼60°C and an amplicon of 150 bp. Primer sequences are listed in Table 1. The University of California, Santa Cruz, Genome Browser in silico PCR tool (http://genome.ucsc.edu) and melt analysis following PCR verified that each primer set amplified only a single product. PCRs were run using Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA) with 2 μl of a 50-fold dilution of cDNA on an ABI 7500 system (Applied Biosystems). Cycle conditions for PCR were as follows: 10 min at 95°C (stage 1), 40 cycles at 95°C for 15 s followed by 60°C for 1 min (stage 2), and melt analysis (stage 3). Cycle threshold (Ct) values were calculated from duplicates of each sample. Raw data were normalized to GAPDH and β2-microglobulin using the geometric mean of these normalization genes (49).
Table 1.
Primer sequences (5′ to 3′) used for quantitative RT-PCR
| Gene |
Primer Sequence |
Accession No. | |
|---|---|---|---|
| Forward | Reverse | ||
| Primary microRNAs | |||
| pri-miR-1-1 | GGCGTCCCGGGGTC | TGTGGAGTGCCCCTCAGT | |
| pri-miR-1-2 | TTTCATGTTTTTACAGCTAACAACTTAGTAA | TAGCTCAGTTTAATTCTCCCTTGG | |
| pri-miR-133a-1 | TAACTGTTTTGGATTCCAAACTAGCA | ACAAATGAAAACGTTGGTTGTCC | |
| pri-miR-133a-2 | CCGACGTCGCTGTTCCT | TGCGGGACCTGCGG | |
| pri-miR-206 | CTTCTGAGATGCGGGCTG | CTCCGGGTGGTACCCTGG | |
| Drosha | ACTGTGGCTGTTTATTTCAAGGG | ATTTTTCAAGCGCATCCATTG | NC000005 |
| Dicer | ATCCCATGATGCGGCC | CTAAATTTGGCAGTTTCTGGTTCC | NC000014 |
| Exportin-5 | GCGCCGAAAGGACCA | AGGGAAGATTCTTAATGTGAACTTCTT | AC000049 |
Primary microRNA sequences were determined from the miRBase sequence data base (http://microrna.sanger.ac.uk/sequences/index.shtml).
Mature miRNA Expression Analysis
miRNAs from the total RNA samples were detected using the mirVana quantitative RT-PCR detection kit and mirVana PCR primer sets for miR-1, -133a, and -206 according to the manufacturer's directions (Ambion). Briefly, an RT reaction was conducted for each RNA sample using specific mirVana RT primers. Each reaction consisted of 30 min of incubation at 37°C and 10 min of enzyme inactivation at 95°C. Samples were immediately placed on ice, and PCR reagents were added to the final RT product using mirVana PCR primers and detected with SYBR Green fluorescence (Invitrogen). Real-time PCR was performed using an iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad) and consisted of a 3-min denaturing step at 95°C followed by 30 cycles at 95°C for 15 s and 60°C for 30 s. A no-template control and a positive control were included with each PCR run. Furthermore, a melt analysis followed each PCR run, and an agarose gel was used to ensure amplification of only a single product. All samples were normalized to an internal control (5S rRNA). 5S, a small nuclear RNA, has shown to be a suitable gene for normalization of miRNA data in skeletal muscle (27).
Calculations
Relative fold changes in mRNA and miRNA within each group and basal differences between groups were determined from the Ct values using the 2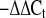 method (29) after normalization to their respective housekeeping genes.
method (29) after normalization to their respective housekeeping genes.
Immunoblotting
Immunoblotting is described elsewhere (10). Briefly, frozen tissue was homogenized and centrifuged for 10 min at 4°C, and supernatant was collected. Total protein concentrations were determined using a modified Bradford assay (Smartspec Plus, Bio-Rad). The supernatant was diluted (1:1) in a 2× sample buffer and then boiled for 3 min at 100°C. Equal amounts of total protein (50 μg) were loaded into each lane, and the samples were separated by electrophoresis (150 V for 60 min) on a 7.5% or 15% polyacrylamide gel (Criterion, Bio-Rad) as determined by the size of the target protein. Each sample was loaded in duplicate, and each gel contained an internal loading control (rodent skeletal muscle) and a molecular weight ladder (Precision Plus, Bio-Rad). After electrophoresis, protein was transferred to a polyvinylidene difluoride membrane (Bio-Rad) at 50 V for 60 min and then incubated in primary and secondary antibodies (see Antibodies). Chemiluminescent solution (ECL Plus, Amersham BioSciences, Piscataway, NJ) was applied to each blot, and optical density was measured with a PhosphoImager (ChemiDoc, Bio-Rad) and subjected to densitometric analysis using Quantity One 4.5.2 software (Bio-Rad). Protein density values were normalized to an internal loading control (which was loaded on each respective blot), and the final protein data were expressed as arbitrary units. Because basal values were not significantly different between the age groups, data are reported as fold change from basal.
Antibodies
The following antibodies were used at 1:500 dilution: Drosha, Dicer, MyoD, myogenin, IGF-I (Santa Cruz Biotechnology, Santa Cruz, CA), Exportin-5 (Abcam, Cambridge, MA), HDAC4, MEF2, and Rheb (Cell Signaling, Danvers, MA). Anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody (Amersham Bioscience) was used at 1:2,000 dilution.
Statistical Analysis
Values are means ± SE. Between- and within-group differences were tested using a repeated-measures ANOVA. Post hoc paired t-tests were used to assess intragroup interaction effects between particular time points when the ANOVA models produced significant main effects. Unpaired t-tests were used to compare baseline differences between groups. Significance level was predetermined to be P ≤ 0.05 unless otherwise indicated. All analyses were done with SPSS version 14 software.
RESULTS
Basal Primary and Mature miRNA Expression
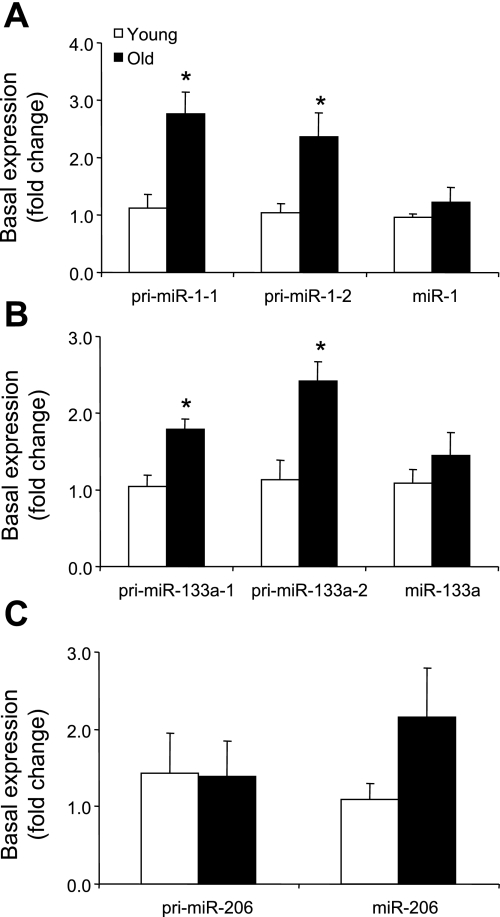
There were noticeable differences in the basal pri-miRNA expression levels between skeletal muscle of young and older men. Specifically, pri-miR-1-1 and -1-2 (Fig. 1A) and pri-miR-133a-1 and -133a-2 (Fig. 1B) expression levels were elevated ∼100–150% in the older compared with the young men (P ≤ 0.05). Although there was a trend for some of the mature miRNAs to be higher in the older men, these differences were not significantly different.
Fig. 1.
Basal expression of primary and mature microRNAs (miRNAs) in young and older men: primary miR-1-1, pri-miR-1-2, and mature miRNA miR-1 (A), pri-miR-133a-1, pri-miR-133a-2, and mature miR-133a (B), and pri-miR-206 and mature miR-206 (C). Values are means ± SE. *Significantly different from young (P ≤ 0.05).
Primary and Mature miRNA Expression After an Anabolic Stimulus
MicroRNA-1.
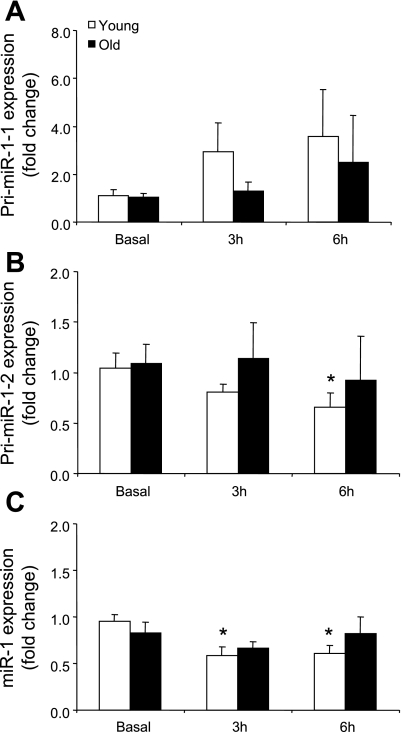
Pri-miR-1-1 mRNA expression was not different in either group after exercise (Fig. 2A). Pri-miR-1-2 mRNA expression (Fig. 2B) was significantly reduced at 6 h after exercise, and miR-1 expression (Fig. 2C) was lower at 3 and 6 h only in the young men compared with basal (P ≤ 0.05).
Fig. 2.
Expression levels of pri-miR-1-1 (A), pri-miR-1-2 (B), and miR-1 (C) in young and older men after a potent anabolic stimulus [resistance exercise + essential amino acid (EAA) ingestion]. Values are means ± SE. *Significantly different from basal (P ≤ 0.05).
MicroRNA-133.
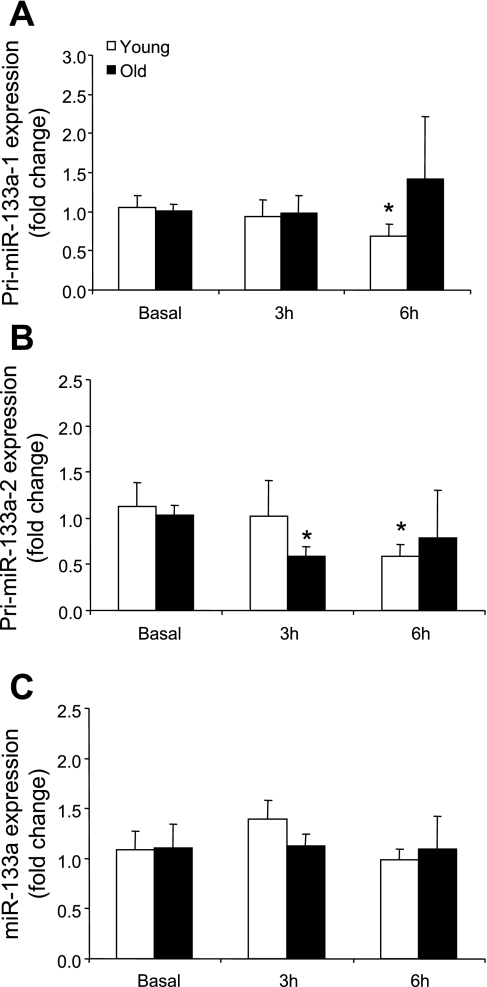
Primary miR-133a-1 mRNA expression (Fig. 3A) was significantly reduced (P < 0.05) at 6 h only in the young men and pri-miR-133a-2 expression (Fig. 3B) was significantly reduced in the older men at 3 h and at 6 h in the young men (P ≤ 0.05) compared with basal. MicroRNA-133a (Fig. 3D) was not different across time or between the age groups compared with basal.
Fig. 3.
Expression levels of pri-miR-133a-1 (A), pri-miR-133a-2 (B), and miR-133a (C) in young and older men after a potent anabolic stimulus (resistance exercise + EAA ingestion). Values are means ± SE. *Significantly different from basal (P ≤ 0.05).
MicroRNA-206.
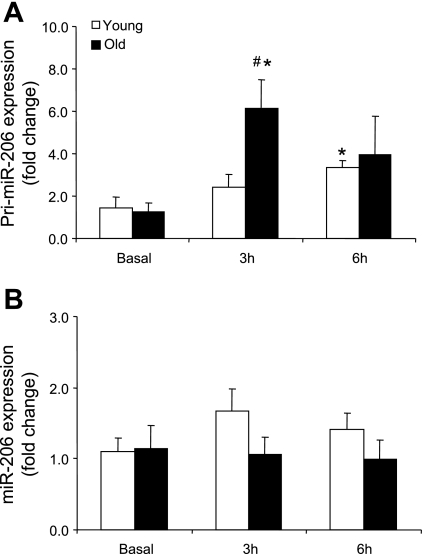
Primary miR-206 expression (Fig. 4A) was significantly increased in the older men at 3 h (with a significant age interaction) and at 6 h in the young men compared with basal (P ≤ 0.05). MicroRNA-206 expression (Fig. 4B) was not different across time or between the age groups compared with basal.
Fig. 4.
Expression levels of pri-miR-206 (A) and miR-206 (B) in young and older men after a potent anabolic stimulus (resistance exercise + EAA ingestion). Values are means ± SE. *Significantly different from basal (P ≤ 0.05). #Significantly different from young at corresponding time point (P ≤ 0.05).
Expression of miRNA Processing and Transporting Factors
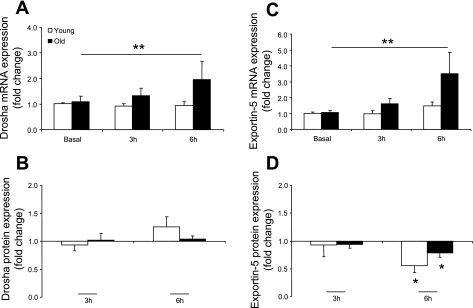
There were no differences at baseline between young and older men for Drosha, Exportin-5, or Dicer mRNA or protein expression (data not shown). There was a main effect for time × age: Drosha mRNA (Fig. 5A) expression was significantly different between the ages across time compared with basal (P ≤ 0.05). Drosha protein expression (Fig. 5B) was unchanged over time or between groups. Furthermore, there was a main effect for time × age: Exportin-5 mRNA (Fig. 5C) expression was different between the age groups over time compared with basal (P ≤ 0.05). On the other hand, Exportin-5 protein (Fig. 5D) was significantly lower in young and older man at 6 h compared with basal (P ≤ 0.05). Dicer mRNA and protein expression was not different across time or between the age groups compared with basal (data not shown).
Fig. 5.
MicroRNA processing and transporting factor expression levels for Drosha mRNA (A), Drosha protein (B), Exportin-5 mRNA (C), and Exportin-5 protein (D) in young and older men after a potent anabolic stimulus (resistance exercise + EAA ingestion). Values are means ± SE. *Significantly different from basal (P ≤ 0.05). **Significant main effect for time × age (P ≤ 0.05).
Upstream Regulators and Downstream miR-1 Targets
The protein expression of MyoD and myogenin was measured in the muscle samples to determine whether these transcription factors have an miR-1 regulatory role (Table 2). We did not determine any statistically significant changes for MyoD or myogenin after the anabolic stimulus or between the age groups.
Table 2.
Upstream regulators and downstream targets of miR-1 after an anabolic stimulus in young and older men
|
Fold Change From Basal |
||
|---|---|---|
| 3 h | 6 h | |
| MyoD | ||
| Young | 1.08±0.04 | 1.07±0.07 |
| Older | 1.25±0.13 | 1.27±0.18 |
| Myogenin | ||
| Young | 1.07±0.10 | 0.99±0.07 |
| Older | 1.43±0.29 | 1.41±0.26 |
| IGF-I | ||
| Young | 0.96±0.07 | 0.87±0.09 |
| Older | 1.12±0.05 | 0.95±0.09 |
| HDAC4 | ||
| Young | 0.87±0.07 | 0.77±0.03* |
| Older | 0.89±0.07 | 0.82±0.10* |
| MEF2 | ||
| Young | 1.00±0.07 | 0.96±0.15 |
| Older | 0.94±0.05 | 0.84±0.08 |
| Rheb† | ||
| Young | 0.99±0.08 | 1.25±0.15 |
| Older | 0.68±0.16 | 1.24±0.28 |
Values are means ± SE. IGF-I, insulin-like growth factor I; HDAC4, histone deacetylase 4; MEF2, myocyte enhancing factor 2; Rheb, Ras homolog enriched in brain.
Significantly different from basal (P ≤ 0.05).
Main effect for time (P ≤ 0.05).
Furthermore, we measured select predicted (IGF-I) and validated (HDAC4, MEF2, and Rheb) miR-1 target protein expression after the anabolic stimulus to determine whether changes in miR-1 after the anabolic stimulus were reflective of changes in protein expression of downstream events (Table 2). HDAC4 protein expression was significantly reduced at 6 h in both age groups (P ≤ 0.05), and there was a main effect for time: Rheb protein expression was significantly higher in young and older men compared with basal (P ≤ 0.05). There were no significant differences across time or between groups for IGF-I or MEF2 protein expression.
DISCUSSION
The two major findings of this study were that, in human skeletal muscle, 1) primary miRNA expression was higher in older than in young men at rest and 2) a maximal protein anabolic stimulus consisting of resistance exercise + EAA ingestion resulted in alterations in primary and mature miRNA expression mainly in the young men, but not in the older men. These data are the first to demonstrate that miRNAs are responsive to physiological challenges in human muscle and suggest that miRNAs may have a role in regulating the acute protein synthesis response in human skeletal muscle. In addition, the differential response of miRNA expression in muscle of older vs. young men may be associated with the altered muscle growth responses known with aging (54) and following a hypertrophic stimulus (22, 52).
There is a gradual loss of muscle mass with advancing age (54). However, the basal rates of human muscle protein synthesis and breakdown in healthy older men are not different from those in young men (50). This suggests that gradual muscle loss occurs by other means (i.e., repeated inadequate responses to a protein anabolic stimulus such as feeding and/or exercise), rather than a reduced rate of basal muscle protein synthesis. Although muscle protein synthesis was not different at rest in our subjects (11), the present data show significant increases in primary miRNA expression in older subjects. Specifically, pri-miR-1-1, -1-2, -133a-1, and -133a-2 were elevated ∼100–150% in the older men compared with their young counterparts (Fig. 1, A and B). Interestingly, these baseline age-related differences were not reflected at the mature miRNA level, thus indicating an altered relationship between the levels of primary miRNA and processing of the mature miRNA. Indeed, we did not detect age-related basal differences in the mRNA or protein for miRNA processing and transporting factors (e.g., Drosha, Exportin-5, and Dicer; data not shown). Thus a lack of change in these components may be a way to compensate for elevations in primary miRNA expression and, therefore, temper potential changes in levels of miRNAs. It is intriguing to speculate that maintenance of the mature muscle-specific miRNAs (despite a dramatic elevation in primary transcripts) may be a way to combat muscle loss associated with aging.
To determine whether a dysregulation of miRNA expression may be limited to the primary transcript level, we measured basal protein expression of select targets of the muscle-specific miRNA, miR-1. We did not detect age-related differences in protein expression of IGF-I, HDAC4, MEF2, or Rheb (data not shown), all of which have been shown to be predicted or validated targets of miR-1. Contrary to our findings, a study in Caenorhabditis elegans showed a decrease in miR-1 expression with advancing age (17). These differences may be due to different stages of life or major complexities between the two species, since miR-1 has been predicted to regulate almost twice as many transcriptional targets in humans as in worms, and worms lack satellite cells and different muscle fiber types. It is important to mention that our experiment was limited to measurement of the muscle-specific miRNAs (e.g., miR-1, -133a, and -206) and select (but not all) miRNA targets and contained only a modest number of human subjects. Therefore, we cannot rule out the possibility that other miRNAs (as well as additional miRNA targets) may be dysregulated at rest, thus contributing to the age-related decline in skeletal muscle mass.
MicroRNA-1 and -133a are well established to be highly enriched in cardiac and skeletal muscle, whereas expression of miR-206 is skeletal muscle specific (18). We chose to study these specific miRNAs and select downstream targets for their role in regulating the transcription of mRNAs responsible for cell growth and satellite cell function (5, 18, 31, 32, 41). Thus a secondary aim of the present study was to determine whether expression of miRNAs and their downstream targets was differentially regulated in skeletal muscle between young and older men during the early recovery hours following a potent anabolic stimulus (resistance exercise + leucine-enriched EAA ingestion). As a result, we found a general relationship between miR-1 (Fig. 2C) and its transcript pri-miR-1-2 (Fig. 2B), such that both were reduced by ∼50% only in young subjects. These results are similar to recent findings in mice, in which miR-1 was reduced by ∼50% after 7 days of functional overload of the plantaris (32). Although miR-133a expression was unaltered in the present study (Fig. 3C) compared with the decrease in miR-133a expression reported by McCarthy and Esser (32), this was only an acute protein anabolic stimulus, and alterations in miR-133a may require repeated bouts of the anabolic stimulus. Indeed, the expression levels of pri-miR-133a-1 and -133a-2 were significantly reduced by an acute anabolic stimulus at 6 h after exercise (Fig. 3, A and B). Thus, with further bouts of exercise and/or EAA supplementation, it is unknown whether alterations in the primary transcript will translate into changes at the mature miRNA level. Together, these findings indicate that expression of miR-1, but not miR-133a, is acutely regulated only in the young men after an acute protein anabolic stimulus of resistance exercise + EAA ingestion.
The primary transcript pri-miR-206 gives rise to the mature miRNA miR-206 (31). In our acute anabolic stimulus model in human skeletal muscle, we found that pri-miR-206 (Fig. 4A) was upregulated in young and older men, albeit at different time points. However, miR-206 was unchanged (Fig. 4B). The absence of a direct relationship between the precursor and the respective miRNA appears to be inconsistent. It is tempting to speculate that the discordant expression may be explained by processing inefficiencies, since Exportin-5 protein expression was lower in both age groups at 6 h after exercise (Fig. 5D). This would limit the export of the precursor miRNA-206 from the nucleus, thus preventing cleavage by Dicer in the cytoplasm. However, similar to other RNAs, we cannot rule out other potential mechanisms that may alter primary and miRNA expression, such as posttranscriptional modification and miRNA half-life (21, 35, 48). Nonetheless, these data are consistent with those of McCarthy and Esser (32), who reported that the primary transcript (pri-miR-206) was elevated with no corresponding change in miR-206 after 7 days of functional overload in mice.
MicroRNA-1 has been validated [e.g., MEF2 (43), Delta (24), HDAC4 (5), Rheb (41)] and predicted [IGF-I, hepatic growth factor, Pax3, and c-Met (http://targetscan.org)] to target many transcripts that are imperative for regulation of muscle growth and function. To test whether alterations in miR-1 expression are associated with changes in expression of target genes, we measured the protein expression of IGF-I, HDAC4, MEF2, and Rheb (Table 2). Contrary to our hypothesis, IGF-I and MEF2 protein expression were unchanged, whereas HDAC4 was decreased, in young and older subjects after the anabolic stimulus. However, we noted a significant increase in skeletal muscle Rheb protein expression (main effect for time) for both age groups after the anabolic stimulus compared with basal. Rheb, a small GTPase, is a positive regulator of the mammalian target of rapamycin and an integral point of control of cell growth (40). Previously, we noted that phosphorylation of mammalian target of rapamycin and protein synthesis were elevated in these same subjects 6 h after the anabolic stimulus (11). Although the majority of the downstream targets for miR-1 were unchanged, it is possible that the time course of muscle sampling was not long enough to allow detection of changes in protein expression. However, because of their small size (∼20 bp), miRNAs are rapidly produced, thus limiting our ability to determine which miR-1 targets may be dysregulated in the older men.
Nevertheless, cell growth and satellite cell function are very important cellular responses when skeletal muscle is challenged by a protein anabolic stimulus. Thus it is not surprising that a downregulation of miR-1 may result in enhancement of such cellular functions. A single bout of resistance exercise results in significant increases in skeletal muscle protein synthesis with concomitant changes in cellular markers of muscle hypertrophy (11), and proliferation of satellite cells are increased in as few as 1–4 days after exercise in young and older humans (6, 8). Furthermore, cyclin D1, a marker of cell cycle progression, is elevated (2, 19) within 12–24 h after resistance exercise in humans. Interestingly, the acute satellite cell response is attenuated in the older subjects after a single bout of resistance exercise, as noted by reduced satellite cell proliferation (8), but is similar to the response in young subjects with resistance exercise training (39). Therefore, we propose that a dysregulated miRNA response in muscle of older men could translate into a disrupted/delayed activation of satellite cells and cell growth after an acute anabolic stimulus.
Previously, we suggested that the blunted/delayed muscle protein synthesis in the older men may be due to limited type II fiber recruitment (as demonstrated by lower blood lactate levels and attenuated extracellular-regulated kinase signaling) (11). We expand on these findings by showing that many of the pri-miRNAs and the mature miRNA, miR-1, measured in the present study were unchanged in the older men during the 6 h after the anabolic stimulus. Because of the limited publications evaluating miRNAs in skeletal muscle and the inability to perform in-depth mechanistic experiments in humans, it is difficult to determine the root cause of the discordant miRNA expression in the older men during the early hours of recovery after the anabolic stimulus. Our data indicate that miRNA processing cannot explain the differences between the age groups over the given time course. Although the mRNA expression levels of Drosha and Exportin-5 were higher in the older than in the young men (Fig. 5, A and C), this difference was not reflected at the protein expression level (Fig. 5, B and D), which may indicate that dysregulation occurs at the stage of transcription. Many transcription factors regulate the fate of miRNA transcription (15). The myogenic regulatory factors myogenin and MyoD (37), as well as serum response factor (55) and MEF2 (28), are bona fide regulators of miR-1 transcription. To test this hypothesis, we measured MyoD and myogenin protein expression (Table 2). Unfortunately, we did not detect any changes across time or between groups that may explain the acute change in miR-1 expression in young subjects shortly after the anabolic stimulus. Therefore, it remains to be determined whether (and which) transcription factors play an integral role in the acute miRNA regulation within human skeletal muscle after an anabolic stimulus. Overall, a downregulation of miRNA-1, in the young men, after an acute protein anabolic stimulus may represent another level of control on transcripts important for muscle growth and satellite cell function but, in the older men, may represent dysregulation.
In conclusion, we identify an upregulation of pri-miRNAs in older men at rest. Furthermore, an acute protein anabolic stimulus, consisting of resistance exercise + EAA ingestion, has the ability to regulate miR-1 skeletal muscle expression in young men, but not in older men. This particular miRNA is suggested to target many transcripts that are responsible for various skeletal muscle characteristics that occur after an anabolic stimulus (i.e., resistance exercise), such as muscle growth and satellite cell function. Further research is needed to determine whether miRNA dysregulation continues to exist between the age groups in response to EAA supplementation and resistance exercise training as well as to investigate the role of the proposed muscle-specific miRNA network (miR-499 and -208b) after an anabolic stimulus (47).
GRANTS
This study was supported by National Institutes of Health Grants AR-049877 (B. B. Rasmussen), AR-053641 (J. J. McCarthy), and AR-45617 (K. A. Esser) and also by Grant P30 AG-024832 and General Clinical Research Grant M01 RR-00073.
Acknowledgments
We thank the General Clinical Research Center and Sealy Center on Aging staff for their valuable efforts in recruiting subjects and assisting with the clinical portion of the study.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Beuvink I, Kolb FA, Budach W, Garnier A, Lange J, Natt F, Dengler U, Hall J, Filipowicz W, Weiler J. A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs. Nucleic Acids Res 35: e52, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bickel CS, Slade J, Mahoney E, Haddad F, Dudley GA, Adams GR. Time course of molecular responses of human skeletal muscle to acute bouts of resistance exercise. J Appl Physiol 98: 482–488, 2005. [DOI] [PubMed] [Google Scholar]
- 3.Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science 310: 1954–1957, 2005. [DOI] [PubMed] [Google Scholar]
- 4.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36, 2003. [DOI] [PubMed] [Google Scholar]
- 5.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38: 228–233, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crameri RM, Langberg H, Magnusson P, Jensen CH, Schroder HD, Olesen JL, Suetta C, Teisner B, Kjaer M. Changes in satellite cells in human skeletal muscle after a single bout of high intensity exercise. J Physiol 558: 333–340, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235, 2004. [DOI] [PubMed] [Google Scholar]
- 8.Dreyer HC, Blanco CE, Sattler FR, Schroeder ET, Wiswell RA. Satellite cell numbers in young and older men 24 hours after eccentric exercise. Muscle Nerve 33: 242–253, 2006. [DOI] [PubMed] [Google Scholar]
- 9.Dreyer HC, Drummond MJ, Pennings B, Fujita S, Glynn EL, Chinkes DL, Dhanani S, Volpi E, Rasmussen BB. Leucine-enriched essential amino acid and carbohydrate ingestion following resistance exercise enhances mTOR signaling and protein synthesis in human muscle. Am J Physiol Endocrinol Metab 294: E392–E400, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dreyer HC, Fujita S, Cadenas JG, Chinkes DL, Volpi E, Rasmussen BB. Resistance exercise increases AMPK activity and reduces 4E-BP1 phosphorylation and protein synthesis in human skeletal muscle. J Physiol 576: 613–624, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drummond MJ, Dreyer HC, Pennings B, Fry CS, Dhanani S, Dillon EL, Sheffield-Moore M, Volpi E, Rasmussen BB. Skeletal muscle protein anabolic response to resistance exercise and essential amino acids is delayed with aging. J Appl Physiol 104: 1452–1461, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, Lidov HG, Kang PB, North KN, Mitrani-Rosenbaum S, Flanigan KM, Neely LA, Whitney D, Beggs AH, Kohane IS, Kunkel LM. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci USA 104: 17016–17021, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3: 87–98, 2006. [DOI] [PubMed] [Google Scholar]
- 14.Fujita S, Dreyer HC, Drummond MJ, Glynn EL, Cadenas JG, Yoshizawa F, Volpi E, Rasmussen BB. Nutrient signalling in the regulation of human muscle protein synthesis. J Physiol 582: 813–823, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobert O Gene regulation by transcription factors and microRNAs. Science 319: 1785–1786, 2008. [DOI] [PubMed] [Google Scholar]
- 16.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA 102: 16961–16966, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibanez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell 5: 235–246, 2006. [DOI] [PubMed] [Google Scholar]
- 18.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 174: 677–687, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JS, Kosek DJ, Petrella JK, Cross JM, Bamman MM. Resting and load-induced levels of myogenic gene transcripts differ between older adults with demonstrable sarcopenia and young men and women. J Appl Physiol 99: 2149–2158, 2005. [DOI] [PubMed] [Google Scholar]
- 20.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 129: 1141–1151, 2007. [DOI] [PubMed] [Google Scholar]
- 21.Kluiver J, van den Berg A, de Jong D, Blokzijl T, Harms G, Bouwman E, Jacobs S, Poppema S, Kroesen BJ. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene 26: 3769–3776, 2007. [DOI] [PubMed] [Google Scholar]
- 22.Kosek DJ, Kim JS, Petrella JK, Cross JM, Bamman MM. Efficacy of 3 days/wk resistance training on myofiber hypertrophy and myogenic mechanisms in young vs. older adults. J Appl Physiol 101: 531–544, 2006. [DOI] [PubMed] [Google Scholar]
- 23.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with “antagomirs.” Nature 438: 685–689, 2005. [DOI] [PubMed] [Google Scholar]
- 24.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci USA 102: 18986–18991, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419, 2003. [DOI] [PubMed] [Google Scholar]
- 26.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20, 2005. [DOI] [PubMed] [Google Scholar]
- 27.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8: 166, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci USA 104: 20844–20849, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2
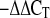 method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar] - 30.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, Filipowicz W, Duchaine TF, Sonenberg N. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 317: 1764–1767, 2007. [DOI] [PubMed] [Google Scholar]
- 31.McCarthy JJ MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta. In press. [DOI] [PMC free article] [PubMed]
- 32.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol 102: 306–313, 2007. [DOI] [PubMed] [Google Scholar]
- 33.McCarthy JJ, Esser KA, Andrade FH. MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am J Physiol Cell Physiol 293: C451–C457, 2007. [DOI] [PubMed] [Google Scholar]
- 34.Mersey BD, Jin P, Danner DJ. Human microRNA (miR29b) expression controls the amount of branched chain α-ketoacid dehydrogenase complex in a cell. Hum Mol Genet 14: 3371–3377, 2005. [DOI] [PubMed] [Google Scholar]
- 35.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA 12: 1161–1167, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 17: 118–126, 2007. [DOI] [PubMed] [Google Scholar]
- 37.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA 103: 8721–8726, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol 175: 77–85, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth SM, Martel GF, Ivey FM, Lemmer JT, Tracy BL, Metter EJ, Hurley BF, Rogers MA. Skeletal muscle satellite cell characteristics in young and older men and women after heavy resistance strength training. J Gerontol A Biol Sci Med Sci 56: B240–B247, 2001. [DOI] [PubMed] [Google Scholar]
- 40.Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol 5: 566–571, 2003. [DOI] [PubMed] [Google Scholar]
- 41.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 100: 416–424, 2007. [DOI] [PubMed] [Google Scholar]
- 42.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5: R13, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon DJ, Madison JM, Conery AL, Thompson-Peer KL, Soskis M, Ruvkun GB, Kaplan JM, Kim JK. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell 133: 903–915, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature 447: 875–878, 2007. [DOI] [PubMed] [Google Scholar]
- 45.Tsuchiya S, Okuno Y, Tsujimoto G. MicroRNA: biogenetic and functional mechanisms and involvements in cell differentiation and cancer. J Pharm Sci 101: 267–270, 2006. [DOI] [PubMed] [Google Scholar]
- 46.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524, 2006. [DOI] [PubMed] [Google Scholar]
- 47.van Rooij E, Liu N, Olson EN. MicroRNAs flex their muscles. Trends Genet 24: 159–166, 2008. [DOI] [PubMed] [Google Scholar]
- 48.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316: 575–579, 2007. [DOI] [PubMed] [Google Scholar]
- 49.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 0034.0031–0034.0012, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volpi E, Sheffield-Moore M, Rasmussen BB, Wolfe RR. Basal muscle amino acid kinetics and protein synthesis in healthy young and older men. JAMA 286: 1206–1212, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang E MicroRNA, the putative molecular control for mid-life decline. Ageing Res Rev 6: 1–11, 2007. [DOI] [PubMed] [Google Scholar]
- 52.Welle S, Totterman S, Thornton C. Effect of age on muscle hypertrophy induced by resistance training. J Gerontol A Biol Sci Med Sci 51: M270–M275, 1996. [DOI] [PubMed] [Google Scholar]
- 53.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci 120: 3045–3052, 2007. [DOI] [PubMed] [Google Scholar]
- 54.Young A Ageing and physiological functions. Philos Trans R Soc Lond B Biol Sci 352: 1837–1843, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436: 214–220, 2005. [DOI] [PubMed] [Google Scholar]