Abstract
The complexity and inter-individual variation of brain signals impedes real-time detection of events in raw signals. To convert these complex signals into results that can be readily understood, current approaches usually apply statistical methods to data from known conditions after all data have been collected. The capability to provide meaningful visualization of complex brain signals without the requirement to initially collect data from all conditions would provide a new tool, essentially a new imaging technique, that would open up new avenues for the study of brain function. Here we show that a new analysis approach, called SIGFRIED, can overcome this serious limitation of current methods. SIGFRIED can visualize brain signal changes without requiring prior data collection from all conditions. This capacity is particularly well suited to applications in which comprehensive prior data collection is impossible or impractical, such as intraoperative localization of cortical function or detection of epileptic seizures.
Introduction
Over the past few decades, increased computing power and novel electrical, magnetic, and metabolic sensor modalities have offered exciting new possibilities for studying brain function and have dramatically increased our understanding of the most complex of our organs. Despite these advances, current approaches to studying brain function with a variety of sensor modalities, e.g., electroencephalography (EEG), electrocorticography (ECoG), magnetoencephalography (MEG), or polysomnography (PSG), are significantly constrained. This is because signals acquired with these sensors are too complex that important information could be readily detected in the raw signals, and because there is often substantial inter-individual variation of task-related brain signal features so that different signal features hold relevant information in different individuals. For these reasons, with few exceptions (Lachaux et al., 2007; Miller et al., 2007a), current techniques are usually limited to retrospective analyses.
In consequence, current methodologies typically require the experimenter to specify (and collect data for) all stimulus/response conditions that are to be evaluated. Subsequently, different mathematical techniques (e.g., linear discriminant analysis) then analyze the brain signal responses by discriminating between the different conditions. Thus, data for all conditions have to be collected before any decision can be made. This is a problem in many applications that interrogate the brain. For example, Crone and others have demonstrated that signal changes associated with movements and language can be passively detected in the electrocorticogram (ECoG) (Crone et al., 2001; Crone et al., 1998a; Crone et al., 1998b; Graimann et al., 2002; Pfurtscheller et al., 2003; Sinai et al., 2005) and thus could be used to augment current methods to locate function, such as electrical stimulation mapping. However, these changes typically involve changes in different frequencies that are subject-specific. Thus, meaningful corresponding analyses can only be performed after the fact, which impedes interactive and exploratory studies. Another limitation is that the time of occurrence of each condition has to be known. In many scenarios, this information is not readily available. Hence, these two limitations typically impede the study of many important problems.
In summary, currently used approaches to studying brain function are based on discrimination of known conditions, which limits their applicability in a number of situations. The capacity to effectively visualize brain signal changes with little a-priori knowledge could be applied to many imaging techniques. It would be a boon to many applications in research and clinical diagnosis such as dynamic localization of brain function, brain-computer interfacing, detection of epileptic seizures, or identification of sleep apneas.
We here present the methodology SIGnal modeling For Real-time Identification and Event Detection (SIGFRIED) that can be used effectively in situations that are not adequately served by these existing approaches. SIGFRIED accomplishes this by implementing a detection-based approach rather than a discrimination-based approach. We demonstrate the capacities of this methodology by showing that it can translate complex brain signals into outputs that can be readily interpreted by an untrained observer with little a-priori knowledge.
Materials and Methods
Subjects
We recorded signals from six patients with intractable epilepsy who underwent temporary placement of intracranial electrocorticographic (ECoG) electrode arrays on the cortical surface to localize seizure foci prior to surgical resection. Data for two of these subjects are presented in this paper. Data for the remaining subjects are presented in the Supplementary Material. Array placement was determined entirely by the patients’ clinical needs. The study was approved by the Human Studies Committee of Washington University Medical Center.
Data Collection
Each patient sat in a hospital bed about 75 cm from a video screen. In all experiments, we recorded electrocorticographic (ECoG) signals from 32 electrodes using the general-purpose brain-computer interface system BCI2000 (Schalk et al., 2004). All electrodes were amplified, bandpass filtered (0.1–220 Hz), sampled at 500 Hz, and stored. First, we collected data while the subject was resting, i.e., awake but not actively engaged in any motor or motor imagery task. We then collected data from several 2-min runs during which the patient was asked to perform different motor or language tasks in response to visual cues. In offline analyses, every 400 ms the time-series ECoG data from the past 400 ms were re-referenced using a common average reference and converted into the frequency domain using the maximum entropy method (Marple, 1987) and a model order of 25. The frequency-domain amplitudes for different frequency bands (e.g., mu/beta (8–12/18–25 Hz) or gamma (i.e., >30 Hz)) were input for SIGFRIED, which is described below.
SIGFRIED
The SIGFRIED procedure first estimated the statistical properties of the brain signals recorded during the resting condition. To do this, for each time point t in the resting data set and for each electrode location, the vector of frequency domain amplitudes Xt = [x1,t,…,xD,t] was input into a modified version of the Competitive-EM (CEM) algorithm (Celeux and Govaert, 1992; Harris et al., 2000). The CEM algorithm is a variant of the Expectation Maximization (EM) algorithm (Dempster et al., 1977). Each iteration of the EM algorithm consists of an Estimation (E) step and a Maximization (M) step. The M step maximizes a likelihood function that is redefined in each iteration by the E step. The CEM algorithm assigns, in a C step, each sample to the most likely cluster. (The EM algorithm requires accumulation of fractional statistics for all clusters, which is slower but more accurate.) This procedure produced a model consisting of Y Gaussian distributions that approximated the statistical distribution of the features at each electrode in the resting data set (Duda et al., 2001). Each of these Gaussian distributions j was characterized by a mean feature vector X̅j a covariance matrix ∑j, and a prior probability wj. As an extension to the standard CEM algorithm, the number of distributions Y was automatically determined by choosing Y such that the resulting model minimized the Akaike Information Criterion (Akaike, 1973). See (Schalk et al., 2008) and the Supplementary Material for further description of this model selection procedure.
Using this statistical model of the resting condition, we then calculated the likelihood that a new data point Xn was produced by the resting signal distribution. To do this, we first calculated the Mahalanobis distance, i.e., essentially a multidimensional z-score between the data point and each Gaussian distribution j. Thus, these distances are essentially normalized and can be compared across different locations. Using these distance measures, we then determined the probability that the D-dimensional data point was created by the model distribution The output of SIGFRIED was the negative log of that probability (so that resulting values were scaled to facilitate visualization). Thus, this output could be expected to be small for samples that are similar to those in the resting signal distribution, and large for samples that are different than that distribution.
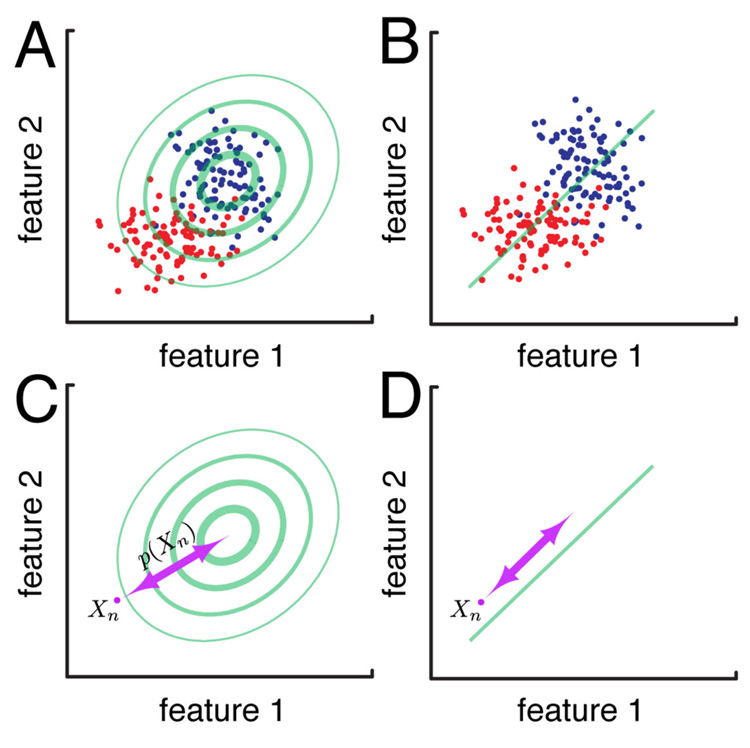
Fig. 1 illustrates the basic principle of the SIGFRIED method for two arbitrary signal features (e.g., amplitudes in two different frequency bands). Panel A: The Gaussian background model (green ellipses) calculated by SIGFRIED for data collected during rest (blue dots) can be used to discriminate between data from the resting condition and a testing condition (red dots), but does not depend on the a priori definition of data from the testing condition. Because it implements a model with multiple Gaussians, the current SIGFRIED implementation supports more complicated distributions and any number of dimensions. Thus, it can master more complicated situations (see Supplementary Material) than those indicated in this simple example. Panel B: Conventional techniques to discriminate between conditions, such as linear regression, which derives a regression function (green line), are calculated between data collected during rest (blue dots) and a testing condition (red dots). In marked contrast to SIGFRIED, they do depend on a priori definition of both data conditions and can thus only be performed after data from both classes have been collected. Panel C: The output of SIGFRIED for a new data sample Xn represents the negative log of the probability p(Xn) that this sample was produced by the description of resting activity. In the example in (C), this probability p(Xn) will be small, and thus the output of SIGFRIED large. In contrast, any data point closer to the description of resting activity (such as one of the blue dots) would result in a higher probability value, and thus a smaller SIGFRIED output value. Panel D: The output of linear regression is determined using a linear function of both features in the direction of the regression line.
Figure 1. SIGFRIED vs. linear regression.
The use of SIGFRIED (symbolized by one Gaussian distribution modelling the blue data points (A)) can be used to discriminate between data from the two conditions (by deriving a distance metric for a new data point Xn (C)). This procedure requires data from only one class (e.g., signals recorded during rest). The use of a traditional technique, such as linear regression (B) can also be used for discrimination (D). In contrast to the detection-based SIGFRIED approach, this procedure requires data from both conditions.
Mapping
We used radiographs and the LOC package (Miller et al., 2007b) to identify the stereotactic coordinates of each electrode in the array. This computer-based procedure implemented the manual procedure described in (Fox et al., 1985). Each patient underwent stimulation mapping to identify motor and speech cortices as part of his/her clinical care. In this mapping, 1-ms 5–10 mA square current pulses were passed through paired electrodes to induce sensation and/or evoke motor responses or speech arrest. This mapping delineated areas important for motor and speech function. These functional areas can be close to the epileptic focus, but typically do not overlap with it. Each subject’s electrode locations were projected on a template model of the brain (http://afni.nimh.nih.gov/afni/suma), and SIGFRIED activation maps were generated using a custom Matlab program.
Real-Time Implementation
To utilize SIGFRIED in real-time operation, we developed a SIGFRIED module for the BCI2000 system (Mellinger and Schalk, 2007; Schalk et al., 2004) (see http://www.bci2000.org). BCI2000 is a general-purpose software system that facilitates development and implementation of a wide array of data acquisition, stimulus presentation, and brain monitoring applications. The system is available for free for academic purposes and to date has been acquired by more than 230 laboratories around the world. The BCI2000 SIGFRIED module can be used in conjunction with any of the more than ten supported data acquisition devices, and with different stimulus presentation or feedback applications. In its current implementation, the output of SIGFRIED at each location is translated into the radius of a circle at the electrode location. The location of each electrode, as well as a number of other display settings, can be parameterized.
Results
We first demonstrate that SIGFRIED can be used to detect changes in brain signals without prior data collection of all conditions. In subject A, we recorded electrocorticographic (ECoG) signals during a 5 minute rest period from 32 electrodes configured in a 8×4 grid. To establish a description of that resting activity, we submitted a comprehensive set (i.e., seven 10-Hz bins from 40–100 Hz) of gamma-band frequencies (which are more spatially specific but less sensitive than mu/beta frequencies) to the SIGFRIED procedure described in the Methods section. This resulted in one resting model for each channel. Each model had an average of 28 Gaussian clusters. We also recorded ECoG signals on a different day while the subject rested or moved her shoulder or hand in response to visual cues. Panel A in Fig. 2 shows the raw ECoG time course (top two traces) and SIGFRIED output time course (bottom two traces) for two adjacent electrodes. Shaded bars in blue and red indicate times cueing for hand and shoulder movements, respectively. The effect of hand or shoulder movement cannot be readily detected in the complex raw signals. In contrast, the blue SIGFRIED output trace detects hand movements while the hand movement cue was presented, but not while the shoulder cue or no cue was presented. The red trace shows the same effect for shoulder movement. Thus, informative results can be gained even without averaging. Panel B illustrates, in color and for each location, the running averages of the SIGFRIED output for the three conditions of rest, shoulder, and hand movement (the three sets of panels were created with identical analysis and display settings). The different locations that are active for shoulder and hand, but not for rest, are evident. Active locations are rapidly defined and remain stable over time. The movie in the Supplementary Material shows the full topography for this data set while the subject is executing these tasks.
Figure 2. Visualization of brain function in subject A.
A: Raw ECoG signal traces (blue and red traces on top corresponding to locations marked by blue and red crosses, respectively). B: Color-coded running averages of the SIGFRIED output for the three conditions of rest, shoulder movement, and hand movement, for each of the 32 locations in the 8×4 grid.
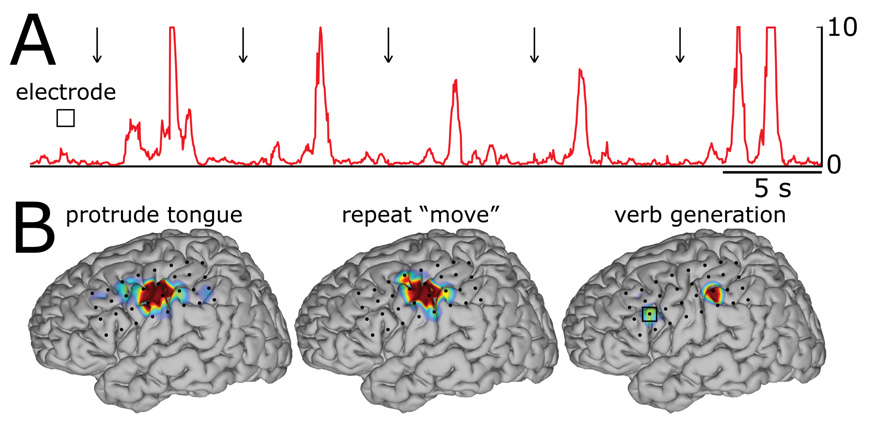
We also show that SIGFRIED can be used to perform sensorimotor and language mapping. We used ECoG signals recorded from subject B and from 32 channels. SIGFRIED first established a description of activity recorded during a 30-min rest period using a comprehensive set (i.e., seven 5-Hz bins from 5–35 Hz) of traditional mu/beta-band frequencies (which are less spatially specific, but more sensitive than gamma frequencies). This resulted in one resting model for each channel. Each model had an average of 18 Gaussian clusters. Then, on a different day, the subject executed two simple motor tasks by protruding the tongue or repeating the word “move” in response to visual cues. In addition, the subject was shown flash cards that contained nouns and was asked to generate verbs corresponding to these nouns. Panel A in Fig. 3 shows the SIGFRIED time course for the electrode that was responsive to the verb generation task. Arrows indicate times of cue presentation. Activations in Broca’s area can be readily identified in the time course. Panel B shows average activations calculated using SIGFRIED. The activation patterns for the motor tasks (i.e., protruding the tongue and repeating the word “move”) are similar and markedly different from those for the verb generation task. These results indicate that, with further verification, these techniques might provide a new clinical tool that could augment existing techniques for functional mapping.
Figure 3. Sensorimotor and language mapping using SIGFRIED in subject B.
A: SIGFRIED output for the electrode marked with a square in the brain figure on the bottom right. B: Average activations calculated using SIGFRIED. Activations are color-coded. Red corresponds to the highest SIGFRIED values and gray to the lowest values. The three brain topographies were created with identical analysis and display settings.
Finally, we show an example of the real-time implementation of SIGFRIED executing within the BCI2000 software framework. In the present implementation, SIGFRIED output values are mapped to the diameter of a circle that is shown at the location of the respective channel. These values are shown on a display and updated in real time (e.g., every 30 ms). In addition, if experimental conditions exist, the system can be configured to show averages or other statistics of SIGFRIED values in additional displays. Fig. 4 shows an example screenshot of this implementation. This combination of SIGFRIED and BCI2000 is now in use by several institutions to explore rapid mapping of cortical function using subdural recordings. In these exploratory tests, meaningful results are typically achieved quickly and after only three minutes of baseline recording. This software system is highly optimized so that it can accommodate a large number of channels at high sampling rates. For example, on a 3.4 GHz Pentium 4 computer, the whole system can provide visualization for 64 channels sampled at 1000 Hz, update this visualization every 30 ms, and still only use about 40% of the processor’s capacity. (In this example, the average number of Gaussian clusters per channel was 15, the window for spectral estimation was 500 ms, and the model order was 120).
Figure 4. Example real-time display of the SIGFRIED module in BCI2000.
The output of SIGFRIED at each location is translated into the diameter of a circle. The left display shows the real-time display (i.e., the SIGFRIED values produced at a particular point in time irrespective of condition). The two displays on the right show SIGFRIED averages for two conditions.
In summary, these results give demonstrations that illustrate that SIGFRIED can produce a dynamic mapping of ongoing brain activity. These examples highlight SIGFRIED’s capacities and illustrate its utility for analyzing brain signals without the benefit of prior collection of data from all classes. The Supplementary Material includes a corresponding video that effectively summarizes these capacities, and also includes additional results from other subjects.
Discussion
This paper describes a novel technique for brain signal analysis (SIGFRIED) that is well suited to situations in which there is little or no a priori knowledge about the nature and/or timing of task-related changes in brain activity. This is accomplished by implementing a measure of novelty rather than discrimination. This technique expands the range of clinical and basic research studies that can be conducted.
One immediate clinical application for the SIGFRIED methodology is passive mapping of cortical function. The use of SIGFRIED could complement existing methods to localize function, such as electrical stimulation, functional magnetic resonance imaging (fMRI), or evoked potentials. In fact, several institutions are now exploring the value of the SIGFRIED/BCI2000 package for this purpose. At the same time, the exact relationship of this novel passive mapping methodology with existing techniques is currently not defined. Thus, comprehensive studies to delineate this relationship and the value of passive mapping are needed.
The SIGFRIED methodology presented here could also expand the range of analysis options available to basic neuroscience research. At present, the typical analysis methodology uses a particular mathematical technique to establish a direct relationship between brain signal features and particular conditions. This limits the complexity of this relationship that can be detected, and also impedes the interpretation of that interpretation. The early field of computer vision faced the same problem when studies attempted to directly classify individual features (e.g., classifying pixels for character recognition). In this field, such problems are now addressed by applying foreground/background detection and segmentation techniques to construct intermediate representations (i.e., visual primitives) prior to categorization and interpretation. The application of SIGFRIED to frequency-domain or other features of brain signals could be a possible approach to identify relevant events that could be grouped into “cortical primitives” (i.e., cortical events of particular structures). In this case, SIGFRIED would be combined with existing classification-based strategies. SIGFRIED would produce different topographical patterns of detected brain signal changes, and conventional techniques would classify these patterns based on prior information (such as the patterns’ anatomical or physiological relevance).
SIGFRIED is of particular value when it is used with complex signals, and in situations in which it is not practical to record data from all conditions ahead of time. While SIGFRIED could be utilized with brain signals acquired with any sensor, simpler signals (such as measurements of the blood oxygen-level dependent (BOLD) response using fMRI) can be adequately analyzed using existing simple techniques such as the subtraction of baseline values. Also, while SIGFRIED requires less a-priori knowledge than traditional techniques, it still needs a subject-specific baseline recording and some understanding which features may carry relevant information (see (Schalk et al., 2008) for more information on the relationship of the number of features with performance).
An initial implementation of SIGFRIED is presented here, but there are clearly several ways in which its performance could be enhanced. For example, in Fig. 2 and 3, we demonstrated that SIGFRIED derived a description of resting datasets that were recorded on a different day, and that this description generalized sufficiently to permit effective analyses on a different day. Adaptation of the resting description to the varying nature of ongoing resting activity may further improve performance. As another example, projections of the typically multidimensional feature space in more compact representations (e.g., using Principal Component Analysis (PCA)) may prove beneficial.
SIGFRIED derives its advantages mainly from a trade-off of sensitivity and specificity. While it can respond to any brain signal change for each description of resting activity (e.g., for each brain location), and thus is very sensitive, it cannot necessarily differentiate among different signal changes within each location (except, as in the examples in Fig. 2 and 3, these changes reflect distinct topographical patterns).
Supplementary Material
Acknowledgements
We are grateful to Dr. Jonathan Carp for his critical comments during the preparation of this manuscript. We also thank Hrvoje Hecimovic for the seizure data in the Supplementary Material. This research was supported by grants from the NIH (EB006356 (NIBIB) (GS), HD30146 (NCMRR/NICHD) (JRW), EB00856 (NIBIB&NINDS) (JRW)) and the US Army Research Office (W911NF-07-1-041 (GS)).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akaike H. Information Theory and an Extension of the Maximum Likelihood Principle. In: Petrov BN, Csaki F, editors. 2nd International Symposium on Information Theory; Budapest. 1973. pp. 267–281. [Google Scholar]
- Celeux G, Govaert G. A Classification EM algorithm for clustering and two stochastic versions. Computational Statistics & Data Analysis. 1992;14:315–332. [Google Scholar]
- Crone NE, Hao L, Hart J, Boatman D, Lesser RP, Irizarry R, Gordon B. 2001 doi: 10.1212/wnl.57.11.2045. [DOI] [PubMed] [Google Scholar]
- Crone NE, Miglioretti DL, Gordon B, Lesser RP. Functional mapping of human sensorimotor cortex with electrocorticographic spectral analysis. II. Event-related synchronization in the gamma band. Brain. 1998a;121(Pt 12):2301–2315. doi: 10.1093/brain/121.12.2301. [DOI] [PubMed] [Google Scholar]
- Crone NE, Miglioretti DL, Gordon B, Sieracki JM, Wilson MT, Uematsu S, Lesser RP. Functional mapping of human sensorimotor cortex with electrocorticographic spectral analysis. I. Alpha and beta event-related desynchronization. Brain. 1998b;121(Pt 12):2271–2299. doi: 10.1093/brain/121.12.2271. [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum Likelihood from Incomplete Data via the EM Algorithm. Journal of Royal Statistical Society B. 1977;39:1–38. [Google Scholar]
- Duda R, Hart P, Stork D. Pattern Classification. 2 ed. New York: Wiley-Interscience; 2001. [Google Scholar]
- Fox PT, Perlmutter JS, Raichle ME. A stereotactic method of anatomical localization for positron emission tomography. J Comput Assist Tomogr. 1985;9:141–153. doi: 10.1097/00004728-198501000-00025. [DOI] [PubMed] [Google Scholar]
- Graimann B, Huggins JE, Levine SP, Pfurtscheller G. Visualization of significant ERD/ERS patterns in multichannel EEG and ECoG data. Clin Neurophysiol. 2002;113:43–47. doi: 10.1016/s1388-2457(01)00697-6. [DOI] [PubMed] [Google Scholar]
- Harris KD, Henze DA, Csicsvari J, Hirase H, Buzsaki G. Accuracy of Tetrode Spike Separation as Determined by Simultaneous Intracellular and Extracellular Measurements. Journal of Neurophysiology. 2000;84:401–414. doi: 10.1152/jn.2000.84.1.401. [DOI] [PubMed] [Google Scholar]
- Lachaux JP, Jerbi K, Bertrand O, Minotti L, Hoffmann D, Schoendorff B, Kahane P. A Blueprint for Real-Time Functional Mapping via Human Intracranial Recordings. PLoS ONE. 2007;2:e1094. doi: 10.1371/journal.pone.0001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marple SL. Digital spectral analysis: with applications. 1987 [Google Scholar]
- Mellinger J, Schalk G. BCI2000: A General-Purpose Software Platform for BCI Research. In: Dornhege G, Millan JdR, Hinterberger T, McFarland DJ, Mueller KR, editors. Brain-Computer Interfaces. Cambridge, MA: MIT Press, NIPS Workshop Series; 2007. [Google Scholar]
- Miller KJ, denNijs M, Shenoy P, Miller JW, Rao RP, Ojemann JG. Real-time functional brain mapping using electrocorticography. Neuroimage. 2007a;37:504–507. doi: 10.1016/j.neuroimage.2007.05.029. [DOI] [PubMed] [Google Scholar]
- Miller KJ, Makeig S, Hebb AO, Rao RP, denNijs M, Ojemann JG. Cortical electrode localization from X-rays and simple mapping for electrocorticographic research: The "Location on Cortex" (LOC) package for MATLAB. J Neurosci Methods. 2007b;162:303–308. doi: 10.1016/j.jneumeth.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Pfurtscheller G, Graimann B, Huggins JE, Levine SP, Schuh LA. Spatiotemporal patterns of beta desynchronization and gamma synchronization in corticographic data during self-paced movement. Clin Neurophysiol. 2003;114:1226–1236. doi: 10.1016/s1388-2457(03)00067-1. [DOI] [PubMed] [Google Scholar]
- Schalk G, Brunner P, Gerhardt LA, Bischof H, Wolpaw JR. Brain-computer interfaces (BCIs): detection instead of classification. J Neurosci Methods. 2008;167:51–62. doi: 10.1016/j.jneumeth.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Schalk G, McFarland DJ, Hinterberger T, Birbaumer N, Wolpaw JR. BCI2000: a general-purpose brain-computer interface (BCI) system. IEEE Trans Biomed Eng. 2004;51:1034–1043. doi: 10.1109/TBME.2004.827072. [DOI] [PubMed] [Google Scholar]
- Sinai A, Bowers CW, Crainiceanu CM, Boatman D, Gordon B, Lesser RP, Lenz FA, Crone NE. Electrocorticographic high gamma activity versus electrical cortical stimulation mapping of naming. Brain. 2005;128:1556–1570. doi: 10.1093/brain/awh491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.