Summary
In Bacillus subtilis, the transcription factor ComA activates several biological processes in response to increasing population density. Extracellular peptide signaling is used to coordinate the activity of ComA with population density. At low culture densities, when the concentration of signaling peptides is lowest, ComA is largely inactive. At higher densities, when the concentration of signaling peptides is higher, ComA is active and activates transcription of at least 9 operons involved in the development of competence and the production of degradative enzymes and antibiotics. We found that ComA binds a degenerate tri-partite sequence consisting of three DNA binding determinants or “recognition elements”. Mutational analyses showed that all three recognition elements are required for transcription activation in vivo and for specific DNA binding by ComA in vitro. Degeneracy of the recognition elements in the ComA binding site is an important regulatory feature for coordinating transcription with population density, i.e., promoters containing an optimized binding site have high activity at low culture density and were no longer regulated in the normal density-dependent manner. We found that purified ComA forms a dimer in solution and we propose a model for how two dimers of ComA bind to an odd number of DNA binding determinants to activate transcription of target genes. This DNA-protein architecture for transcription activation appears to be conserved for ComA homologs in other Bacillus species.
Keywords: quorum sensing, transcription, response regulator, DNA binding, Bacillus subtilis
Introduction
Bacteria often coordinate physiological processes using quorum or diffusion sensing 1–3. Cells communicate with one another using small diffusible signaling molecules that are secreted into the environment and sensed by neighboring cells. In gram negative bacteria, signaling molecules are typically acylated homoserine lactone derivatives. In contrast, signaling molecules used by gram positive bacteria are typically peptides (see reviews 4,5). Responding to population density enables bacteria to coordinate responses when sufficient numbers of cells are present.
In Bacillus subtilis, quorum sensing contributes to a variety of physiological processes including the development of genetic competence, the decision to sporulate, and the production of degradative enzymes and antibiotics 5–8. The ComX-ComP-ComA signaling pathway controls the quorum response in B. subtilis. ComX pheromone is a farnesylated 10 amino acid peptide that is secreted into the growth medium and accumulates extracellularly as the culture density increases 9–12. ComX binds to its cognate receptor kinase ComP, resulting in autophosphorylation of ComP at a conserved histidine residue 13. As with other two-component systems, phosphorylated ComP donates phosphate to its cognate response regulator, ComA, on a conserved aspartate. ComA is present and appears to be expressed continuously during exponential growth (unpublished observations) and once phosphorylated, ComA~P functions to activate transcription of target genes 6–8,14,15.
At least four other density-dependent signaling pathways influence the activity of ComA. PhrC (also known as the competence and sporulation stimulating factor or CSF), PhrF, PhrH, and PhrK are pentapeptides that are secreted into the growth medium. The pentapeptides are transported back into the cell through the oligopeptide permease Opp (a.k.a., Spo0K) where they bind to and inhibit the activity of their cognate Rap proteins, RapC, RapF, RapH, and RapK. RapC, RapF, RapH, and, presumably, RapK inhibit ComA binding to its target sites 16–21. Thus, the activity of ComA is highly regulated resulting in little or no ComA-dependent activation of target genes at low culture densities when the concentration of signaling peptides is low. At higher culture densities, when the concentration of signaling peptides increases, ComA becomes activated and stimulates transcription of target genes (reviewed in 22,23).
The combined work from several groups led to the identification of 20 genes in 9 operons whose expression appears to be directly regulated by ComA 14,16,24–29. Sequence alignments, mutational analyses, and DNA footprinting studies led to the model that the binding site for ComA is an inverted repeat containing two 6 bp recognition elements separated by a 4 bp spacer 14,26,28,29. Several genes regulated by ComA have a single inverted repeat, whereas others have multiple inverted repeats.
We describe experiments indicating that the ComA binding site contains three distinct sequence recognition elements. For simplicity, we refer to these as RE1, RE2, and RE3 throughout. RE1 and RE2 comprise the inverted repeat previously characterized as part of the ComA binding site. RE3 is a previously un-recognized sequence downstream from RE1 and RE2 with a consensus sequence identical to that of RE1. We analyzed the relative contributions of all three sequence elements (RE1, RE2, and RE3) and the spacing between the three elements in transcription activation by ComA. Based on our results, we conclude that: 1) all three recognition elements are critical for activation by ComA in vivo and for DNA binding by ComA in vitro, 2) that the spacing between recognition elements is important for transcription activation by ComA, 3) that there is some sequence-dependent information in the spacer regions, and 4) that the overall sequence context and the degeneracy of the binding site is critical for the population density-dependent regulation of genes controlled by ComA.
Results
A third potential recognition element in ComA binding sites
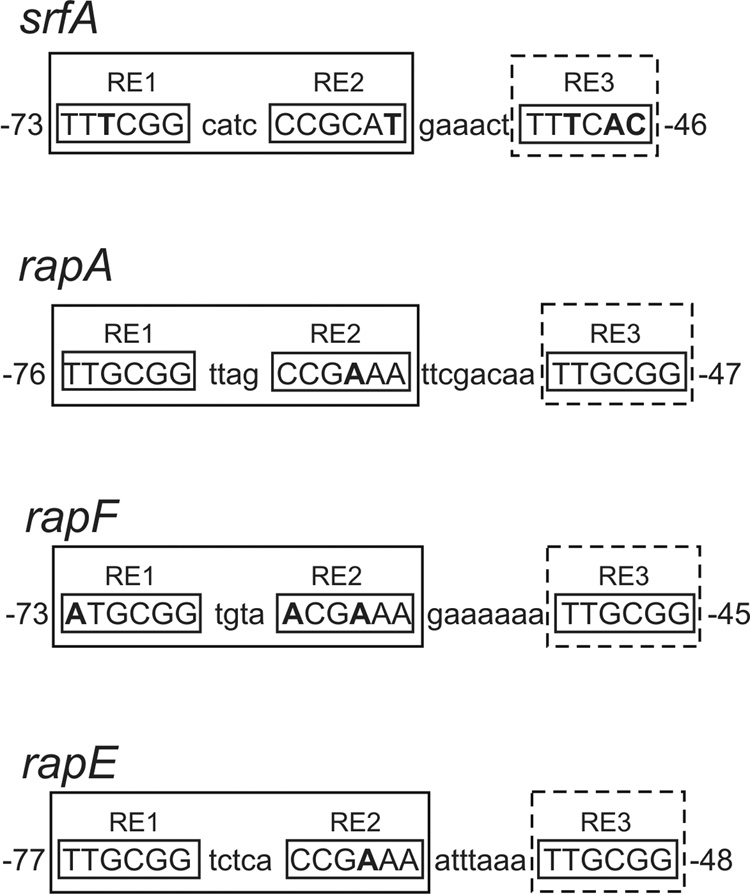
The previously proposed consensus ComA binding site (5'-TTGCGGnnnnCCGCAA) is an inverted repeat comprised of two 6 bp half-sites (consensus 5’-TTGCGG) separated by a 4 bp spacer 14,26,28,29. For characterized promoter regions, the ComA binding site is located upstream of the −35 recognition element for RNA polymerase. We refer to the promoter distal half-site of the inverted repeat (5'-TTGCGG) as “Recognition Element 1” (RE1) and the promoter-proximal half-site of the inverted repeat (5'-CCGCAA) as “Recognition Element 2” (RE2) (Fig. 1).
Figure 1. Alignment of ComA DNA binding sites used in this study.
The promoter-proximal ComA binding sites of target genes used in this study are shown. The numbering represents the position of the binding site relative to the start of transcription as determined by primer extension for each gene (unpublished results). rapA and rapF have two transcription start sites separated by 2 bp. The most abundant (upstream) transcript was used in each case to determine the position of the ComA binding site. Three DNA sequence determinants, referred to as recognition elements 1–3 (RE1-3) make up a single ComA binding site. srfA, rapE, and rapA have two ComA binding sites and only the promoter-proximal site is shown. RE1 and RE2 form a palindrome comprised of two half-site sequences (consensus 5’-TTGCGG) separated by a 4 bp spacer; the exception being rapE which has a 5 bp spacer. The inverted repeat formed by RE1 and RE2 is depicted as a square with solid lines while the newly identified RE3 is depicted as a dashed square. Nucleotides shown in bold represent mismatches from the consensus sequence.
During the course of analyzing ComA function, DNA binding, and in vivo target genes, we noticed that all nine targets known or thought to be directly activated by ComA contained a conserved sequence with a consensus identical to RE1 (consensus 5'-TTGCGG) located downstream of the inverted repeat (Fig. 1 and data not shown). We refer to this sequence element as “Recognition Element 3” (RE3). There is not a recognizable fourth recognition element upstream of RE1 or downstream of RE3, which would be expected if two complete inverted repeats are required for transcription activation by ComA. This arrangement of three putative DNA binding determinants, consisting of an inverted repeat and a half-site, is found for some LysR-type transcriptional regulators 30,31, but otherwise seems to be unusual for transcriptional activators. Results described below demonstrate that a single ComA binding site includes all three recognition elements.
RE3 can function in activation of srfA
srfA is one of the most widely characterized target operons of ComA due to its involvement in the production of the antimicrobial agent surfactin and the development of genetic competence, i.e., the ability to take up DNA 32–35. The srfA promoter has two RE1-RE2 inverted repeats separated by 28 bps. ComA binds to both inverted repeats in the srfA regulatory region 14,26. Deletion of the promoter-distal inverted repeat greatly reduces transcription from the srfA promoter 26. However, transcription is restored by compensatory mutations in the promoter-proximal inverted repeat that make it closer to consensus 26.
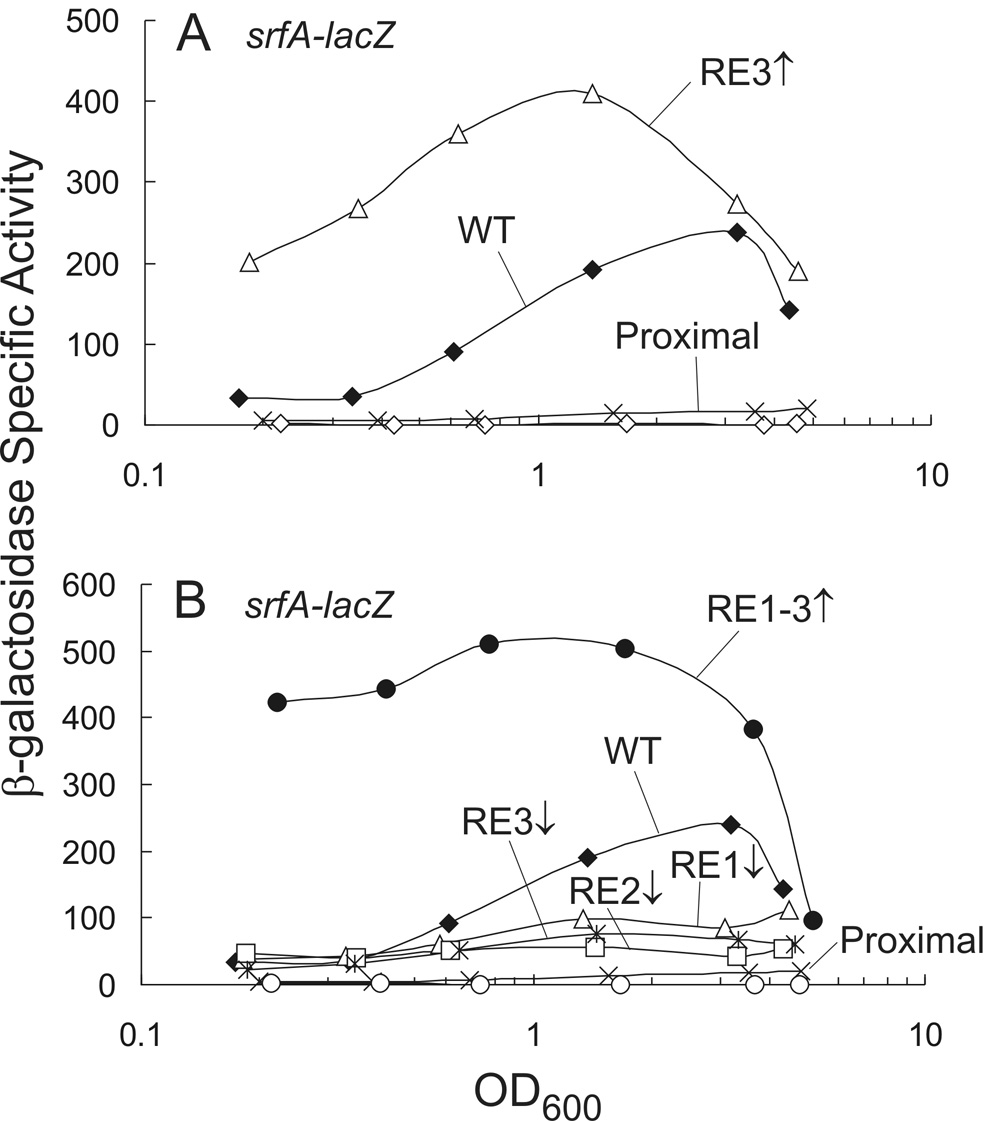
A third sequence element, RE3, is present downstream of each of the previously characterized inverted repeats (RE1 + RE2) in the srfA regulatory region. We characterized the downstream ComA binding site (including the inverted repeat and RE3) and found that RE3 was important for transcription. As observed previously 26, removal of the upstream inverted repeat reduced transcription of a srfA-lacZ transcriptional fusion ~8-fold (Fig. 2A). However, three substitutions in the promoter-proximal RE3 (5'-TTTCAC to 5'-TTGCGG), making it match the consensus sequence, compensated for the lack of the upstream inverted repeat (Fig. 2A). Furthermore, expression was greater than that of the complete wild type promoter containing both inverted repeats (Fig. 2A). This increased expression was dependent on ComA (data not shown) and was most obvious at low culture density where there was a >5-fold increase in β-galactosidase specific activity relative to that of wild type (Fig. 2A). From these results, we conclude that RE3 can function to promote transcriptional activation of srfA and this activation depends on ComA.
Figure 2. Role of RE3 in transcription activation of srfA.
Cultures containing PsrfA–lacZ fusions were grown in defined minimal medium and samples removed throughout growth for determination of β-galactosidase specific activity. β-galactosidase specific activity is plotted as a function of cell density (OD600). Mutation to a consensus recognition element (5’-TTGCGG for RE1 and RE3; 5’-CCGCAA for RE2) is depicted as an up arrow while the down arrow represents mismatches from consensus in all 6 positions of a single recognition element (5’-GCATAT for RE1 and RE3; 5’-ATATGC for RE2).
A. KG125 wild type (filled diamonds); KG102 promoter-proximal binding site only (X); KG160 promoter-proximal binding site with RE3 consensus (open triangles); and KG150 comA null mutant with the wild type reporter (open diamonds).
B. KG125 wild type (filled diamonds) and KG102 promoter-proximal site only (X) are the same as in Panel A for comparison; KG158 promoter-proximal site only with RE1-3 consensus (filled circles); KG780 promoter-proximal site only with consensus RE2 and RE3 and non-consensus RE1 (5’-GCATAT) (open triangles); KG567 promoter-proximal site only with consensus RE1 and RE3 and non-consensus RE2 (5’-ATATGC) (open squares); KG565 promoter-proximal site only with consensus RE1 and RE2 and non-consensus RE3 (5’-GCATAT) (asterisk); and KG464 promoter-proximal site only with consensus RE1-3 in ΔcomA background (open circles)
Analysis of a mutant srfA promoter containing consensus sequences in all three recognition elements
To determine the relative contribution of all three recognition elements in transcription activation, we modified the srfA-lacZ promoter fusion containing only the promoter-proximal ComA binding site such that all three recognition elements matched the consensus sequence (5'-TTGCGG for RE1 and RE3 and the reverse complement, 5'-CCGCAA, for RE2). We also made constructs in which each of the consensus recognition elements was individually replaced with 5’-GCATAT (containing changes away from consensus at every position) and measured the effects on expression.
Expression of the srfA-lacZ fusion with all three recognition elements matching the consensus sequence for ComA binding was quite high (~11-fold greater than wild type), especially at low culture densities (Fig. 2B). Replacement of any of the consensus recognition elements with 5’-GCATAT caused a significant decrease in transcription and the magnitude of the decrease was similar for mutations in each element (Fig. 2B). These results indicate that in the context of three consensus recognition elements, each one is equally important for transcription activation of the srfA promoter.
All three recognition elements contribute to DNA binding by ComA in vitro
To determine if RE3 is important for ComA to bind DNA, we measured the binding of purified ComA (with a his6 tag on the amino-terminus) to DNA using gel mobility-shift assays. his6-ComA was active in vivo based on the ability of the tagged gene to complement a comA null mutation (data not shown). his6-ComA was purified by Ni-affinity chromatography and gel mobility shift assays were performed using a 32P-labeled DNA fragment constructed from two oligonucleotides annealed together to form a 33 bp DNA fragment (Materials and Methods).Purified protein and DNA were allowed to equilibrate prior to separation by native polyacrylamide gel electrophoresis (Materials and Methods).
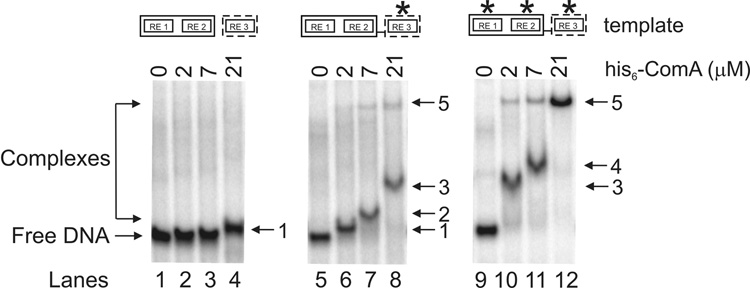
Purified his6-ComA was able to bind to a DNA fragment containing RE1, RE2, and RE3 from the wild type promoter-proximal ComA binding site in the srfA regulatory region (from −73 bp to −46 bp from the start of the annotated coding sequence), although a relatively high concentration of protein was required. We detected a single shifted DNA species with 21 µM his6-ComA and no shift with ≤7 µM his6-ComA (Fig. 3; lanes 1–4). The shifted DNA (species 1; lane 4) migrates very close to the free DNA so a high percentage acrylamide gel (15%) was required to separate the two species. Consistent with the weak binding of his6-ComA to this DNA fragment in vitro, the promoter-proximal ComA binding site in srfA (the sequences used here) is not sufficient to activate transcription of srfA-lacZ unless there are mutations that make RE2 (data not shown) or RE3 closer to consensus (Fig. 2A), or the upstream binding site is included (Fig. 2 and 26).
Figure 3. Gel mobility shift assays using purified ComA and short DNA templates.
Gel mobility shift assays were performed using µM quantities of purified his6-ComA and 5–10 nM 32P-labeled select DNA templates in the presence of 10 nM poly(dI-dC). Binding conditions are described in Materials and Methods. A representative gel is shown. Lanes 1–4: wild type (5’- TTTCGGcatcCCGCATgaaactTTTCAC). Lanes 5–8: consensus RE3 (5’- TTTCGGcatcCCGCATgaaactTTGCGG). Lanes 9–12: consensus RE1-3 (5’- TTGCGGcatcCCGCAAgaaactTTGCGG). Amounts of his6-ComA in each lane (groups of 4 going from left to right): 0, 2 µM, 7 µM, and 21 µM. The numbers to the right of each gel represent different ComA-DNA complexes. The asterisk represents a consensus recognition element (5’-TTGCGG for RE1 and RE3; 5’-CCGCAA for RE2).
We found that alterations in RE3 that increased transcription activation by ComA in vivo (Fig. 2A) significantly enhanced ComA DNA binding in vitro. Using a DNA template that contained the consensus sequence in RE3 (the triple substitution 5’-TTTCAC to 5’-TTGCGG), we observed four shifted species depending on the concentration of his6-ComA (Fig. 3; lanes 5–8). With 2 µM his6-ComA, there was a single shifted species that appeared to correspond to species 1 observed with the wild type sequence and 21 µM his6-ComA (Fig. 3; lane 4). At 7 µM his6-ComA, a single slower-migrating complex was observed (species 2), and at 21 µM his6-ComA a still slower complex (species 3) was present. An additional slower-migrating complex (species 5) was barely visible at 7 µM and 21 µM his6-ComA (Fig. 3; lanes 7–8).
Like the changes in RE3, changes in both RE1 and RE2 toward the consensus sequence (in the context of a consensus RE3) greatly stimulated transcription activation by ComA in vivo (Fig. 2B) and had significant effects on DNA binding in vitro. Using a DNA template that contained the consensus sequences in all three recognition elements, we observed three shifted species depending on the concentration of his6-ComA (Fig. 3; lanes 9–12). With 2 µM his6-ComA, there was a single prominent shifted species (Fig. 3; lane 10) that appeared to correspond to species 3 seen above with consensus mutations in RE3 and 21 µM his6-ComA (Fig. 3; lane 8). With 7 µM his6-ComA, a slightly slower-migrating species (species 4) was observed (Fig. 3; lane 11). Finally, with 21 µM his6-ComA, there was a single slower-migrating species observed (species 5) (Fig. 3; lane 12). Species 5 was also present with 2 µM and 7 µM his6-ComA and the optimal DNA binding sequence (Fig. 3; lanes 10–11). The abrupt transitions to more slowly migrating complexes in vitro indicate that multiple molecules of his6-ComA are probably binding cooperatively to DNA.
The correlation between the binding of his6-ComA in vitro and the extent of transcription activation in vivo indicates that the in vivo phenotypes are due to effects of the DNA sequence on ComA binding. Furthermore, the in vitro results indicate that all three recognition elements contribute directly to ComA binding to DNA. Taken together, the in vivo and in vitro analyses of the srfA regulatory region indicate that a functional ComA binding site includes all three recognition elements.
The proposed tri-partite ComA DNA binding site is consistent with previous in vitro footprinting experiments analyzing ComA binding to the srfA promoter region 14,36. The published footprinting gels indicate that ComA protects the inverted repeat and 4–6 bp upstream and downstream of the inverted repeat from cleavage by DNaseI. Weaker protection of the DNA is observed from RE3 extending into the −35 promoter hexamer. Several hyper-sensitive sites are present within the inverted repeat region and the RE2-RE3 spacer, indicating that conformational changes occur to the DNA when bound by ComA 14,36.
We are not able to directly compare the concentrations of ComA needed for binding DNA in vitro to those needed for transcription activation in vivo because the fraction of purified his6-ComA that is active in vitro is not known. In addition, the DNA templates used for in vitro binding assays are short linear fragments, whereas the templates in vivo are contained in supercoiled chromosomal DNA coated with many DNA binding proteins.
Relative contribution of RE1, RE2, and RE3 in transcription activation of rapA
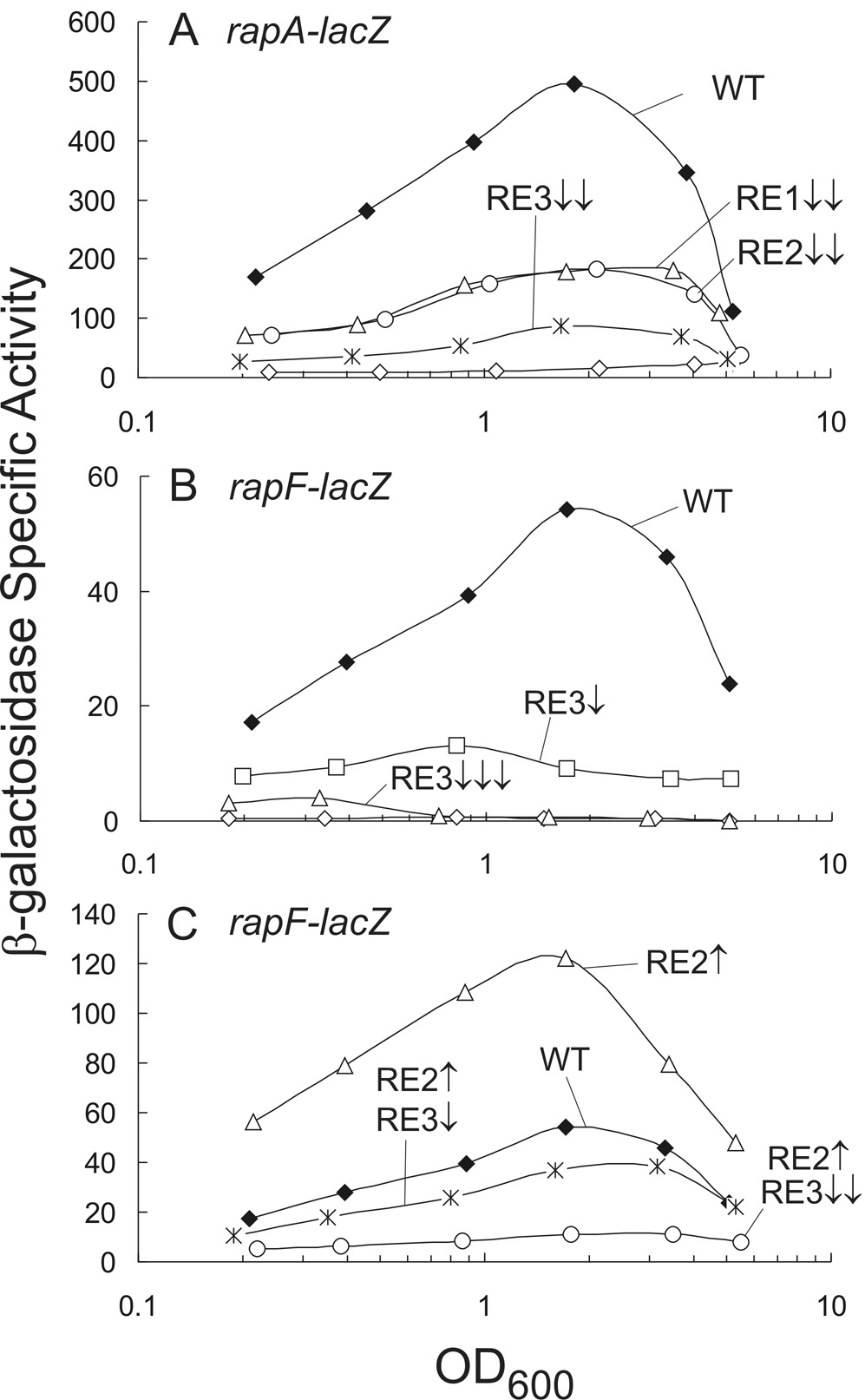
rapA has two potential ComA binding sites 29 and the promoter-proximal site (Fig. 1) is close to consensus in all three recognition elements, containing only a single change in RE2 (5’-CCGAAA) away from consensus (5'-CCGCAA). In contrast to srfA, which requires both the proximal and distal ComA binding sites (Fig. 2 and 26), the promoter-proximal ComA binding site from rapA was sufficient for regulated expression of a rapA-lacZ transcriptional fusion (Fig. 4A; data not shown). Transcription of rapA-lacZ was relatively high at low culture density (Fig. 4A). There was a small, but reproducible 2–2.5–fold increase in β-galactosidase specific activity as the culture density increased (Fig. 4A). In contrast, there was an ~10-fold increase in β-galactosidase specific activity in cultures containing a srfA-lacZ fusion (Fig. 2).
Figure 4. Roles of recognition elements in transcription activation of rapA and rapF.
Cultures containing PrapA-lacZ (A) or PrapF-lacZ (B and C) fusions were grown in defined minimal medium and aliquots taken throughout growth for determination of β-galactosidase specific activity. Each arrow represents a single base substitution: up arrows indicate substitutions toward consensus and down arrows away from consensus. Underlined nucleotides represent mismatches from the consensus sequence.
A. KG112 wild type PrapA-lacZ (filled diamonds); KG544 RE1 5’-TTTCGA and RE2 consensus (triangles); KG513 RE2 5’-TCGAAA (circles); KG545 RE3 5’-TTTCGA and RE2 consensus (asterisk), and KG148 wild type reporter ΔcomA (open diamonds).
B. KG277 wild type PrapF-lacZ (filled diamonds); KG556 1 mismatch in RE3 from consensus (5’-TTTCGG) (squares); KG266 3 mismatches in RE3 from consensus (5’-GTGTCG) (triangles); and KG239 wild type reporter ΔcomA (open diamonds).
C. KG277 wild type PrapF-lacZ (filled diamonds) is the same as in Panel B; KG555 1 mismatch in RE2 toward consensus (5’-CCGAAA) (triangles); KG566 1 mismatch in RE2 toward consensus (5’-CCGAAA) 1 mismatch in RE3 from consensus (5’-TTTCGG) (asterisk); and KG557 1 mismatch in RE2 toward consensus (5’-CCGAAA) 2 mismatches in RE3 from consensus (5’-TTTCGT) (circles).
Like srfA, we found that each of the recognition elements contributes to transcription of rapA-lacZ. Substitutions were made in each recognition element so that two positions varied away from consensus (5’-TTTCGA). Mutations in RE1 and RE2 reduced β-galactosidase specific activity ~2–2.5-fold (Fig. 4A). Mutations in RE3 had a larger effect, reducing β-galactosidase specific activity ~5-fold compared to wild type (Fig. 4A). Based on these results, we conclude that all three recognition elements are required for optimal expression of rapA and that in the context of this promoter, RE3 is most important.
Recognition Element 3 is required for transcriptional activation of rapF
rapF has a single ComA binding site in its regulatory region (Fig. 1). Transcription of rapF-lacZ increased as the culture density increased and maximal β-galactosidase specific activity occurred near the end of exponential growth (Fig. 4B). Like the other target genes tested, transcription of rapF was dependent on ComA as very little β-galactosidase activity was observed in a comA null mutant (Fig. 4B).
We found that RE3 is required for transcription activation of rapF-lacZ. A single G to T mutation at position 3 of RE3 (5’-TTGCGG to 5’-TTTCGG) caused a significant decrease in β-galactosidase specific activity (Fig. 4B). A triple mutation in RE3 (5’-TTGCGG to 5’-GTGTCG) away from consensus reduced β-galactosidase specific activity of rapF-lacZ to levels similar to those in a comA null mutant (Fig. 4B). The decrease in transcription correlates with the severity of mismatches away from consensus and indicates that RE3 is required for transcriptional activation of rapF.
Compensatory substitutions within the ComA binding site restore transcription activation of rapF
We found that substitutions in RE2 toward consensus could compensate for mutations in RE3 away from consensus in the rapF regulatory region. The single mutation away from consensus in RE3 (5’-TTGCGG to 5’-TTTCGG), described above, was significantly suppressed by an A to C substitution toward consensus in position 1 of RE2 (5’-ACGAAA to 5’-CCGAAA) as expression of rapF-lacZ was restored to near wild type levels (Fig. 4C). An additional mutation in RE3 (5’-TTGCGG to 5’-TTTCGT) away from consensus (again in the context of the A1C mutation in RE2) further decreased rapF-lacZ expression to ~3–5-fold below that of wild type (Fig. 4C). In the context of the wild type RE3, the RE2 A1C mutation caused an ~2–3-fold increase in expression throughout the growth cycle, as compared to wild type (Fig. 4C).
In combination, our results indicate that substitutions in one recognition element toward the consensus sequence can compensate for substitutions in the other element(s) away from consensus. The different nucleotide combinations within the three binding determinants influence transcription activation in vivo by affecting expression at low culture density and the amount of induction that occurs when cultures are grown to high density.
Determination of the oligomeric state of ComA in solution
We found that ComA interacts with itself in a yeast two hybrid assay (data not shown). Furthermore, when native ComA and his6-ComA were over-expressed together in E. coli from compatible plasmids, the two forms of ComA appeared to stably associate with each other. Purification of his6-ComA by Ni-affinity chromatography resulted in the recovery of both his6-ComA and un-tagged ComA (data not shown). These results indicate that ComA is able to interact with itself, and that it probably does not function as a monomer.
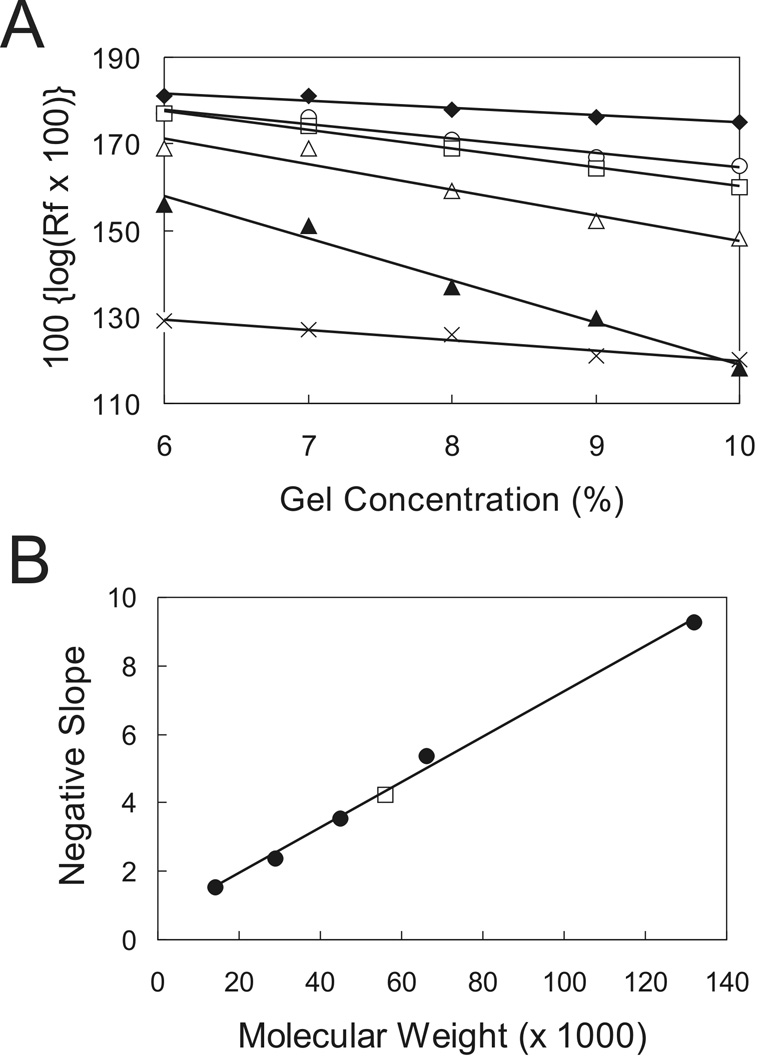
ComA is thought to function as a dimer based on its binding to an inverted repeat 14,26. Based on our findings that three recognition elements are required for ComA-mediated transcriptional activation in vivo and contribute to DNA binding in vitro, we sought to determine the oligomeric state of ComA. We used a method based on mobility in gels with different polyacrylamide concentrations 37. Briefly, purified his6-ComA and a set of protein standards of predetermined molecular weight were subjected to native gel electrophoresis in different concentrations of polyacrylamide. The migration distance of each protein was plotted against the acrylamide concentration with the slope of the line representing the retardation coefficient (Fig. 5A). A standard curve was generated from the retardation coefficients of the protein standards and the molecular weight of ComA was interpolated from the graph (Fig. 5B). Based on the average of three individual experiments, the molecular weight of his6-ComA appeared to be 53 KDa. Since the theoretical molecular weight of his6-ComA is 25 KDa, it appears that ComA functions as a dimer in solution.
Figure 5. Determination of the oligomeric state of ComA.
Purified his6-ComA was separated by native gel electrophoresis in different concentrations of polyacrylamide along with protein standards of known molecular weight. The migration distance (Rf) was determined by measuring the distance each protein traveled in the gel and dividing this value by the distance traveled by the bromophenol blue dye.
A. Representative experiment showing the migration distances of purified his6-ComA and protein standards in gels with different polyacrylamide concentrations. α-lactalbumin (filled diamonds); chicken egg albumin (circles); his6-ComA (squares); bovine serum albumin (BSA) monomer (open triangles); BSA dimer (filled triangles); and carbonic anhydrase from bovine erthrocytes (X).
B. Representative standard curve of the slope of the migration distances of each protein standard. The negative slope of each protein standard was determined from the experiment in Panel A and plotted as a function of known molecular weight for that particular protein: α-lactalbumin (14.2 KDa), carbonic anhydrase (29 KDa), egg albumin (45 KDa), BSA monomer (66 KDa), and BSA dimer (132 KDa). Protein standards (filled circles) and his6-ComA (square). The molecular weight of his6-ComA was 53 KDa based on the average of three independent experiments. The theoretical molecular weight of his6-ComA is 25 KDa. We conclude that his6-ComA functions as a dimer in solution.
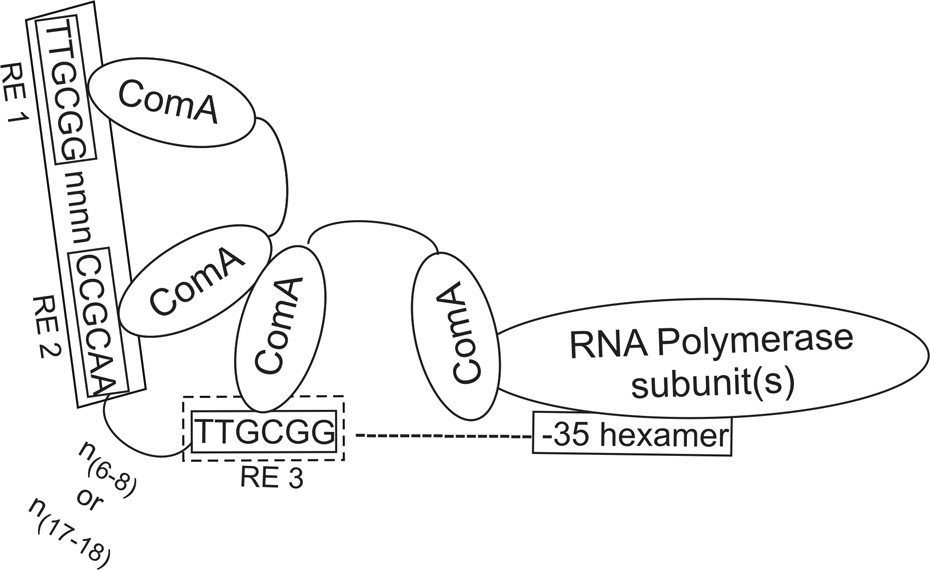
The question remains how does ComA bind to the tri-partite ComA binding site to activate transcription? We postulate that two dimers of ComA occupy a single binding site consisting of RE1, RE2, and RE3. Two possible models for the binding configuration seem most plausible and results described below support the first model. 1) One dimer of ComA binds RE1 and RE2 and a second dimer binds RE3 and perhaps non-specific sequences downstream of RE3. Alternatively, 2) a dimer of ComA could bind RE2 and RE3 (themselves an inverted repeat) and a second dimer could bind RE1 and perhaps non-specific sequences upstream of RE1. It is also possible that the fourth ComA subunit in these putative complexes is not bound to DNA. In either model, it seems most likely that the spacing between the two recognition elements that are bound by a single ComA dimer would be most severely restricted in length, while the spacer separating two dimers of ComA might be more accommodating to alterations in length. We tested the effects of altering the lengths of the spacer between RE1 and RE2 and also between RE2 and RE3.
Spacer length separating recognition elements 1 and 2 is important for transcription activation
The 4 bp spacer length separating RE1 and RE2 is conserved among the known ComA binding sites, except for rapE which has a 5 bp spacer (see below). We tested the importance of the spacer distance in the context of rapF and found that any deviation from 4 bps severely disrupted transcription. We created insertions and deletions of varying lengths (−1, −2, −3, +1, +2, and +3 bps) between RE1 and RE2 of the rapF regulatory region. We also introduced half and full helical turns of DNA (i.e., 5, 6, 10, or 11 consecutive adenines) within the spacer. All of the mutations severely reduced transcription of rapF in vivo (data not shown) indicating that a 4 bp spacer separating RE1 and RE2 is the optimal length for transcription activation of rapF by ComA and that changes are not well-tolerated.
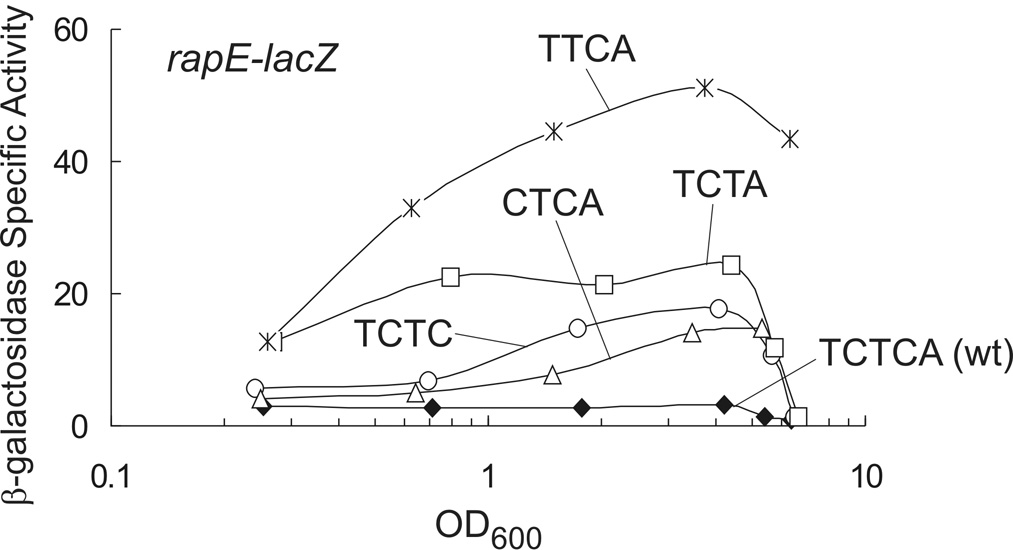
Since rapE has an unusual 5 bp spacer (TCTCA) separating RE1 and RE2 (Fig. 1), we sought to determine whether or not this atypically long spacer had an effect on transcription. A fragment of the rapE promoter containing only the promoter-proximal ComA binding site and encoding the first 10 codons of rapE was fused to lacZ and used to monitor transcription. Expression of rapE-lacZ was low and relatively constant throughout growth with little obvious increase in β-galactosidase specific activity at high culture density (Fig. 6). Moreover, the low level of β-galactosidase specific activity was reduced to background levels in a comA null mutant indicating that transcription of rapE is dependent on comA (data not shown). We constructed four single nucleotide deletions in the spacer separating RE1 and RE2 (resulting in spacer sequences: TTCA, TCTA, TCTC, and CTCA) and measured the effects on expression of rapE. Removal of a single nucleotide significantly increased transcription of rapE in all cases, albeit to different extents depending on the sequence (Fig. 6). All of the four nucleotide spacers, with the exception of TCTA, allowed the density-dependent increase in transcription typically observed with other genes activated by ComA. The spacer sequence TTCA had the largest effect on transcription; i.e., a 6-fold increase in β-galactosidase specific activity was observed at low culture density and expression at high density was increased ~17-fold relative to wild type (Fig. 6). The spacer sequence TCTA also increased β-galactosidase specific activity at low culture density 6-fold compared to wild type, but resulted in the smallest induction in expression (~1.5-fold) at high culture density of all the spacer mutants tested (Fig. 6). The spacer sequences TCTC and CTCA had similar effects increasing β-galactosidase specific activity ~6-fold at high culture density compared to wild type, while no effect was observed on expression at low culture density (Fig. 6). From this, we conclude that a 4 bp spacer separating RE1 and RE2 is optimal for transcriptional activation of target genes by ComA. Moreover, the sequence of the spacer affects both the level of expression observed at low culture density and the amount of induction during the response to high culture density.
Figure 6. Role of the spacer separating RE1 and RE2 in transcription activation of rapE.
Cultures containing PrapE-lacZ fusions were grown in defined minimal medium and aliquots taken throughout growth for determination of β-galactosidase specific activity. KG522 wild type TCTCA spacer (filled diamonds); KG841 TTCA spacer (asterisk); KG521 TCTA spacer (squares); KG268 TCTC spacer (circles); and KG852 CTCA spacer (triangles).
DNA sequence determinants in the spacer between RE1 and RE2
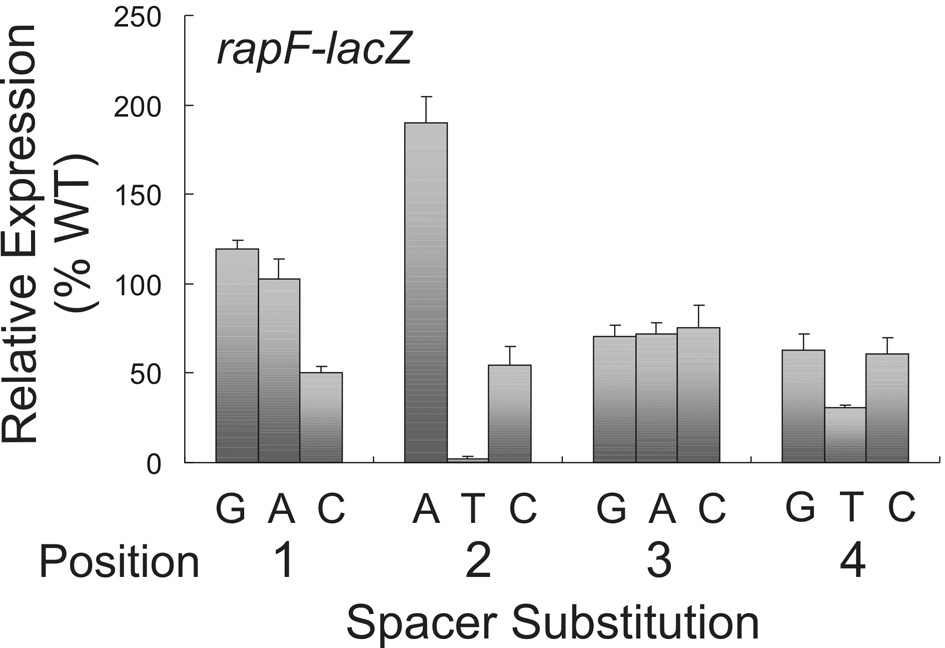
The sequence of the spacer separating RE1 and RE2 is not well conserved among the known ComA binding sites (consensus N(a/t)T(g/c) 29. However, the results with the spacer mutations in rapE (above) indicate there might be additional sequence-specific information in this region. To determine if sequence specific information does exist in the spacer separating RE1 and RE2, we altered each base in the ComA binding site of rapF-lacZ to the other three bases and measured effects on β-galactosidase specific activity. In general, substitutions at positions 1 and 3 were well tolerated and had little or no effect on expression of rapF; the exception being a T to C substitution at position 1 which decreased transcription ~2-fold (Fig. 7). Substitutions at positions 2 and 4 had more dramatic effects on transcription of rapF. Specifically, a T at either position had a negative effect virtually eliminating transcription when present at position 2 and reducing it >2-fold when present at position 4 (Fig. 7). Interestingly, a G to A substitution at position 2 had a stimulatory effect on transcription increasing it ~2-fold relative to wild type (Fig. 7).
Figure 7. Effect of the spacer sequence separating RE1 and RE2 in transcription activation of rapF.
Cultures containing PrapF-lacZ fusions were grown in defined minimal medium and aliquots taken throughout growth for determination of β-galactosidase specific activity. The time point containing the maximal β-galactosidase activity which typically peaks at OD600 1–1.5 is shown. The wild type RE1-RE2 spacer is 5’-TGTA and defined as 100%. Substitutions in position 1 of the spacer: KG331 (T-G); KG282 (T-A); and KG283 (T-C). Position 2: KG284 (G-A); KG285 (G-T); and KG305 (G-C). Position 3: KG311 (T-G); KG312 (T-A); and KG286 (T-C). Position 4: KG325 (A-G); KG310 (A-T); and KG287 (A-C). The data are an average of three independent experiments with standard deviation shown.
Taken together, our results indicate that there is sequence-specific information in the spacer region between RE1 and RE2. However, it appears that this information is largely context-specific; analyses of other spacer regions indicate that the effects of specific nucleotide changes differ among the various regulatory regions (unpublished results). A more detailed combinatorial analysis would be necessary to make general rules about the sequence information in the RE1-RE2 spacer region.
Role of the spacer separating RE2 and RE3 in transcription activation
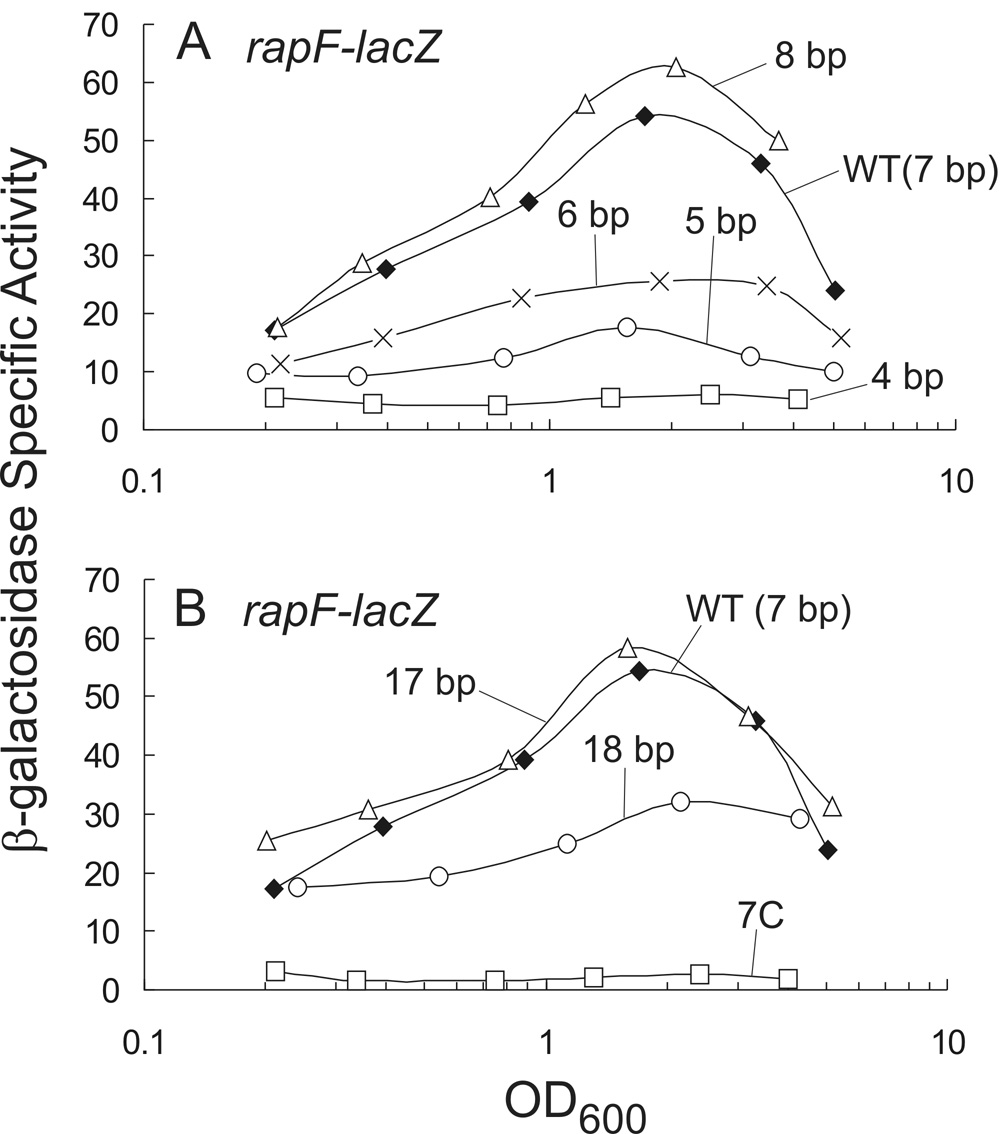
Of the known ComA binding sites present within target gene promoters, the spacer region separating RE2 from RE3 varies in length from 4–8 nucleotides, with a median length of 7 bp (Fig. 1; data not shown). We sought to determine the role of the spacer separating RE2 and RE3 in transcription by altering the length of the spacer in the rapF regulatory region. We found that a 7 bp or 17 bp spacer (an added helical turn of DNA) is optimal for transcription. Moreover, the sequence composition is important as A/T-rich residues are required for transcription activation by ComA.
We altered the spacer separating RE2 and RE3 in rapF-lacZ from 7 bp to 8, 6, 5, and 4 bp and monitored β-galactosidase activity throughout growth. An 8 nucleotide spacer (GAAAAAAA, with an extra A) had no effect on transcription of rapF-lacZ (Fig. 8A). Removal of additional nucleotides within the spacer to 6, 5, and 4 bp resulted in a progressive decrease in transcription of rapF-lacZ. For example, removal of 1 or 2 nucleotides (resulting in GAAAAA or GAAAA) decreased β-galactosidase specific activity ~2-fold or ~3-fold, respectively, while a 4 bp spacer (GAAA) reduced expression of rapF-lacZ to levels of a comA null mutant (Fig. 4B and Fig 8A).
Figure 8. Effects of the spacer separating RE2 and RE3 in transcription activation ofrapF.
Cultures containing PrapF-lacZ fusions were grown in defined minimal medium and aliquots taken throughout growth for determination of β-galactosidase specific activity.
A. KG277 wild type 7 bp spacer G(A)6 (filled diamonds); KG531 8 bp spacer G(A)7 (triangles); KG324 6 bp spacer G(A)5 (X); KG532 5 bp spacer G(A)4 (circles); and KG314 4 bp spacer G(A)3 (squares).
B. KG277 wild-type 7 bp spacer G(A)6 (filled diamonds) is the same as in Panel A; KG309 17 bp spacer G(A)16 (triangles); KG472 18 bp spacer G(A)17 (circles), and KG473 7 bp spacer comprised entirely of cytosines (C)7 (squares).
To further refine the spacer length separating RE2 and RE3 and to investigate any helical phasing that might exist, we introduced a half helical turn (5 or 6 adenines) and a full helical turn (10 or 11 adenines) of DNA within this region of the ComA binding site. Addition of a full helical turn of DNA (10 adenines) for a spacer length of 17 bp had no effect on transcription of rapF-lacZ and addition of 11 adenines had a small effect reducing β-galactosidase activity ~2- fold compared to wild type (Fig. 8B). Addition of a half-helical turn of DNA reduced transcription of rapF 60–80% of wild type (data not shown).
To determine if the spacing requirements separating RE2 and RE3 identified for rapF are also true for other target genes of ComA, we altered this region of the ComA binding site in rapA and rapC and determined the effects on transcription. rapA has an 8 bp spacer separating RE2 and RE3 (TTCGACAA). Changing this spacer to 7 bp (TTCACAA) caused a small increase in transcription of rapA-lacZ (data not shown). In contrast, rapC normally has a 7 bp spacer (ACAAAGA). Changing that to 8 bp caused a small decrease in transcription of rapC-lacZ (data not shown). These results indicate that a 7 bp spacer separating RE2 and RE3 is optimal for activation by ComA, at least for the three promoters tested (rapF, rapA, and rapC).
To determine if the same helical phasing exists between RE2 and RE3 as was determined for rapF, we introduced 9 and 10 consecutive adenines between RE2 and RE3 of the rapA and rapC ComA binding sites, respectively, to yield a total spacer length of 17 bps. Like rapF, introduction of DNA to the optimal 17 bp spacer length had no effect on transcription of rapA nor rapC (data not shown). Taken together, we conclude that a 7 bp or 17 bp spacer separating RE2 and RE3 is optimal for transcription activation of target genes by ComA and that the helical phasing is important.
The base composition of the spacer separating RE2 and RE3 is conserved among the known ComA binding sites. The average A/T composition within this region of all known ComA binding sites is 74%, unusually high even for the low G+C B. subtilis. We found that changing the base composition of the A/T-rich spacer to G/C caused a decrease in transcription of rapF-lacZ. The seven adenines were replaced with cytosines and the β-galactosidase specific activity of rapF-lacZ was reduced to levels similar to those in a comA null mutant (Fig. 4B and Fig 8B).
Taken together, our findings indicate that within the context of a minimal ComA binding site, the optimal spacer length separating RE2 and RE3 appears to be 7 bp or 17 bp. Finally, the composition of the spacer appears to be critical for transcription activation of rapF and, by inference, other known target genes.
Estimation of the number of ComA binding sites present in the B. subtilis genome
Based on the mutagenesis of the regulatory regions of several ComA-dependent target genes, we propose a refined consensus ComA binding site (5'-TTGCGGnnnnCCGCAA -n(6–8 or 17–18)-TTGCGG). Although our results indicate the optimal RE2-RE3 spacer length is 7 bp or 17 bp, spacer lengths of 6 bp, 8 bp, and 18 bp are still functional for activation by ComA and are included in our consensus sequence. The identification of a revised ComA binding site should aid in our understanding of how ComA functions to activate transcription of target genes and the identification of additional target genes, should they exist. The nine known target genes contain an average of 3.6 mismatches from the consensus sequence. Search of the B. subtilis genome using this refined sequence as query and allowing for 3 and 4 mismatches, revealed 37 and 208 hits, respectively. This is significantly less than the number of predicted sites (~700) expected with the old consensus sequence.
The ComA-dependent regulatory regions of all known target genes were identified in our search, except for yvfH and pel which contains a 4 bp and 5 bp RE2-RE3 spacer, respectively. We excluded sites containing a spacer < 6 bp in our refined consensus sequence because the mutagenesis with rapF showed that a spacer of this length was deleterious for transcription (Fig. 8A). In addition to a small spacer, the yvfH ComA binding site has 6 mismatches away from consensus with half of them residing in RE3. Our mutagenesis of the srfA regulatory region indicates that 3 mismatches in a single recognition element virtually eliminate transcription activation by ComA unless additional upstream sites are present (Fig. 2). With no obvious upstream regulatory sequences present in the yvfH promoter, we presume the combination of a degenerate RE3 and a suboptimal RE2-RE3 spacer explains why ComA has such a small effect (<2-fold) on transcription activation of yvfH 29. pel, on the other hand, has additional upstream regulatory sequences that presumably compensate for an unusually small RE2-RE3 spacer (unpublished data).
Without considering the effects of additional regulatory elements, e.g., those present in pel, we may have under-estimated the number of potential ComA binding sites. Nonetheless, our genomic analysis revealed a total of 33 additional genes (many of which have no known function) that contain a putative ComA binding site located within 500 bp upstream of the coding sequence. None of these genes were found to be affected by ComA using DNA microarray analyses in two independent studies 28,29. It is possible that the ComA binding site for some of these genes is non-functional for activation of transcription because it is not in the proper position relative to the binding site of RNA polymerase. Alternatively, some of these genes may require regulatory proteins in addition to ComA to activate transcription, as is the case with degU 24. Another possibility is that some genes are negatively regulated by transcription factors under the growth conditions examined. We suspect there are regulatory regions corresponding to each of these possibilities.
Discussion
In this work, we found that the promoter regions for genes activated by ComA contain three recognition elements: RE1, RE2, and RE3. RE1 and RE2 comprise an inverted repeat and the RE3 consensus sequence is identical to that of RE1. Each one of these sequence elements is required for ComA-dependent transcriptional activation of target genes. Each element influences binding by purified his6-ComA in vitro and previous footprinting studies 14,36 indicate some protection of all three elements by ComA. The simplest interpretation of these findings is that the ComA binding site is composed of all three recognition elements.
Model for ComA binding DNA
In addition to the three recognition elements, the sequences separating them are important for transcription activation of target genes by ComA, presumably because they function to properly position the recognition elements for ComA DNA binding. As a result, permutations in the length of the spacers typically have deleterious effects on transcription (Fig. 6 and Fig 8).
Effects of altering the spacers length strongly support a model in which a dimer of ComA binds to RE1 and RE2 and a second dimer binds to RE3 (Fig. 9). The strict 4 bp spacing requirement separating RE1 and RE2 is consistent with a single dimer of ComA occupying these recognition elements. Moreover, the flexibility in the spacer separating RE2 from RE3 (i.e., spacer length of 6–8 bp or 17–18 bp) indicates that another dimer of ComA probably binds RE3 and interacts with the dimer bound at RE1 and RE2. In this model, the RE2-RE3 spacer serves as a flexible bridge allowing the two dimers to interact resulting in cooperative binding observed in the gel-mobility assays (Fig. 3). Replacement of the A/T rich spacer separating RE2 and RE3 with G-C bps virtually eliminated transcription activation of rapF (Fig. 8B), presumably because the DNA could not bend properly to allow for functional alignment of the recognition elements.
Figure 9. Model for ComA binding DNA.
We propose that 2 dimers of ComA bind DNA with one dimer occupying RE1 and RE2 and a second dimer occupying RE3 and non-specific sequence downstream. Protein-protein interactions between the two dimers probably help stabilize the complex resulting in cooperative binding observed in the gel-shift assays (Fig. 3). The A/T-rich tract separating RE2 and RE3 probably facilitates DNA bending and the proper positioning of the recognition elements for ComA DNA binding.
Prevalence of a tri-partite ComA regulatory sequence in other Bacillus species
Homologs of ComA are present in other Bacillus species including B. licheniformis ATCC 14580, B. amyloliquefaciens FZB42, and B. pumilus SAFR-032. In B. licheniformis, ComA directly regulates transcription of lchA, involved in the production of the lipopeptide lichenysin A 38,39. The lchA regulatory region of B. licheniformis resembles that of srfA in B. subtilis with two inverted repeats (5’-TTTCGGtatcACGCAT and 5’-ATTCGGcatcCCGCAT) separated by 17 bp. Mutational analyses of the promoter-proximal inverted repeat revealed the importance of RE1 and RE2 in transcription of lchA by ComA 38. The existence of RE3 was not known at the time of that analysis; however, closer examination of the lchA promoter region reveals a putative RE3 (5’-TTTCAC) located 6 bp downstream of RE2 in the promoter-proximal ComA binding site.
In B. amyloliquefaciens, ComA was postulated to directly regulate transcription of degQ 40. Analysis of the degQ promoter reveals a well conserved RE1 and RE2 inverted repeat (5'- TTGCGGtgtcACGCAG) with a putative RE3 (5’-TTTCGG) positioned 17 bp downstream of RE2. It appears that ComA likely utilizes a similar tri-partite binding site to activate transcription of target genes in other Bacillus species.
Degeneracy of the ComA binding site is required for normal cell density-dependent regulation
We found that degeneracy of the ComA binding site is important for the regulation of genes in a population density-dependent manner. ComA binding sites in gene regulatory regions average 3.6 mismatches away from consensus. Promoters with a near consensus ComA binding site(s) have elevated transcription at low culture density compared to promoters with a degenerate site. This elevated activity, in turn, depresses the magnitude of the response (fold induction) observed at high culture density, thus lessening the ability to coordinate transcription with population density. On the other hand, promoters with a degenerate site have low transcription at low culture density and respond to increased population density with significantly increased transcription resulting in a larger induction ratio.
These trends are most obvious when comparing transcription of rapA and srfA. rapA has a near consensus binding site with high expression at low culture density and a modest 2.5-fold induction as the population density increases (Fig. 4A). In contrast, srfA has two degenerate binding sites resulting in low expression at low culture density and an ~10-fold increase in expression at high culture density (Fig. 2), i.e., srfA is regulated in a population density-dependent manner. Mutations in the promoter-proximal ComA binding site of the srfA regulatory region toward the consensus sequence increased expression of srfA at low culture density so much that no further increase in transcription was observed at high culture density (Fig. 2B). This result could indicate that there is no regulation by population density and expression is always at a high level. Alternatively, there could still be some regulation by culture density, but we are technically unable to go to a low enough density to see the effect. In either case, the normal pattern of cell density regulation is abolished and although a better binding site enhances ComA DNA binding and transcription activation of target genes, it is detrimental for the regulation of genes in a population density-dependent manner.
The affinity of ComA for its DNA binding site provides a mechanism to control the temporal expression of regulon genes and fine tune the response to population density. Transcription of the ComA regulon is dependent on the concentration of ComA~P. At low culture density, ComA is predominately in the non-phosphorylated, inactive state. The small amounts of ComA~P present at low culture density probably bind to high affinity sites (e.g., the regulatory region of rapA) resulting in enhanced transcription of target genes at low population densities (Fig. 4A). In contrast, we presume that low affinity degenerate sites (e.g., those present in srfA) are largely unoccupied by the small amounts of ComA~P present at low culture densities resulting in low levels of transcription (Fig. 2).
Degeneracy in transcription factor binding sites is analogous to degeneracy in bacterial promoter sequences. There exists tremendous variation in the sequences of bacterial promoters recognized by a given form of RNA polymerase. Many weak promoters require activator proteins that stimulate transcription initiation, often by recruiting RNA polymerase to the promoter 41–43. In the well studied example of the lac operon promoter, mutations in the promoter toward consensus can bypass the need for the activator CAP-cAMP, thereby reducing some of the regulation normally associated with the activator. Thus, sequence degeneracy in certain DNA binding sites is critical to their regulatory function. Definitions of consensus sequences can be misleading and often fail to capture the importance of weaker binding sites for regulation.
Materials and Methods
Bacterial strains and growth media
Routine cloning was performed in E. coli strain DH5α. B. subtilis strains (Table 1) were all derived from the parental strain JH642 (trpC2 pheA1) 44. Liquid cultures of B. subtilis were grown in S7 defined minimal medium salts 45 containing 50 mM MOPS instead of 100 mM (S750) and supplemented with 1% glucose, 0.1% glutamate, tryptophan (40 µg/ml), phenylalanine (40 µg/ml), and threonine (120 µg/ml), where appropriate. B. subtilis was grown on solid medium containing Spizizen’s minimal salts 46 supplemented with 1% glucose, 0.1% glutamate and the appropriate individual amino acids as described above. LB agar plates were used for routine cloning and growth of B. subtilis and E. coli. The following concentrations of antibiotics were used: ampicillin (100µg/ml), neomycin (2.5 µg/ml), and chloramphenicol (5 µg/ml).
Table 1.
Strains used.
| Strain | Genotypea |
|---|---|
| srfA-lacZ fusions | |
| KG102 | amyE::{srfA (−372 – +10)-lacZ neo} |
| KG125 | amyE::{srfA (−434 – +10)-lacZ neo} |
| KG150 | amyE:{srfA (−434 – +10)-lacZ neo} ΔcomA::cat |
| KG158 | amyE::{srfA (−372 – +10; −362T-G; −349T-A; −340T-G; −338A-G; −337C-G)-lacZ neo} |
| KG160 | amyE::{srfA (−372 – +10; −340T-G; −338A-G; −337C-G)-lacZ neo} |
| KG464 | amyE::{srfA (−372 – +10; −362T-G; −349T-A; −340T-G; −338A-G; −337C-G)-lacZ neo} ΔcomA::cat |
| KG565 | amyE::{srfA (−372 – +10; −362T-G; −349T-A; −342 –−337GCATAT)-lacZ neo} |
| KG567 | amyE::{srfA (−372 – +10; −362T-G; −354 –−349ATATGC; −340T-G; −338A-G; −337C-G)-lacZ neo} |
| KG780 | amyE::{srfA (−372 – +10; −364 –−359GCATAT; −349T-A; −340T-G; −338A-G; −337C-G)-lacZ neo} |
| rapA-lacZ fusions | |
| KG112 | amyE::{rapA (−126 – +10)-lacZ neo} |
| KG148 | amyE::{rapA (−126 – +10)-lacZ neo} ΔcomA::cat |
| KG513 | amyE::{rapA (−126 – +10; −96C-T)-lacZ neo} |
| KG544 | amyE::{rapA (−126 – +10; −93A-C; −101G-A; −104G-T)-lacZ neo} |
| KG545 | amyE::{rapA (−126 – +10; −93A-C; −80A-T; −77G-A)-lacZ neo} |
| rapF-lacZ fusions | |
| KG239 | amyE::{rapF (−731 – +10)-lacZ neo} ΔcomA::cat |
| KG266 | amyE::{rapF (−108 – +10; −83T-G; −80C-T; −79G-C)-lacZ neo} |
| KG277 | amyE::{rapF (−108 – +10)-lacZ neo} |
| KG282 | amyE::{rapF (−108 – +10); −100T-A)-lacZ neo} |
| KG283 | amyE::{rapF (−108 – +10; −100T-C)-lacZ neo} |
| KG284 | amyE::{rapF (−108 – +10; −99G-A)-lacZ neo} |
| KG285 | amyE::{rapF (−108 – +10; −99G-T)-lacZ neo} |
| KG286 | amyE::{rapF (−108 – +10; −98T-C)-lacZ neo} |
| KG287 | amyE::{rapF (−108 – +10; −97A-C)-lacZ neo} |
| KG305 | amyE::{rapF (−108 – +10; −99G-C)-lacZ neo} |
| KG309 | amyE::{rapF (−108 – +10; −84[+10A])-lacZ neo} |
| KG310 | amyE::{rapF (−108 – +10; −97A-T)-lacZ neo} |
| KG311 | amyE::{rapF (−108 – +10; −98T-G)-lacZ neo} |
| KG312 | amyE::{rapF (−108 – +10; −98T-A)-lacZ neo} |
| KG314 | amyE::{rapF (−108 – +10; Δ[−86 – −84])-lacZ neo} |
| KG324 | amyE::{rapF (−108 – +10; Δ-84A)-lacZ neo} |
| KG325 | amyE::{rapF (−108 – +10; −97A-G)-lacZ neo} |
| KG331 | amyE::{rapF (−108 – +10; −100T-G)-lacZ neo} |
| KG472 | amyE::{rapF (−108 – +10; −84[+11A])-lacZ neo} |
| KG473 | amyE::{rapF (−108 – +10; −90G-C; [−89 –−84]A-C)-lacZ neo} |
| KG531 | amyE::{rapF (−108 – +10; −84[+1A])-lacZ neo} |
| KG532 | amyE::{rapF (−108 – +10;Δ [−85–−84])-lacZ neo} |
| KG555 | amyE::{rapF (−108 – +10; −96A-C)-lacZ neo} |
| KG556 | amyE::{rapF (−108 – +10; −81G-T)-lacZ neo} |
| KG557 | amyE::{rapF (−108 – +10; −96A-C; −81G-T; −78G-T)-lacZ neo} |
| KG566 | amyE::{rapF (−108 – +10; −96A-C; −81G-T)-lacZ neo} |
| rapE-lacZ fusions | |
| KG268 | amyE::{rapE (−112 – +10; Δ-98A)-lacZ neo} |
| KG521 | amyE::{rapE (−112 – +10; Δ-99C)-lacZ neo} |
| KG522 | amyE::{rapE (−112 – +10)-lacZ neo} |
| KG841 | amyE::{rapE (−112 – +10; Δ-101C)-lacZ neo} |
| KG852 | amyE::{rapE (−112 – +10; Δ-102T)-lacZ neo} |
All strains are derived from JH642 and contain trpC2 and pheA1 alleles (not indicated). The position of DNA relative to the start of the coding sequence and alterations in the ComA binding site are indicated in parentheses.
Oligonucleotides
All oligonucleotides used in this study were synthesized by Integrated DNA Technologies (IDT) and sequences are available upon request.
Cloning and mutagenesis
Transcriptional fusions to lacZ were first created by amplifying the promoter of interest from B. subtilis genomic DNA using the polymerase chain reaction (PCR) with Taq DNA polymerase (Roche). Fusions are indicated in the strain table and numbers in the regulatory regions represent the number of base pairs from the start of the open reading frame. EcoRI and BamHI restriction enzyme recognition sites were engineered into the 5’ and 3’ ends of each PCR product, respectively. PCR products were digested with EcoRI and BamHI restriction enzymes (NEB) and ligated into pKS2 9 which was also digested with the same two enzymes. Ligation reactions were transformed into strain DH2α and plated on LB with ampicillin. Plasmid DNA was isolated from transformants by the alkaline lysis method according to the manufacturer (Qiagen and Invitrogen). Clones were verified by DNA sequencing (MIT Biopolymers Lab and MGH Sequencing Facility). Plasmid DNA was transformed into B. subtilis strain JH642 and plated on LB with neomycin. All lacZ fusions contained the first 10 amino acids of the coding sequence of the gene of interest followed by a termination codon. Mutations in the ComA binding sites were created by add-on PCR or PCR SOEing 47, where appropriate.
An over-expression construct was made to express ComA as a hexa-histidine tagged fusion protein (his6-ComA) for purification in E. coli. Briefly, an N-terminal hexa-histidine tag was introduced between codons 1 and 2 of comA by add-on PCR of B. subtilis genomic DNA using Taq polymerase and the following primers (5’-GCTTAGTGGGTACCAAGGAGATATACATATGcatcaccatcaccatcacAAAAAGATACTAGT GATTGA-3’ and 5’-TGCTACGAGCATGCTTAAAGTACACCGTCTGA-3’) where KpnI and SphI restriction enzymes are in bold, the ribosome binding site is underlined, the hexa-histidine tag is in lowercase, and the termination codon is in italics. The PCR product was digested with KpnI and SphI and ligated into pBAD-Ap18 which was also digested with the same two restriction enzymes. The ligation reaction was transformed into strain DH5α and plated on LB with ampicillin. Plasmid DNA was isolated and the correct identity verified by sequencing.
Growth conditions and assay of β-galactosidase activity
Overnight cultures were grown as light lawns on minimal medium plates incubated at 37°C. Three ml of Spizizen salts was used to flood each plate and the OD600 determined using a spectrophotometer. Shaker flasks containing S750 minimal medium were inoculated to OD600 ~0.02 and incubated with vigorous aeration at 37°C. One ml aliquots were taken at specified times throughout the growth cycle and placed in a 2.2 ml 96-well polypropylene block (Qiagen) which was stored at −20°C until time to assay β-galactosidase activity. A second aliquot was taken to determine the OD600.
β-galactosidase specific activity was determined as described 48 with some modifications. Briefly, cells in the 96-well blocks were thawed to room temperature and 20 µl of toluene was added to each well. Cells were permeabilized directly in the blocks by vigorous pipetting up and down using a multi-channel pipettor. Permeabilized cells were transferred to a second block containing 1 ml Z-Buffer 48. A 100 µl aliquot of the cell suspension was transferred to a microtiter plate and the β-galactosidase assay initiated with the addition of 20 µl freshly prepared ONPG (4 mg/ml) and terminated with the addition of 40 µl 1M Na2CO3. Cell debris was pelleted in the microtiter plate by centrifugation at 3,000 g for 10 min. A 100 µl aliquot of each supernatant was transferred to a new plate using a multi-channel pipettor. The A420 was determined using a SpectraMax plate reader (Molecular Dynamics) and data analysis performed using Microsoft Excel. β-galactosidase specific activity was calculated as follows: 1000 × {(ΔA420/min/ml) / OD600 of culture}.
Purification of his6-ComA
A fresh overnight of strain DH5α containing pBAD-his6-ComA was diluted 1:200 into LB with ampicillin (300 µg/ml) and grown to OD600 0.5–0.8 at 37°C with vigorous aeration. L-arabinose (Sigma) was added (0.2% final concentration) to induce expression from pBAD. Cells were harvested 4–6 hrs later by centrifugation at 5,000 g for 5 min at 4°C. The cell pellet from 1 L of culture was resuspended in 10 ml Sonication Buffer (20 mM Tris pH 8, 0.3 M NaCl, 5% glycerol, 5 mM imidazole, 5 mM β-mercaptoethanol, 5 mM MgCl2) and cells were lysed by sonication (8 cycles of 20 sec on and 40 sec off on setting 4–5). The culture was cleared by centrifugation at 10,000 rpm for 20 min at 4°C and the cell extract was passed over 2 ml of Ni-NTA (Qiagen). After 10 washes with 10 ml Sonication buffer, his6-ComA was eluted from the column in 10 ml Sonication buffer with increasing concentrations of imidazole (15 mM, 50 mM, 120 mM, and 300 mM). The fractions were analyzed for purity by SDS-PAGE followed by coomassie staining. Typically, the fraction eluted in 120 mM imidazole was >95% pure (data not shown) and was dialyzed to remove imidazole against 5 buffer changes of 2 L dialysis buffer (20 mM Tris pH 8, 0.3 M NaCl, 5% glycerol, 10 mM β-mercaptoethanol, 5 mM MgCl2). Dialyzed protein was concentrated to >10 mg/ml using a Centricon-10 (Amicon). Glycerol was added to a final concentration of 40% and the protein concentration determined by Bradford assay using BSA as the protein standard. Purified his6-ComA was stored at −20°C until further use.
Gel mobility shift assays
DNA for the gel mobility shift assays was prepared by annealing two complementary oligonucleotides containing the promoter-proximal ComA binding site in the srfA regulatory region from −73bp to −46 bp from the start of the annotated coding sequence. Briefly, one of the oligonucleotides from each pair was labeled on its 5’end using (32P)gamma-ATP (NEN) and T4 polynucleotide kinase (NEB). A 1.3-fold molar excess of its complement was added to the mixture and heated to 95°C for 5 min followed by slow cooling to room temperature to facilitate annealing of the oligonucleotides. Duplex DNA was purified away from unincorporated label using a G-25 Centrispin 10 column (Princeton Separations). DNA templates contained the bases 5’-TCA preceding the ComA binding sequence and TC-3’ following it.
In vitro binding reactions contained 10 mM HEPES pH 7.6, 2 mM MgCl2, 0.1 mM EDTA, 0.2 M KCl, 10% glycerol, 5 mM DTT, 5–10 nM labeled DNA, 10 nM poly(dI-dC), and µM of purified his6-ComA. Protein-DNA complexes were allowed to equilibrate at 37°C for 30 min prior to the addition of 5µl 5X agarose gel loading dye. Samples were loaded into the wells of 15% polyacrylamide gels containing 5% glycerol and electrophoresed into the gel at 300 volts. Once the loading dye entered the gel, the voltage was reduced to 120 volts and gels were run for 5–6 hrs at 4°C. Gels were dried and analyzed using a PhosphorImager (Molecular Dynamics).
Determination of the oligomeric state of ComA
The oligomeric state of ComA was determined using native gels as previously described 37. Briefly, purified his6-ComA and native protein standards including bovine milk α-lactalbumin (MW 14.2 kDa), carbonic anhydrase from bovine erythrocytes (MW 29 kDa), chicken egg albumin (MW 45 kDa), bovine serum albumin (BSA) monomer (66 kDa), and BSA dimer (132 kDa) were subjected to electrophoresis in native gels containing 6, 7, 8, 9, and 10% acrylamide. The relative mobility (Rf) of each protein was determined by dividing its migration distance from the top of the gel to the center of the protein band by the migration distance of the bromophenol blue tracking dye from the top of the gel. A standard curve was generate by plotting 100 × {Log(Rf × 100)} versus the gel concentration. The negative slopes generated from the standard curve were plotted against the known molecular weights of the protein standards. The approximate native molecular weight of his6-ComA was estimated. The experiments were performed three times with an average native molecular weight of 53 kDa for his6-ComA.
BLAST searches of the B. subtilis genome
BLAST pattern searches of the B. subtilis genome were performed using the Subtilist website (http://genolist.pasteur.fr/SubtiList/). The previously proposed consensus sequence (5'-TTGCGGnnnnCCGCAA) and the refined consensus sequence (5'-TTGCGG-nnnn-CCGCAA -n(6–8 or 17–18)- TTGCGG) were used as query sequences. Using the previously proposed sequence as query and allowing for 2 and 3 mismatches, 372 and 3,210 hits, respectively, were revealed compared to just 37 and 208 hits using the refined consensus sequence as query and allowing for 3 and 4 mismatches, respectively. All previously known ComA binding sites present in target gene promoters were identified in both BLAST searches, except for yvfH and pel which has a 4 bp and 5 bp spacer separating RE2 and RE3, respectively.
Acknowledgments
We thank C.A. Lee and R.B. Weart for comments on the manuscript. This work was supported, in part, by the NIH Ruth L. Kirschstein NRSA GM071224 to KLG and Public Health Service grant GM50895 from the NIH to ADG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10:365–370. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- 3.Winans SC, Bassler BL. Chemical Communication among Bacteria. ASM Press; 2008. [Google Scholar]
- 4.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 5.Auchtung JM, Grossman AD. Extracellular peptide signaling and quorum responses in development, self-recognition, and horizontal gene transfer in Bacillus subtilis. In: Winans SC, Bassler BL, editors. Chemical Communication Among Microbes. Washington DC: ASM Press; 2007. in press. [Google Scholar]
- 6.Lazazzera B, Palmer T, Quisel J, Grossman AD. Cell density control of gene expression and development in Bacillus subtilis. In: Dunny GM, Winans SC, editors. Cell-Cell Signaling in Bacteria. Washington DC: ASM Press; 1999. pp. 27–46. [Google Scholar]
- 7.Msadek T. When the going gets tough: survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 1999;7:201–207. doi: 10.1016/s0966-842x(99)01479-1. [DOI] [PubMed] [Google Scholar]
- 8.Tortosa P, Dubnau D. Competence for transformation: a matter of taste. Curr Opin Microbiol. 1999;2:588–592. doi: 10.1016/s1369-5274(99)00026-0. [DOI] [PubMed] [Google Scholar]
- 9.Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 10.Tortosa P, Logsdon L, Kraigher B, Itoh Y, Mandic-Mulec I, Dubnau D. Specificity and genetic polymorphism of the Bacillus competence quorum-sensing system. J. Bacteriol. 2001;183:451–460. doi: 10.1128/JB.183.2.451-460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ansaldi M, Marolt D, Stebe T, Mandic-Mulec I, Dubnau D. Specific activation of the Bacillus quorum-sensing systems by isoprenylated pheromone variants. Mol Microbiol. 2002;44:1561–1573. doi: 10.1046/j.1365-2958.2002.02977.x. [DOI] [PubMed] [Google Scholar]
- 12.Bacon Schneider K, TM P, Grossman AD. Characterization of comQ and comX, two genes required for the production of ComX pheromone in Bacillus subtilis. J. Bacteriol. 2002;184:410–419. doi: 10.1128/JB.184.2.410-419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinrauch Y, Penchev R, Dubnau E, Smith I, Dubnau D. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 1990;4:860–872. doi: 10.1101/gad.4.5.860. [DOI] [PubMed] [Google Scholar]
- 14.Roggiani M, Dubnau D. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J. Bacteriol. 1993;175:3182–3187. doi: 10.1128/jb.175.10.3182-3187.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossman AD. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Ann. Rev. Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 16.Solomon JM, Lazazzera BA, Grossman AD. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 17.Core LaP, M TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol Microbiol. 2003;49:1509–1522. doi: 10.1046/j.1365-2958.2003.03659.x. [DOI] [PubMed] [Google Scholar]
- 18.Bongiorni C, Ishikawa S, Stephenson S, Ogasawara N, Perego M. Syngeristic regulation of competence development in Bacillus subtilis by two Rap-Phr systems. 2005;187:4353–4361. doi: 10.1128/JB.187.13.4353-4361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auchtung JM, Lee CA, Grossman AD. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J Bacteriol. 2006;188:5273–5285. doi: 10.1128/JB.00300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi H, Kensuke T, Kobayashi K, Ogasawara N, Ogura M. Bacillus subtilis RghR (YvaN) represses rapG and rapH, which encode inhibitors of expression of the srfA operon. Mol Microbiol. 2006;59:1714–1729. doi: 10.1111/j.1365-2958.2006.05059.x. [DOI] [PubMed] [Google Scholar]
- 21.Smits WK, Bongiorni C, Veengin JW, Hamoen LW, Kuipers OP, Perego M. Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol Microbiol. 2007;65:103–120. doi: 10.1111/j.1365-2958.2007.05776.x. [DOI] [PubMed] [Google Scholar]
- 22.Lazazzera BA. Quorum sensing and starvation: signals for entry into stationary phase. Curr Opin Microbiol. 2000;3:177–182. doi: 10.1016/s1369-5274(00)00072-2. [DOI] [PubMed] [Google Scholar]
- 23.Pottathil M, Lazazzera BA. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Frontiers in Bioscience. 2003;8:32–45. doi: 10.2741/913. [DOI] [PubMed] [Google Scholar]
- 24.Msadek T, Kunst F, Klier A, Rapoport G. DegS-DegU and ComP-ComA modulator-effector pairs control expression of the Bacillus subtilis pleiotropic regulatory gene degQ. J. Bacteriol. 1991;173:2366–2377. doi: 10.1128/jb.173.7.2366-2377.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mueller JP, Bukusoglu G, Sonenshein AL. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: Control of gsiA by the ComP-ComA signal transduction system. J. Bacteriol. 1992;174:4361–4373. doi: 10.1128/jb.174.13.4361-4373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakano MM, Zuber P. Mutational analysis of the regulatory region of the srfA operon in Bacillus subtilis. J. Bacteriol. 1993;175:3188–3191. doi: 10.1128/jb.175.10.3188-3191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang M, Grau R, Perego M. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J Bacteriol. 2000;182:303–310. doi: 10.1128/jb.182.2.303-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogura M, Yamaguchi H, Yoshida K, Fujita Y, Tanaka T. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B.subtilis two-component regulatory systems. Nucleic Acids Res. 2001;29:3804–3813. doi: 10.1093/nar/29.18.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comella N, Grossman AD. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum sensing transcription factor ComA in Bacillus subtiilis. Mol. Micriobiol. 2005;57:1159–1174. doi: 10.1111/j.1365-2958.2005.04749.x. [DOI] [PubMed] [Google Scholar]
- 30.Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 31.Jourlin-Castelli C, Mani N, Nakano MM, Sonenshein AL. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J Mol Biol. 2000;295:865–878. doi: 10.1006/jmbi.1999.3420. [DOI] [PubMed] [Google Scholar]
- 32.Nakano MM, Marahiel MA, Zuber P. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J. Bacteriol. 1988;170:5662–5668. doi: 10.1128/jb.170.12.5662-5668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaacks KJ, Healy J, Losick R, Grossman AD. Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J. Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Sinderen D, Withoff S, Boels H, Venema G. Isolation and characterization of comL, a transcription unit involved in competence development of Bacillus subtilis. Mol Gen. Genet. 1990;224:396–404. doi: 10.1007/BF00262434. [DOI] [PubMed] [Google Scholar]
- 35.Nakano MM, Magnuson R, Meyers A, Curry J, Grossman AD, Zuber P. srfA is an operon required for surfactin production, competence development, and efficient sporulation in Bacillus subtilis. J. Bacteriol. 1991;173:1770–1778. doi: 10.1128/jb.173.5.1770-1778.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Nakano S, Choi SY, Zuber P. Mutational analysis of the Bacillus subtilis RNA polymerase alpha C-terminal domain supports the interference model of Spx-dependent repression. J Bacteriol. 2006;188:4300–4311. doi: 10.1128/JB.00220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryan JK. Molecular weights of protein multimers from polyacrylamide gel electrophoresis. Anal Biochem. 1977;78:513–519. doi: 10.1016/0003-2697(77)90111-7. [DOI] [PubMed] [Google Scholar]
- 38.Yakimov MM, Golyshin PN. ComA-dependent transcriptional activation of lichenysin A synthetase promoter in Bacillus subtilis cells. Biotechnol. Prog. 1997;13:757–761. doi: 10.1021/bp9700622. [DOI] [PubMed] [Google Scholar]
- 39.Yakimov MM, Kroger A, Slepak TN, Giuliano L, Timmis KN, Golyshin PN. A putative lichenysin A synthetase operon in Bacillus licheniformis: initial characterization. Biochimica et Biophysica Acta. 1998:141–153. doi: 10.1016/s0167-4781(98)00096-7. [DOI] [PubMed] [Google Scholar]
- 40.Koumoutsi A, Chen XH, Vater J, Borriss R. DegU and YczE positively regulate the synthesis of Bacillomycin D by Bacillus amyloliquefaciens strain FZB42. Appl Environ Microbiol. 2007;73:6953–6964. doi: 10.1128/AEM.00565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 42.Reznikoff WS, Abelson JN. The lac promoter. In: Miller JH, Reznikoff WS, editors. The Operon. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1978. pp. 221–243. [Google Scholar]
- 43.Busby S, Ebright RH. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 44.Perego M, Spiegelman GB, Hoch JA. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 1988;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 45.Vasantha N, Freese E. Enzyme changes during Bacillus subtilis sporulation caused by deprivation of guanine nucleotides. J. Bacteriol. 1980;144:1119–1125. doi: 10.1128/jb.144.3.1119-1125.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harwood CR, Cutting SM. Molecular Biological Methods for Bacillus. Chichester, England: John Wiley & Sons; 1990. [Google Scholar]
- 47.Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 48.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1972. [Google Scholar]