Abstract
In this issue of Molecular Cell, Yamagata et al. (2008) identify a role for the protein methyltransferase, PRMT1, in Akt-dependent regulation of the FOXO1 transcription factor and demonstrate another level of control in this critical metabolic and cell survival signaling pathway.
The mammalian forkhead class O (FOXO) transcription factors are key regulators of cell fate that modulate the expression of a wide range of genes involved in apoptosis, cell-cycle transitions, DNA repair, oxidative stress, aging, muscle growth, cell differentiation, and glucose metabolism. Forkhead transcription factors utilize a winged helix domain to locate consensus FOXO-recognized elements that are present within the promoters of proapoptotic genes such as the Bcl-2 family member, bim, and fas ligand (fasL) (Brunet et al., 1999; Gilley et al., 2003). In addition to roles in cell survival, FOXO proteins direct metabolism and oxidative stress resistance by inducing the expression of genes such as glucose-6-phosphatase (G6pc) and manganese superoxide dismutase (MnSOD) (Kops et al., 2002; Schmoll et al., 2000). FOXO factors also regulate proliferation by modulating the expression of cell-cycle proteins, including the cyclin-dependent kinase inhibitor p27, Ink4, and Arf (Salih and Brunet, 2008). Thus, FOXO transcription factors clearly play critical roles in a wide range of important biological processes.
Because of these diverse functions in cell-fate decisions, FOXOs are subject to multiple levels of regulation through phosphorylation, ubiquitination, and acetylation. Upon growth factor or insulin stimulation, phosphatidyl-inositol-3 kinase activation induces Akt-mediated phosphorylation of FOXOs to promote the association of FOXOs with 14-3-3 chaperone proteins. This sequestration of FOXO proteins in the cytoplasm prevents FOXO-dependent gene regulation. Ultimately, FOXO proteins can be ubiquitinated and subject to proteasomal degradation. FOXO proteins can also be regulated by acetylation via p300 and CREB-binding protein (CBP) to promote nuclear accumulation and inhibit transactivation activity by reducing FOXO-DNA binding (Huang and Tindall, 2007; Salih and Brunet, 2008).
Yamagata et al. (2008) have now revealed yet another layer of complexity in the regulation of FOXO transcription factors (Figure 1). Using in vitro and in vivo assays, the authors demonstrate that protein arginine methyltransferase 1 (PRMT1)-mediated methylation of Arg248 and Arg250 in FOXO1 regulates its phosphorylation by Akt and, therefore, its nuclear exclusion, ubiquitination, and proteasomal degradation. PRMTs catalyze methylation of terminal nitrogens of guanidinium side chains within arginine residues. There are eleven PRMT isoforms found in nearly all eukaryotes that have demonstrated roles in the modulation of histones, DNA repair, transcriptional regulation, mRNA stability, assembly of splicing complexes, signal transduction, and protein-protein interactions. Arginine methylation can modulate the activity of transcription factors including CBP/ p300, PGC-1α, and HNF4, as well as the localization of Stat1 (Krause et al., 2007). Through methylation, PRMT1 prevents Akt-mediated FOXO1 phosphorylation, induces FOXO1 nuclear accumulation, and enhances oxidative stress-induced apoptosis. In support of this model, the addition of hydrogen peroxide promoted the interaction of PRMT1 and FOXO1 and facilitated methylation of FOXO1 at multiple residues. This FOXO1 methylation prevented Akt-mediated phosphorylation and augmented the ability of FOXO1 to induce target gene expression, including that of the proapoptotic gene bim. Among the many PRMTs, PRMT1 was critical to this process as PRMT1 silencing simultaneously reduced the levels of FOXO1 and Bim proteins and protected against oxidative stress-induced apoptosis.
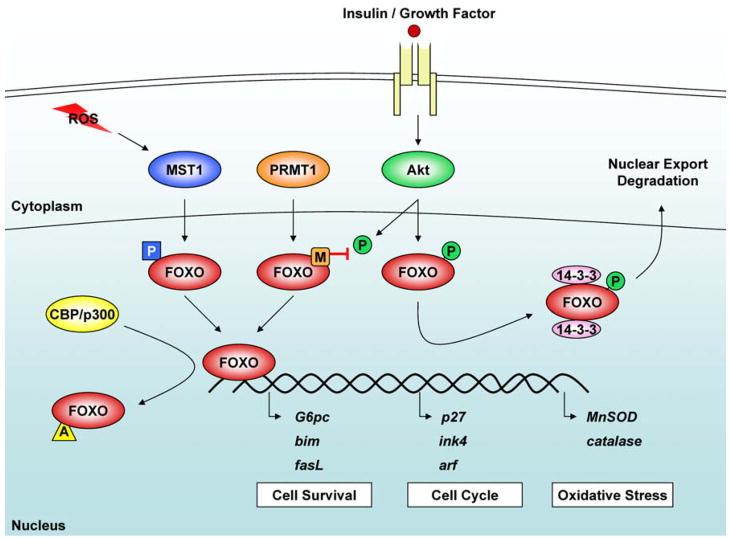
Figure 1. Multiple Pathways for FOXO Regulation.
FOXO transcription factors can be regulated through several mechanisms. Phosphorylation by Akt or MST1 can either promote or inhibit, respectively, FOXO nuclear localization. Akt-mediated phosphorylation induces FOXO association with 14-3-3 and promotes nuclear export and ubiquitination for degradation. By contrast, MST1-mediated phosphorylation promotes nuclear accumulation and gene regulation. Additionally, FOXO acetylation by acetyltransferases such as p300 and CREB-binding protein reduce FOXO DNA binding. Yamagata et al. (2008) now demonstrate that FOXO methylation by PRMT1 prevents Akt-mediated phosphorylation and enhances FOXO1-dependent transcription.
These findings raise important questions concerning the regulation of FOXO-mediated gene expression. First, the physiological signals that promote PRMT1 activity and subsequent FOXO1 methylation are uncertain. One stimulus that can regulate PRMT1 is oxidative stress. Treatment of cells with hydrogen peroxide increased PRMT1 activity and FOXO1 methylation. Indeed, increased PRMT1 expression and activity was also recently correlated with the accumulation of intracellular reactive oxygen species (ROS) in endothelial cells, suggesting that cellular redox status may affect FOXO1 activity through PRMT1-mediated methylation (Jia et al., 2006). Mechanistically, however, it remains uncertain how PRMT1 might be regulated by ROS or if other stimuli could also affect PRMT1-mediated FOXO1 methylation.
Second, although PRMT1 methylation inhibits the ability of Akt to phosphorylate FOXO1, it is unclear if or how methylation might regulate FOXO phosphorylation by other kinases, such as mammalian Ste20-like kinase (MST1). MST1 phosphorylation of FOXOs occurs in response to oxidative stress and promotes FOXO nuclear accumulation and subsequent cell death (Salih and Brunet, 2008). By analogy with inhibition of Akt phosphorylation, PRMT1-mediated FOXO1 methylation might promote MST1-directed phosphorylation. Intriguingly, Yamagata et al. (2008) identified several additional methylation sites on FOXO1 that appeared to have no role in Akt-mediated phosphorylation. Methylation at these residues could potentially alter the accessibility of FOXO1 to other modifications, such as phosphorylation, acetylation, or ubiquitination. In this light, methylation could, therefore, play a very general role in the regulation of FOXO1 activity.
Finally, Yamagata et al. (2008) found that in addition to FOXO1, PRMT1 could also methylate other FOXO subfamily members: FOXO3, FOXO4, and FOXO6. As these proteins perform an array of individual functions, it remains to be determined how methylation alters the activity and posttranslational regulation of these proteins. Methylation of FOXOs might, therefore, be a master regulatory mechanism to dictate cell-fate decisions such as proliferation and survival. It is also possible that methylation is a common mechanism to regulate the ability of Akt to phosphorylate its substrates. It will be interesting to address these possibilities as the roles of methylation become apparent in the many functions of FOXO transcription factors and other Akt targets.
As an essential component of diverse growth and survival signaling pathways, FOXOs serve as a link between aging and age-related diseases such as diabetes and cancer. FOXO dysregulation has been reported in multiple tumor types, including rhabdomyosarcoma and leukemia (Huang and Tindall, 2007). By contrast, FOXO1 deletion in hepatocytes protects against excessive glucose production and diabetes in insulin receptor null mice (Matsumoto et al., 2007). Therefore, proteins that regulate FOXOs form attractive targets for therapeutic intervention. The findings from Yamagata et al. (2008) suggest that PRMT1 could be a novel target for modulating the activity of FOXO proteins and, therefore, cell-fate decisions. This study demonstrates that PRMT1 can enhance FOXO activity to promote cell death, whereas decreased PRMT1 activity can promote cell survival. As FOXO proteins elicit such an array of effects on cells, pharmacologically modulating PRMT1 activity or expression might uncover novel approaches to target multiple FOXOs and alter survival and growth of cells in a variety of pathological conditions.
References
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Gilley J, Coffer PJ, Ham J. J Cell Biol. 2003;162:613–622. doi: 10.1083/jcb.200303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Tindall DJ. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- Jia SJ, Jiang DJ, Hu CP, Zhang XH, Deng HW, Li YJ. Vascul Pharmacol. 2006;44:143–148. doi: 10.1016/j.vph.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S. Pharmacol Ther. 2007;113:50–87. doi: 10.1016/j.pharmthera.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Salih DA, Brunet A. Curr Opin Cell Biol. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoll D, Walker KS, Alessi DR, Grempler R, Burchell A, Guo S, Walther R, Unterman TG. J Biol Chem. 2000;275:36324–36333. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, Mukai H, Kasuya Y, Fukamizu A. Mol Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. this issue. [DOI] [PubMed] [Google Scholar]