Summary
Precise regulation of the formation, maintenance, and remodeling of the vasculature is required for normal development, tissue response to injury, and tumor progression. How specific microRNAs intersect with and modulate angiogenic signaling cascades is unknown. Here we identified microRNAs that were enriched in endothelial cells derived from mouse embryonic stem (ES) cells and in developing mouse embryos. We found that miR-126 regulated the response of endothelial cells to VEGF. Additionally, knockdown of miR-126 in zebrafish resulted in loss of vascular integrity and hemorrhage during embryonic development. miR-126 functioned in part by directly repressing negative regulators of the VEGF pathway, including the Sprouty-related protein, SPRED1, and phosphoinositol-3 kinase regulatory subunit 2 (PIK3R2). Increased expression of Spred1 or inhibition of VEGF signaling in zebrafish resulted in defects similar to miR-126 knockdown. These findings illustrate that a single miRNA can regulate vascular integrity and angiogenesis, providing a new target for modulating vascular formation and function.
Keywords: microRNA, angiogenesis, MAP kinase, SPRED1, endothelial, post-transcriptional, VEGF
Introduction
The vascular network is composed of an intricate series of vessels that serve as conduits for blood flow, regulate organ growth, and modulate the response to injury. The vasculature is also required for the expansion of tumor masses, as inhibition of new vessel formation prevents tumor growth. The signaling and transcriptional pathways that govern vascular formation are well studied, and that knowledge has formed the basis for novel therapeutic approaches (Lien and Lowman, 2008).
Vascular endothelial cells initially differentiate from angioblastic precursors and proliferate and migrate to form the primitive vascular plexus through the process of vasculogenesis. This network is further remodeled by angiogenesis and stabilized by recruitment of pericytes and vascular smooth muscle cells to form a functioning circulatory system. Several angiogenic stimuli are essential to establish the circulatory system during development and to control physiologic and pathologic angiogenesis in the adult. For example, secreted growth factors, including members of the vascular endothelial growth factor (VEGF), platelet-derived growth factor (PDGF), and fibroblast growth factor (FGF) families, bind to membrane-bound receptors and transmit signals through kinase-dependent signaling cascades. These signals ultimately result in gene expression changes that affect the growth, migration, morphology, and function of endothelial cells.
How endothelial cells selectively respond to the wide variety of angiogenic signals present in their microenvironment is poorly understood. For example, exposure to VEGF induces the proliferation of stalk cells in an angiogenic sprout, while inducing directed migration, rather than proliferation, of cells at the tip. One mechanism for modulating the endothelial response to growth factors is through inhibition of downstream kinase signaling cascades (Hellstrom et al., 2007; Jones et al., 2008). Sprouty and sprouty-related proteins are negative regulators of growth factor signaling (reviewed in (Mason et al., 2006)). In particular, Sprouty-related EVH1 domain-containing protein 1 (SPRED1) negatively regulates the activation of the MAP kinase pathway by binding and inactivating RAF1, an upstream kinase of the pathway (Nonami et al., 2004; Taniguchi et al., 2007; Wakioka et al., 2001).
Another potential mechanism by which endothelial cells may regulate growth factor responses may be through the use of microRNAs. microRNAs are a class of small RNAs (~20–25 nt) that regulate expression of target mRNAs post-transcriptionally, either through translational inhibition or destabilization of target mRNAs (reviewed in (Wu and Belasco, 2008; Zhao and Srivastava, 2007)). A high degree of sequence complementarity between the 5′ end of the microRNA, known as the “seed” sequence, and the mRNA is an important determinant of microRNA targeting, as is the accessibility of the mRNA at the potential binding site (Kertesz et al., 2007; Zhao et al., 2005).
Mice homozygous for a hypomorphic allele of Dicer, an enzyme essential for the biogenesis of most microRNAs, develop gross abnormalities during blood vessel development in the embryo and in the yolk sac (Yang et al., 2005). While several broadly expressed microRNAs regulate in vitro endothelial cell behavior, including proliferation, migration and the ability to form capillary networks (Chen and Gorski, 2008; Kuehbacher et al., 2007; Lee et al., 2007a; Poliseno et al., 2006; Suarez et al., 2007), the in vivo functions of endothelial-specific microRNAs, or their targets, have yet to be described. By microRNA profiling of ES cell-derived endothelial cells, we identified a group of endothelial-enriched microRNAs, including miR-126, miR-146, miR-197, and miR-625. These microRNAs were also enriched in endothelial cells of developing mouse embryos. Through modulation of the expression of miR-126 in vitro and in vivo, we demonstrate that miR-126 positively regulates the response of endothelial cells to VEGF. Furthermore, we show that miR-126 regulates VEGF-dependent PI3 kinase and MAP kinase signaling by directly targeting PI3KR2 (p85-β) and SPRED1, respectively, two negative regulators of the VEGF signaling pathway.
Results
miR-126 is the most highly enriched microRNA in endothelial cells
To determine when the endothelial lineage first appears in differentiating mouse ES cells in the embryoid body (EB) model, extensive mRNA expression profiling was performed for endothelial marker expression using quantitative reverse transcriptase PCR (qRT-PCR). Oct4, a marker of pluripotent ES cells, was rapidly down-regulated during differentiation (data not shown). At day 4 (d4) of EB formation, Flk1/Vegfr2 was dramatically induced, but other endothelial markers, such as Tie2/Tek and CD31/Pecam1, were unchanged (Fig. 1A). This population of Flk1-positive cells at d4 contains vascular precursor cells, as evidenced by the ability of isolated Flk1-positive cells to differentiate into the endothelial lineage in the presence of VEGF (data not shown). By d6 of EB formation, endothelial markers, including CD31 and Tie2, were robustly expressed and remained elevated even after 14 days of differentiation.
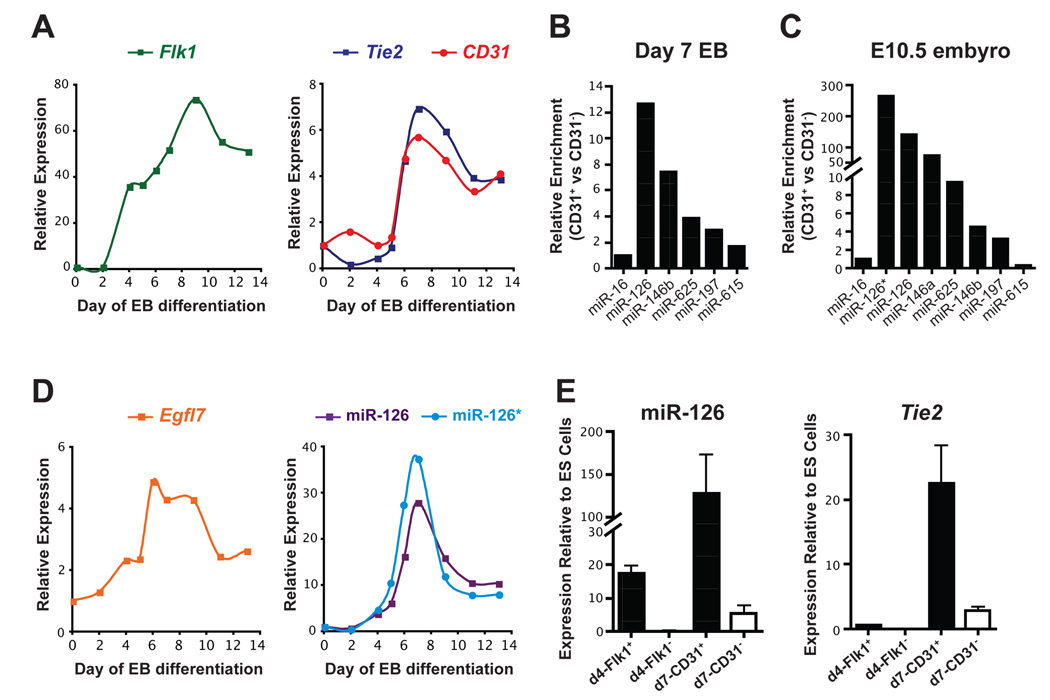
Figure 1. Identification of microRNAs enriched in endothelial cells.
(A) Gene expression changes were monitored by qRT-PCR during differentiation of mouse ES cells in an embryoid body (EB) model. Flk1 is expressed in vascular progenitors and mature endothelial cells, while CD31 and Tie2 are markers of mature endothelial cells. Expression was normalized to Tata-binding protein (Tbp) levels. The average of multiple experiments is shown. (B) Endothelial cells were isolated from day 7 (d7) EBs by cell sorting with anti-CD31 antibodies and microRNA arrays were performed. microRNAs enriched more than 1.5-fold relative to miR-16 in CD31+ cells compared to CD31− cells are shown. (C) Enrichment of microRNAs identified in (B) in CD31+ endothelial cells sorted from E10.5 mouse embryos assayed by qRT-PCR. (D) Expression of Egfl7, miR-126 and miR-126* in EBs assayed by qRT-PCR. (E) miR-126 was enriched in sorted vascular progenitors (Flk1+) from d4 EBs and in endothelial cells (CD31+) from d7 EBs compared to ES cells. Tie2 expression was used to assess mature endothelial gene expression in sorted cells used in (B) and (E).
We isolated CD31-positive and CD31-negative cells from d7 EBs by fluorescence-activated cell sorting (FACS) and profiled microRNA expression by microarray. While several microRNAs, including miR-146b, miR-197, miR-615, and miR-625, were enriched more than 1.5-fold in CD31-positive cells, miR-126 was the most highly enriched microRNA (Fig. 1B). microRNA arrays were also performed at d14 of EB formation, and the same subset of microRNAs was enriched (data not shown), suggesting that relatively few microRNAs are enriched in endothelial cells.
To determine if these microRNAs were also enriched in endothelial cells in vivo, we used FACS to isolate CD31-positive cells from E10.5 mouse embryos. qRT-PCR with RNA from these cells confirmed the enrichment of the above microRNAs, with the exception of miR-615, in CD31-positive cells compared to CD31-negative cells from the same embryos (Fig. 1C). Additionally, miR-126*, expressed from the opposite strand of the miR-126 pre-miRNA, was also highly enriched in endothelial cells in vivo, as was miR-146a, which differs from miR-146b by only two nucleotides near the 3′ end of the mature microRNA (Fig. 1C).
miR-126 is located in an intron of Egfl7, a gene that is highly expressed in endothelial cells (Parker et al., 2004). The expression of Egfl7 largely mirrored that of endothelial markers during EB formation (Fig. 1D). Interestingly, expression of Egfl7, miR-126 and miR-126* was induced at d4 of EB formation and further increased at d6, when endothelial markers were robustly expressed. miR-126 (Fig. 1E) and miR-126* (data not shown) were highly enriched in Flk1-positive vascular progenitors sorted at d4 and were also enriched in mature CD31-expressing endothelial cells at d7.
miR-126 does not control endothelial lineage commitment
We have previously shown that the muscle-specific microRNA, miR-1, controls cell fate decisions of multipotent cells (Ivey et al., 2008). Because of the early induction of miR-126 in vascular progenitors, we investigated whether this microRNA might regulate differentiation towards the endothelial lineage. We created stable mouse ES cell lines that expressed miR-126 under control of the ubiquitously-expressed EF1-α promoter (mESmiR-126) and confirmed miR-126 over-expression (Suppl. Fig. 1A). The expression of several endothelial genes, including Flk1, eNOS/NOS3, Tie2 and CD31, was not affected by miR-126 over-expression during ES cell differentiation (Suppl. Fig. 1B), and the number of CD31-positive endothelial cells at d7 was not altered (Suppl. Fig. 1C). This suggests that while miR-126 is enriched in vascular progenitors, it is not sufficient to promote differentiation of pluripotent cells toward the endothelial lineage.
miR-126 modulates endothelial phenotype in vitro
To study the loss-of-function of miR-126 in endothelial cells, a morpholino (MO) antisense to miR-126 that spanned the miR-126 5′ Dicer cleavage site of the miR-126 pri-cursor was introduced into human umbilical vein endothelial cells (HUVECs). These cells expressed high levels of miR-126 (Supp. Fig. 2A). Introduction of the MO resulted in decreased levels of both mature miR-126 and miR-126* and an increase in miR-126 pri-cursor, beginning at 24 hour (h) post-transfection (Fig. 2A and Suppl Fig. 2B). While both miR-126 and miR-126* were reduced to a similar extent, the absolute basal level of miR-126 was much higher than miR-126* in endothelial cells (Suppl. Fig. 2C). Importantly, levels of spliced EGFL7 mRNA, detected by qRT-PCR with primers surrounding the intron containing miR-126, and protein levels of EGFL7 were unaffected by introduction of this MO (Fig. 2A).
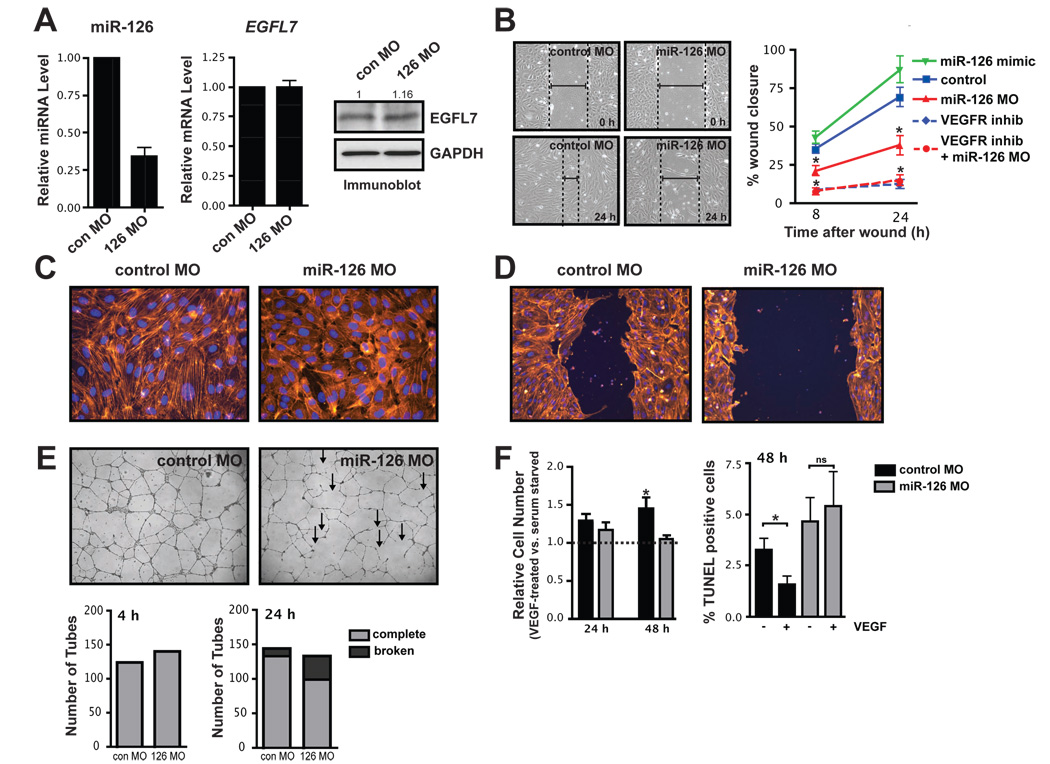
Figure 2. miR-126 regulates endothelial phenotype in vitro.
(A) Relative levels of mature miR-126, spliced EGFL7 mRNA across the miR-126-containing intron, and EGFL7 protein (immunoblot) were measured 72 h after HUVECs were electroporated with miR-126 morpholinos (MOs) or control (con) MO. Densitometric analysis of EGFL7 protein levels is indicated above. (B) The effect of miR-126 knockdown (MO) or over-expression (mimic) on endothelial cell migration in response to VEGF in the presence or absence of a VEGF receptor inhibitor was determined by generating a “scratch” in a confluent monolayer of endothelial cells and measuring the degree of “wound closure” after 8 and 24 h. Dashed lines indicate width of “wound”. Percent wound closure is shown as the mean ± SEM of five scratches from one representative experiment. * p < 0.05 compared to control. (C) Actin cytoskeletal structure was observed by phalloidin staining of control and miR-126 knockdown HUVECs. (D) Actin staining revealed a decrease in cytoskeletal reorganization and cytoplasmic extensions into the denuded area in a scratch assay. (E) Capillary tube formation of endothelial cells transfected with control or miR-126 MOs and seeded onto Matrigel. Endothelial cells transfected with miR-126 MOs formed capillary tubes, but these tubes were unstable, exhibiting frequent dissociation of tubular structures compared to control. Shown is a representative image at 24 h after plating. Arrows indicate examples of broken tubes. Quantification of the average number of tubes per field of view at 4 h and the number of complete and broken tubes at 24 h from a representative experiment is shown below. (F) VEGF-mediated changes in cell number and cell death by TUNEL were assessed in control and miR-126 knockdown cells. VEGF treatment increased total cell number and decreased the percent of serum-starved cells that were TUNEL positive, but this effect was absent in cells with reduced levels of miR-126. * denotes a significant difference between VEGF treated and non-treated cells (p<0.05).
Endothelial cells with reduced levels of miR-126 were phenotypically similar to to control MO-transfected cells, but had an elevated rate of proliferation (Suppl. Fig. 3A). The endothelial phenotype was further studied in an in vitro wound closure or scratch assay, in which the rate of migration of cells into a denuded area of a confluent monolayer was monitored. Modulating miR-126 levels had no effect on cell migration when complete medium was used (Suppl. Fig. 3B). However, VEGF- (Fig. 2B) and bFGF-induced (Suppl. Fig. 3C) migration was inhibited in miR-126 knockdown cells compared to control MO-transfected cells. Conversely, in cells transfected with miR-126 mimic, which express 50-fold more miR-126, there was a trend towards increased migration in response to VEGF stimulation (Fig. 2B). Examination of the actin cytoskeleton revealed a reduction in large actin filaments and more diffuse actin staining in many miR-126 knockdown cells (Fig. 2C). Stimulation of endothelial cells with VEGF normally results in the reorganization of the cytoskeleton, but this rearrangement was defective in miR-126 knockdown cells (Suppl. Fig. 3D). Consequently, in scratch assays there was a reduction in cell protrusions into the denuded area in knockdown cells (Fig. 2D). These data suggest that endothelial cell migration is regulated by miR-126 abundance.
The effect of miR-126 on the formation and stability of capillary tubes on matrigel was also assessed. While initial formation of tubes appeared normal, the capillary tubes were less stable and appeared thin, with dissociation of many tubes after 24 h (Fig. 2E). Assessment of cell adhesion by measuring the kinetics of endothelial cell attachment (Suppl. Fig. 3E), and cell-cell junction formation by VE-cadherin immunostaining (data not shown), revealed no obvious defects in miR-126 knockdown cells, suggesting that the instability of tubes was not attributable to defects in cell adhesion. To determine if there were survival defects in cells with reduced levels of miR-126, cells were serum starved in the presence or absence of VEGF. The number of control serum-starved cells increased with addition of VEGF, but miR-126 knockdown cells were refractory to this increase in cell number in response to VEGF (Fig. 2F). TUNEL staining demonstrated that VEGF treatment of control serum-starved cells resulted in a decrease in apoptosis, but this effect was absent in miR-126 knockdown cells (Fig. 2F).
miR-126 regulates blood vessel integrity in vivo
Considering the dramatic effects of miR-126 on the behavior of human endothelial cells in vitro, we assessed the in vivo function of miR-126. For this purpose we used zebrafish as a model system, in which a functioning cardiovascular system is not required for viability through relatively advanced stages of embryogenesis. The mature forms of zebrafish miR-126 and miR-126* are identical to their human orthologues. FACS isolation of GFP-positive cells from the endothelial cell-specific zebrafish reporter line, Tg(flk1:GFP)s843 (Jin et al., 2007), demonstrated that miR-126 and miR-126* were highly enriched in zebrafish endothelial cells (Fig. 3A). As in human endothelial cells, miR-126 was more abundant than miR-126* in zebrafish embryos (Fig. 3B). miR-126 expression was induced in 24 hpf embryos, was further increased at 52 hpf, and remained elevated between 72–96 hpf (Fig. 3C). We decreased miR-126 expression during zebrafish development by injecting two unique morpholinos targeting pri-miR-126 (miR-126 MO1 and MO2 (Suppl. Fig. 4A)), into fertilized eggs. Injection of these MOs blocked processing of pri-miR-126, resulting in a profound decrease in mature levels of miR-126 and miR-126* (Fig. 3D). Importantly, levels of egfl7, which hosts one of the two copies of zebrafish miR-126, and regulates tubulogenesis in zebrafish (Parker et al., 2004), were not appreciably altered by the miR-126 MOs (Fig. 3D).
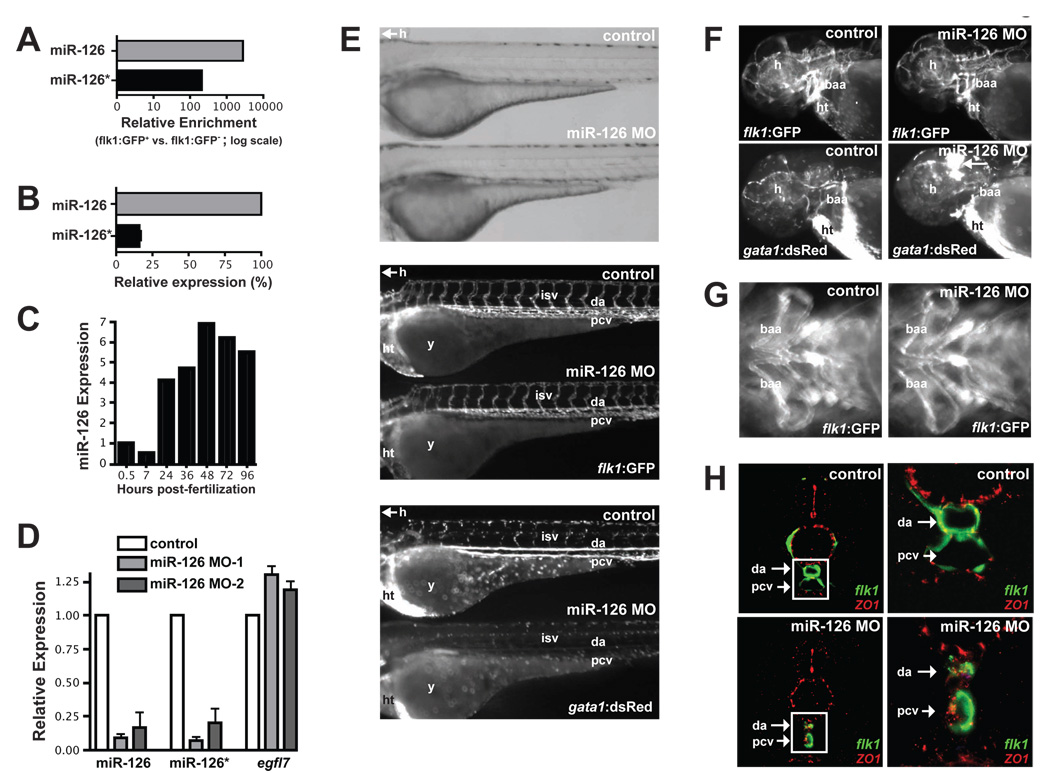
Figure 3. miR-126 regulates vascular integrity and lumen maintenance in vivo.
(A) miR-126 and miR-126* enrichment (qRT-PCR) in GFP+ endothelial cells from 72 hpf Tg(flk1:GFP)s843 zebrafish compared to GFP− cells. (B) Relative levels of miR-126 and miR-126* in 72 hpf zebrafish embryos. (C) miR-126 expression monitored by qRT-PCR during zebrafish development. (D) Levels of miR-126/126* or egfl7 (across intron containing miR-126) quantified by qRT-PCR in 72 hpf zebrafish injected with miR-126 MOs relative to control. (E) Lateral views of control and miR-126 MO-injected Tg(flk1:GFP)s843; Tg(gata1:dsRed)sd2 zebrafish (72 hpf). Brightfield microscopy (top) revealed no major changes in gross morphology, while flk1:GFP showed normal blood vessel patterning (middle). Presence of blood cells (gata1:dsRed) in the intersomitic vessels (isv), dorsal aorta (da) and primary cardinal vein (pcv) was greatly reduced in morphants (bottom). y, yolksac; h, head; ht, heart. (F) miR-126 morphants had normal vessel patterning (flk1:GFP), but developed cranial hemorrhages (gata1:dsRed; arrow) in the head. baa, branchial arch arteries. (G) Ventral view of baa suggested smaller luminal size (indicated by arrow) in miR-126 morphants. (H) Transverse section of control or miR-126 MO-treated zebrafish revealed that the da and pcv of morphants had a smaller lumen than controls; higher magnification (right panels) of boxed area shows collapsed da and small pcv in morphants. flk1:GFP, green; ZO-1, an epithelial marker, red.
Through the use of Tg (flk1:GFP)s843; Tg(gata1:dsRed)sd2 zebrafish, which express GFP in the vasculature, and dsRed in blood cells, we assessed the effect of miR-126 knockdown on vascular development and circulation. No differences in gross morphology (Fig. 3E, top panel) or vascular patterning (Fig. 3E, middle panel) were evident between control and miR-126 morpholino-injected (morphant) embryos. Additionally, FACS quantification revealed no significant difference in the percentage of Tg(flk1:GFP)s843-expressing endothelial, or Tg(gata1:dsRed)sd2-expressing blood cells, in control, miR-126 MO1 or miR-126 MO2-injected zebrafish (Suppl. Fig. 4B). However, several abnormalities were evident in the circulation and vessel morphology. These defects occurred in 70±5% (n=60 per experiment, 3 independent experiments) of the miR-126 MO-injected embryos, and were similar with either MO. While circulation of blood occurred normally between 24–36 hpf, the presence of Tg(gata1:dsRed)sd2-expressing blood cells in the head vasculature, intersomitic vessels (ISVs), dorsal aorta (DA), and primary cardinal vein (PCV) was reduced between 36 and 72 hpf (Fig. 3E (bottom panel), Fig. 3F and data not shown). Blood cells were made and were visible in the heart (ht) at these time points (Fig. 3E, F). Severe cranial hemorrhages were also present in 20±3% (n=60 per experiment, 3 independent experiments) of embryos, evidenced by the accumulation of dsRed-positive cells (Fig. 3F). Importantly, cardiac contractile function was not grossly affected by miR-126 MO at either 48 or 72 h.
We observed that some Tg(gata1:dsRed)sd2-expressing cells were trapped in the ISVs of miR-126 morphants (data not shown), indicating that blood vessel integrity might be compromised. Indeed, branchial arch vessels (Fig. 3F,G) appeared to have a reduced lumen diameter in morphants. To better characterize these defects we analyzed the integrity of endothelial tubes by examining the DA and PCV in miR-126 morphants by confocal microscopy (Fig. 3H). These experiments revealed collapsed lumens and compromised endothelial tube organization in miR-126 morphants, suggesting that miR-126 expression was required to maintain vessel integrity and caliber during zebrafish vascular development.
Identification of genes regulated by miR-126 by microarray
Although miR-126 morphants had severe defects in vessel integrity, the number of endothelial cells was not altered. We took advantage of this phenotype and isolated Tg(flk1:GFP)s843-expressing endothelial cells by FACS from control and miR-126 MO1-and MO2-injected fish and analyzed mRNA expression by microarray. Since similar genes were altered in miR-126 MO1 and MO2 injected fish, the data sets were combined to identify dysregulated genes in miR-126 morphants (see Suppl. Table I and II for up- and down-regulated genes, respectively). By Gene Ontology (GO) statistical analysis the most highly dysregulated class of genes in the endothelium of miR-126 morphants encoded transcription factors (Suppl. Table III). The homeobox (HOX) and forkhead box (FOX) family of genes were especially affected in miR-126 morphants, many of which regulate endothelial cell biology, including angiogenesis (Bruhl et al., 2004; Dejana et al., 2007; Myers et al., 2000).
Microarray analysis was also performed with RNA from human endothelial cells (HUVECs) in which miR-126 was knocked-down for 72 h (see Suppl. Table IV and V for up- and down-regulated genes, respectively). The most over-represented GO terms were related to the cell cycle and the cytoskeleton (Suppl. Table VI). This observation supports our finding that cells with reduced levels of miR-126 proliferated more rapidly than control cells and had altered cytoskeletal structures (Suppl. Fig. 3A and Fig 2C,D). Platelet-derived growth factors (PDGF) A, B, C and D, which are important in endothelial biology, were all significantly down-regulated in cells with reduced levels of miR-126 (Suppl. Table VI and data not shown). In addition, genes categorized as important for vascular development were highly dysregulated (Suppl. Table VI). A total of 61 genes were similarly altered (p < 0.05) in zebrafish and human miR-126 knockdown expression arrays (data not shown), suggesting a high conservation in the gene repertoire regulated by miR-126. To determine how many of the upregulated genes in miR-126 knockdown endothelial cells might be direct miR-126 targets, we performed bioinformatic analyses of miR-126 and miR-126* seed matches in the 3′ UTRs of genes upregulated by >1.5-fold in human cells with reduced levels of miR-126. miR-126 seed matches were highly enriched in the upregulated genes, while seed matches for miR-126* or an unrelated microRNA, miR-124, were not enriched (Suppl. Fig. 5). Surprisingly genes containing both miR-126 and miR-126* seed matches were also statistically over-represented, while the combination of miR-126 and miR-124 seed matches were not. This suggests that miR-126 and miR-126*, which are derived from the same pri-miRNA, may coordinately regulate target genes.
miR-126 regulates EGFL7 expression in a negative feed-back loop
EGFL7 mRNA was highly upregulated on the human array despite our earlier finding that levels of spliced EGFL7 mRNA and protein were unchanged (Fig. 2A). To understand this discrepancy, we used qRT-PCR with primer sets specific for the transcriptional start sites of the three EGFL7 isoforms (named here EGFL7 isoform-A, -B and -C, which all contain the same open reading frame (ORF)), as well as several primer sets that were common to all three isoforms. EGFL7 mRNA levels were increased throughout the EGFL7 transcriptional unit (Suppl. Fig. 6A), except for the spliced EGFL7 mRNA surrounding the miR-126-containing intron, as we noted earlier (Fig. 2A). Thus, EGFL7 was upregulated in miR-126 MO-treated cells, but the MO apparently inhibited processing of the intron containing miR-126, resulting in no net change in EGFL7 protein levels. Only EGFL7 isoform B was induced by miR-126 MO (Suppl. Fig. 6A). Since all three isoforms contain the same 3′ UTR, miR-126 may regulate isoform B in a 3′ UTR-independent fashion. By performing RNA polymerase II (Pol II) chromatin immunoprecipitation (ChIP) experiments we noted an increase in Pol II density at the promoter of isoform B and in the coding region (which is common to all three isoforms), but not at the promoter of isoform A, which was not induced by miR-126 MO (Suppl. Fig. 6B). Thus, miR-126 may indirectly regulate EGFL7 isoform B at the transcriptional level.
miR-126 represses SPRED1, VCAM1 and PIK3R2 post-transcriptionally
To understand the mechanisms by which miR-126 regulates endothelial biology we searched for potential direct mRNA targets of miR-126. Several miRNA target prediction algorithms were employed, including one developed in our lab that incorporates sequence complementarity and mRNA target site accessibility (K. Ivey and D. Srivastava, unpublished). Portions of the 3′ UTR of several potential targets were cloned into the 3′ UTR of a luciferase construct, and the ability of miR-126 to affect luciferase expression was determined in HeLa cells, which do not normally express miR-126. Six potential targets were initially chosen based on binding sites (Suppl. Fig. 7) and a known role in endothelial cell signaling or vascular function. These included: regulator of G-protein signaling 3 (RGS3) (Bowman et al., 1998; Lu et al., 2001); SPRED1 (Wakioka et al., 2001); PIK3R2 (also known as p85-β) (Ueki et al., 2003); CRK (Park et al., 2006); integrin alpha-6 (ITGA6); and vascular cell adhesion molecule 1 (VCAM1). miR-126, but not a control miRNA, miR-1, significantly repressed the activity of luciferase derived from RNAs containing the 3′ UTR of SPRED1, VCAM1, and PIK3R2 (Fig. 4A).
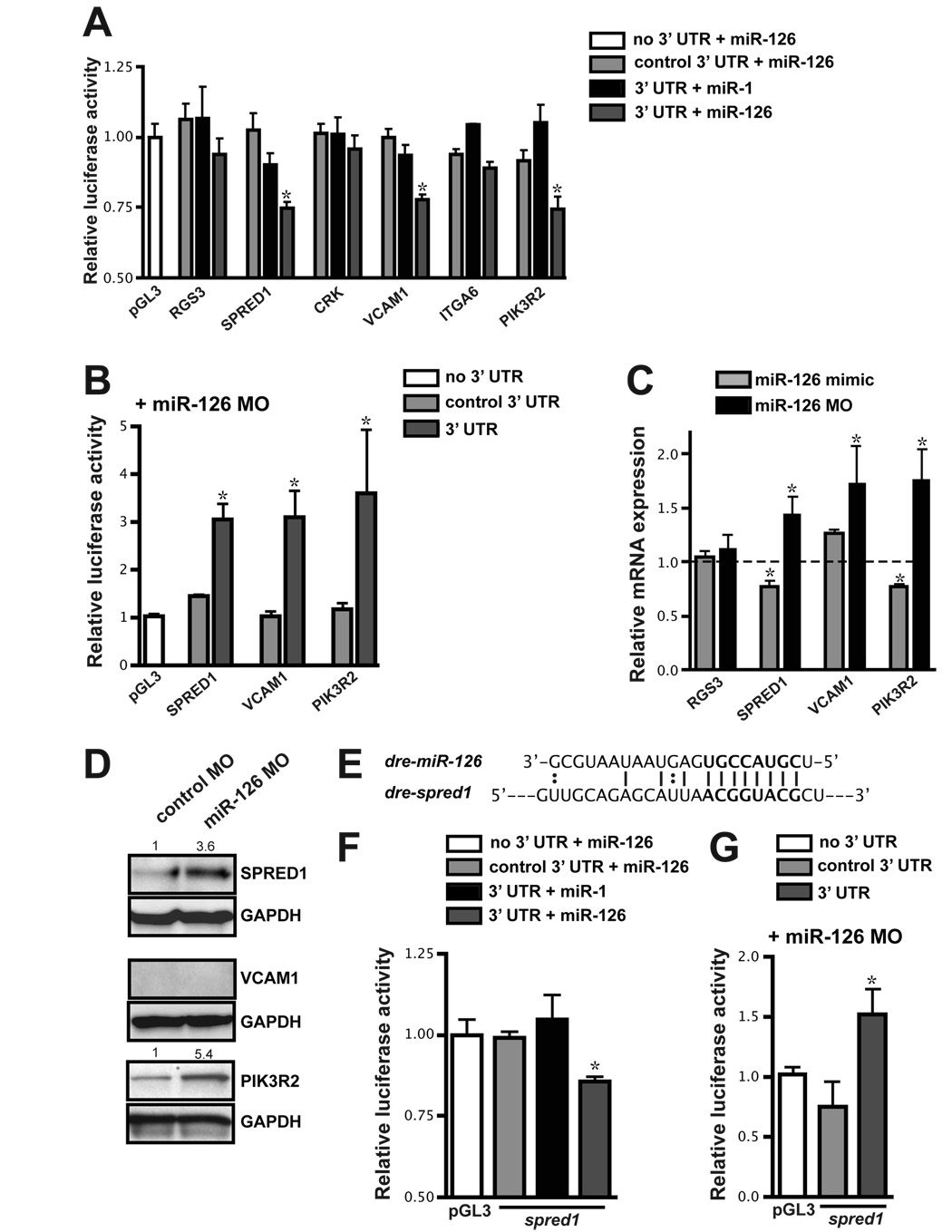
Figure 4. Identification of miR-126 mRNA targets.
(A) Relative luciferase activity of constructs containing the 3′ UTR of potential miR-126 targets introduced into HeLa cells in the presence of miR-1 or miR-126. The 3′ UTR was also inserted in the antisense orientation as a control (control 3′ UTR). Firefly luciferase activity for each construct was normalized to the co-transfected Renilla luciferase construct and then normalized to the change in pGL3 luciferase in the presence of microRNA. For each 3′ UTR construct, normalized luciferase activity in the absence of microRNA was set to 1. * p < 0.05 compared to pGL3. (B) Relative luciferase activity of select constructs in (A) in HUVECs upon inhibition of miR-126 with MOs. Normalization was performed as in (A). (C) mRNA levels of potential targets in HUVEC cells transfected with miR-126 mimic or MO quantified by qRT-PCR. Values are relative to transfection controls. (D) Immunoblot of SPRED1, VCAM1 and PIK3R2 protein in control or miR-126 MO-transfected HUVECs 72 h post-transfection. GAPDH is shown as a loading control. (E) Sequence complementarity of a potential miR-126 binding site in the zebrafish spred1 3′ UTR. (F and G) Luciferase assays (as in (A) and (B), respectively) using the zebrafish spred1 3′ UTR.
Luciferase experiments were also performed in endothelial cells in which endogenous miR-126 levels were knocked down by antisense MO. The activity of luciferase from constructs that included portions of the SPRED1, VCAM1 or PIK3R2 3′ UTR was increased upon knockdown of miR-126 (Fig. 4B). In contrast, a MO directed to miR-21, which is also expressed in endothelial cells, had no effect on the activity of the constructs tested (data not shown).
MicroRNAs can regulate mRNA stability or translation of target mRNAs. We quantified mRNA expression of potential miR-126 targets by qRT-PCR in HUVECs that had been transfected with antisense miR-126 MO or a miR-126 mimic (Fig. 4C). While SPRED1 and PIK3R2 mRNA levels were reciprocally regulated by miR-126 abundance, VCAM1 mRNA levels were elevated upon miR-126 inhibition, but were not decreased in the presence of miR-126 mimic. As a control, we examined levels of RGS3, since the 3′ UTR of this gene did not affect luciferase activity in the presence of miR-126. RGS3 expression was unchanged when miR-126 levels were modulated (Fig. 4C). We also assessed the expression of SPRED1, PIK3R2 and VCAM1 protein by western blot after introduction of control or miR-126 MOs (Fig. 4D). SPRED1 and PIK3R2 protein were increased when miR-126 levels were decreased (Fig. 4D). However, we could not detect VCAM1 protein in either control or miR-126 MO-transfected endothelial cells. This was not due to an ineffective antibody, since VCAM1 protein was readily detectible in TNF-α-treated endothelial cells (data not shown). Indeed, VCAM1 has recently been identified as a miR-126 target in TNF-α-treated endothelial cells (Harris et al., 2008).
The 3′ UTR of zebrafish spred1 contains a highly conserved 8-mer that is perfectly complementary to nucleotides 2–9 of miR-126 (Fig. 4E). Addition of miR-126 specifically repressed the activity of luciferase reporters containing this 3′ UTR (Fig. 4F). Conversely, knockdown of miR-126 led to an increase in luciferase activity of the spred1 3′ UTR luciferase construct when transfected into HUVECs (Fig. 4G). This suggests that miR-126 targeting of SPRED1 is conserved in zebrafish.
miR-126 modulates VEGF-dependent events by targeting SPRED1 and PIK3R2
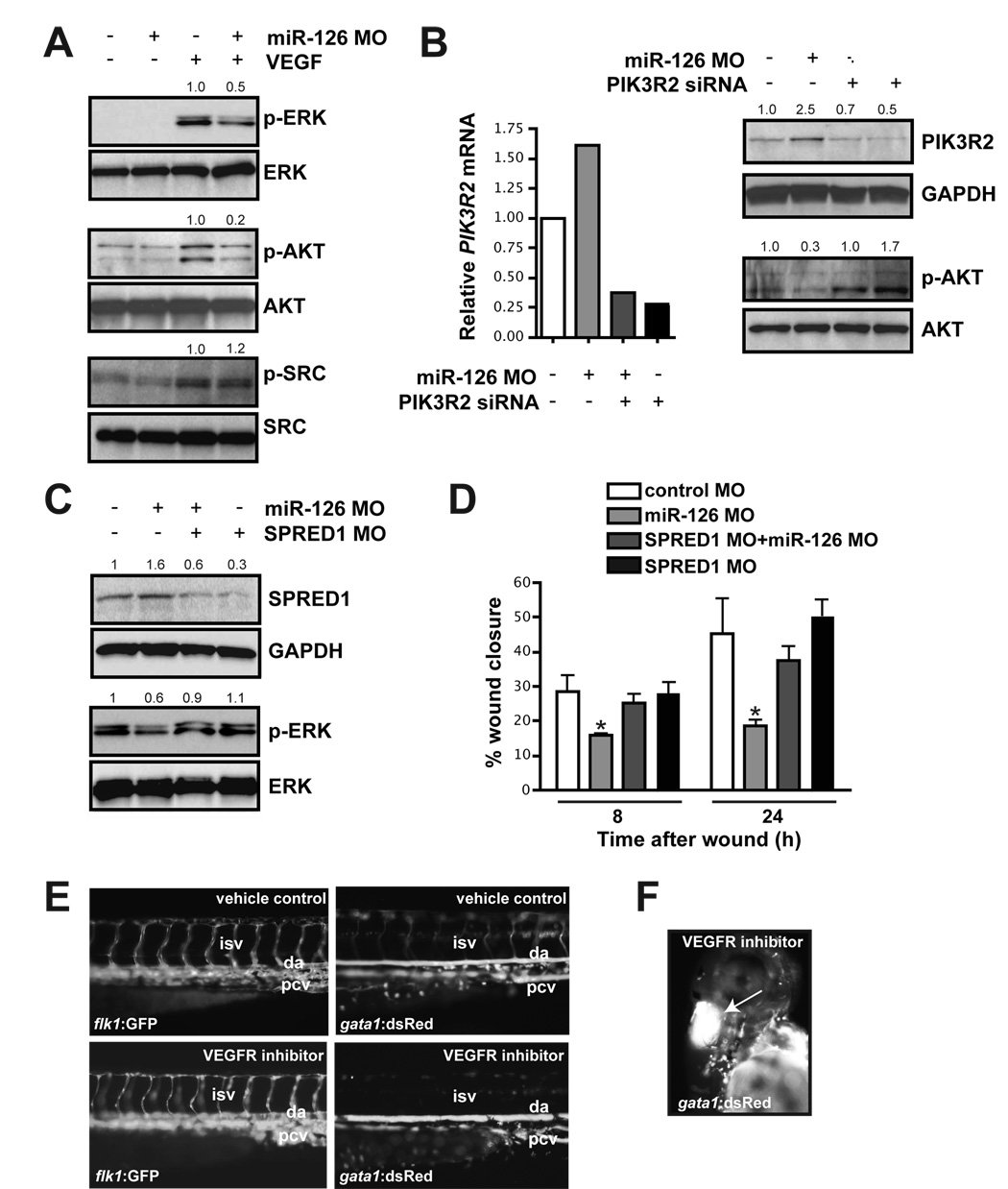
SPRED1 and PIK3R2 negatively regulate growth factor signaling via independent mechanisms. SPRED1 functions by inhibiting growth factor-induced activation of the MAP kinase pathway (Taniguchi et al., 2007), while PIK3R2 is thought to negatively regulate the activity of PI3 kinase (Ueki et al., 2003). Activation of the MAP and PI3 kinase pathways by growth factor stimulation can be assessed by measuring the phosphorylation status of ERK and AKT, two respective downstream targets of these pathways. We found that the VEGF-induced phosphorylation of ERK and AKT was attenuated in miR-126 knockdown cells (Fig. 5A). In contrast, phosphorylation of SRC was unaffected by modulation of miR-126, suggesting that some arms of the VEGF signaling pathway were unaffected by miR-126 knockdown (Fig. 5A). Activation of ERK and AKT in response to EGF and bFGF-stimulation was also reduced in miR-126 knockdown cells in vitro (Suppl. Fig. 8A). However, the defects in signaling downstream of EGF and bFGF were less pronounced than signaling downstream of VEGF. In contrast, MAP kinase signaling downstream of TNF-α was unaffected (Suppl. Fig. 8B).
Figure 5. miR-126 positively regulates VEGF signaling in endothelial cells by repressing SPRED1 and PIK3R2.
(A) Immunoblot of lysates from HUVECs transfected with control or miR-126 MOs in the presence or absence of VEGF (10 ng/mL, 10 min). VEGF induced phosphorylation of ERK (p-ERK) and AKT (p-AKT), which was blocked by miR-126 inhibition. Total levels of ERK and AKT were not affected. Phosphorylation of SRC was not affected by miR-126 knockdown. Densitometric analysis of normalized protein levels are indicated above. (B) PIK3R2 mRNA was knocked-down by RNAi in HUVECs transfected with control or miR-126 MOs (qRT-PCR). Western analysis indicates a decrease in PIK3R2 protein by introduction of siRNA, even in the presence of miR-126 MOs. Knockdown of PIK3R2 rescued the defect in VEGF-dependent phosphorylation of AKT in miR-126 MO-treated cells. (C) Western analysis shows reduced SPRED1 levels by transfection of a MO that blocks SPRED1 translation, even in the presence of miR-126 MO. SPRED1 MO rescued the defect in VEGF-induced phorphorylation of ERK in miR-126 MO-transfected cells. (D) Quantification of percent (%) wound closure of endothelial cells in a “scratch” assay reveals rescue of miR-126 MO effects by knocking down SPRED1. * p < 0.05 compared to control MO. (E) Treatment of 48 hpf embryos with a VEGF receptor inhibitor (Vatalanib, 5 µM, 18 h) resulted in reduced circulation of blood cells (gata1:dsRed). ISVs (flk1:GFP) appeared to lack a lumen after VEGF inhibition. (F) Hemorrhages also occurred in some embryos treated with VEGF receptor inhibitor (arrow).
To determine if SPRED1 and PIK3R2 might be involved in miR-126-dependent signaling defects, we knocked these genes down in cells with reduced levels of miR-126. The defect in VEGF-dependent AKT phosphorylation was rescued by siRNA-mediated knockdown of PI3KR2 (Fig. 5B), while inhibition of SPRED1 rescued the defect in ERK phosphorylation (Fig. 5C). We also investigated whether decreasing SPRED1 expression in miR-126 knockdown cells could rescue the VEGF-dependent migration defect described earlier (Fig. 2B). While SPRED1 MO alone had no effect on VEGF-induced endothelial cell migration, the knockdown of SPRED1 protein largely rescued the migration defect in cells with decreased miR-126 expression (Fig. 5D).
To test whether decreased VEGF signaling in vivo would result in a defect in vascular maintenance similar to miR-126 knockdown, we treated 48 hpf embryos, which have a fully functioning circulatory system, with a VEGF receptor inhibitor (Chan et al., 2002). After 18 h, >90% of treated embryos displayed severe circulatory defects, including collapsed vessels (Fig. 5E and Suppl. Fig. 9A). The amount of blood in the embryos was the same as that of vehicle-treated controls (data not shown). Circulation in the ISVs (Fig. 5E) and in the head vasculature (data not shown) was absent or severely diminished, and more than 15% of the embryos also developed hemorrhages (Fig. 5F). This phenotype was similar in many respects to miR-126 morphant embryos.
Increased Spred1 in zebrafish disrupts vascular integrity similar to miR-126 knockdown
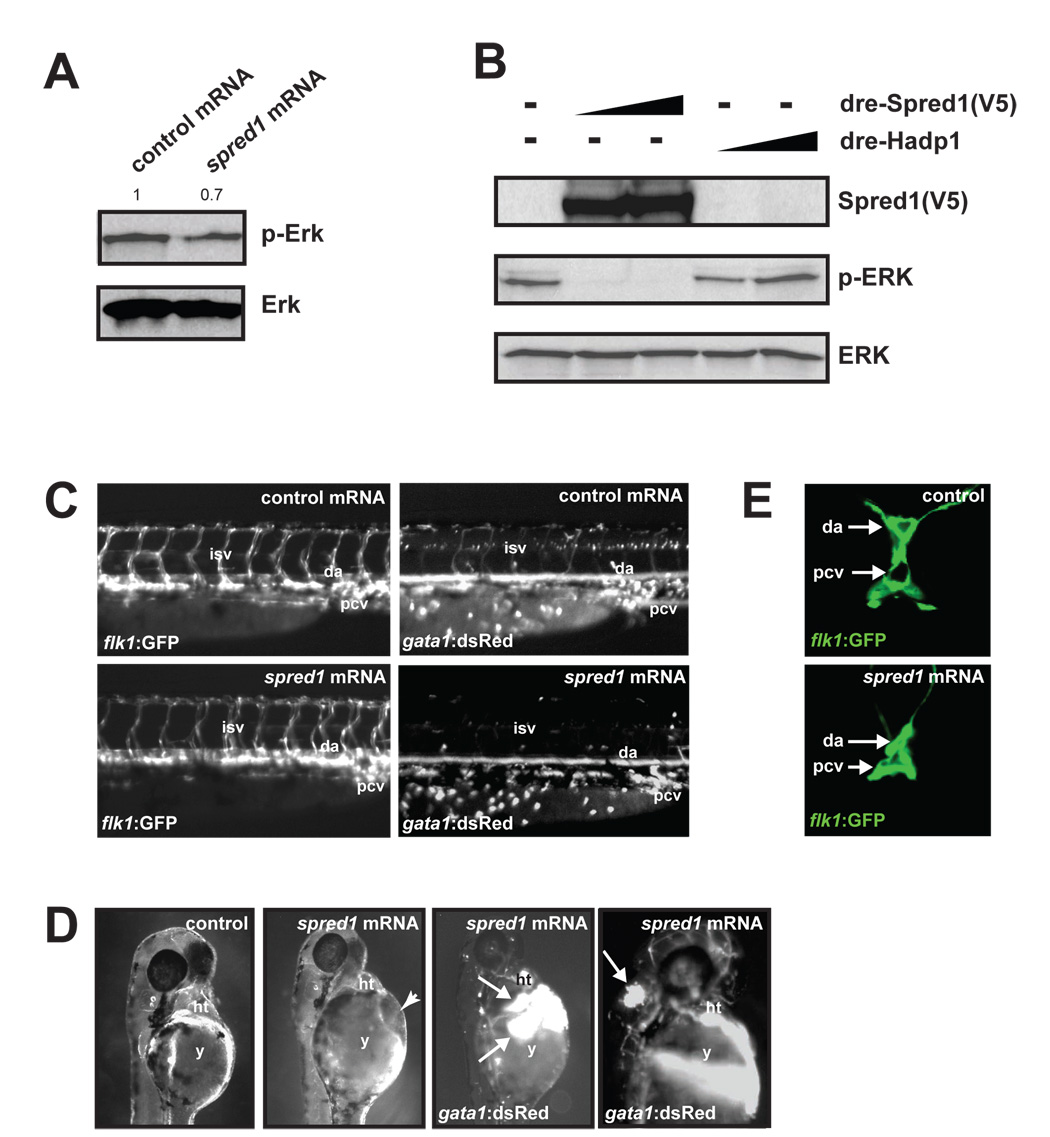
To determine whether excess Spred1 in zebrafish could cause vascular defects similar to miR-126 inhibition, we injected spred1 mRNA, which is expressed in endothelial cells (Supp. Fig. 9B), into zebrafish embryos. Consistent with the known function of Spred1, spred1 mRNA-injected embryos had decreased levels of phosphorylated ERK compared to control (hapd1) mRNA-injected fish, suggesting diminished growth factor signaling (Fig. 6A). Additionally, transfection of COS-1 cells with an expression construct containing zebrafish spred1 cDNA resulted in dramatically reduced levels of phosphorylated ERK, confirming that zebrafish Spred1, like its mammalian counterpart, negatively regulates the MAP kinase pathway (Fig. 6B). Gastrulation defects occurred in many of the spred1- and control mRNA-injected embryos. However, the remaining embryos appeared grossly normal and were analyzed further. Vascular patterning as assessed by Tg(flk1:GFP)s843 expression was relatively normal in the majority of spred1 mRNA-injected embryos (Fig. 6C, left panels). However, the presence of blood cells marked by Tg(gata1:dsRed)sd2 expression was markedly decreased in the ISVs, DA and PCV (Fig. 6C, right panels). Greater than 20% of the embryos developed cranial and pericardial hemorrhages, indicating the presence of blood cells but the lack of vascular integrity (Fig. 6D). Additionally, confocal analysis revealed collapsed blood vessels, similar to miR-126 morphants (Fig. 6E). The phenotypic and functional similarities in embryos with increased expression of Spred1 compared to those with increased Spred1 secondary to miR-126 inhibition suggests that Spred1 may be a major mechanism by which miR-126 regulates vascular integrity.
Figure 6. Increased Spred1 causes vascular instability and hemorrhage similar to miR-126 knockdown.
(A) Levels of phosphorylated Erk were reduced in zebrafish embryos (8 hpf) injected with spred1 mRNA compared to control mRNA (hadp1). Densitometric analysis is shown above. (B) Zebrafish Spred1, but not Hadp1, expression also reduced p-ERK in COS cells as observed by immunoblot. (C) Lateral view of trunk region of 48 hpf embryos after injection of control (hadp1) or 100 pg of spred1 mRNA. flk1:GFP reveals normal vascular patterning but gata1:dsRed shows diminished blood cells in the dorsal aorta (da), primary cardinal vein (pcv) and intersomitic vessels (isv). (D) Injection of spred1 mRNA also resulted in pericardial (2nd panel from right) and cranial (far right) hemorrhages (arrows) visualized by gata1:dsRed marking of blood cells, and edema (2nd panel from left). (E) Confocal analysis of transverse sections of 48 hpf embryos revealed collapsed lumens of da and primary cardinal vein (pcv) in spred1-injected embryos.
We attempted to test whether knockdown of Spred1 (Suppl. Fig. 9C) could rescue the vascular defects in miR-126 morphants, but observed severe consequences of Spred1 inhibition. Embryos injected with a spred1 splice-blocking (sb) MO (spred1 MOsb) developed cranial hemorrhages and pericardial edema, even at low doses of MO (Suppl. Fig. 9D). A second, non-overlapping translation-blocking morpholino (spred1 MOATG) also resulted in pericardial edema (data not shown).
Discussion
Here we have shown that miR-126 regulates many aspects of endothelial cell biology, including cell migration, reorganization of the cytoskeleton, capillary network stability and cell survival. and demonstrate that miR-126 is required for the maintenance of vascular structure in vivo. miR-126 directly targets SPRED1, VCAM1 and PIK3R2 for repression and functions to promote VEGF signaling by inhibiting SPRED1 and PIK3R2. The identification of an endothelial-specific microRNA that regulates angiogenic signaling and vascular integrity represents an advance that has implications for many aspects of biology including development, cancer and tissue response to injury.
miR-126 (Kloosterman et al., 2006; Wienholds et al., 2005) and its host transcript, Egfl7 (Parker et al., 2004), are highly expressed in endothelial cells. Our analysis of miR-126 function suggests that Egfl7 and miR-126 play somewhat related, yet temporally and functionally distinct, roles in zebrafish vascular development. The initial patterning of the zebrafish vasculature appeared unaffected in miR-126 morphants, yet miR-126 morphants displayed a profound loss of lumenized blood vessels by 48 hpf. While Egfl7 morphants also have defects in lumenization, these defects occur very early and involve the inability of angioblasts to initially form a lumen during tubulogenesis (De Maziere et al., 2008; Parker et al., 2004). While it is not known whether knockdown of Eglf7 also affects miR-126 levels, it is unlikely that miR-126 dysregulation contributes significantly to the Egfl7 morphant phenotype, since rescue can be achieved by injection of egfl7 mRNA, which lacks miR-126 sequences (Parker et al., 2004). It is interesting that the egfl7 transcriptional unit may have multiple roles in regulating lumenization, with Egfl7 regulating the formation, and miR-126 the maintenance, of the vascular lumen.
Our finding that miR-126 directly targets SPRED1 and PIK3R2 provides important evidence that the VEGF pathway can be regulated at multiple levels by a microRNA (Fig. 7). Cells with reduced levels of miR-126 were less responsive to VEGF and other growth factors. SPRED1 inhibits activity of the RAF1 kinase (Wakioka et al., 2001), but its function in the vasculature is only partially understood. For example, Spred1 and Spred2 redundantly control lymphangiogenesis during mouse development by negatively regulating Vegfr-3 signaling (Taniguchi et al., 2007). miR-126-dependent defects in ERK phosphorylation and cell migration were partially rescued by MO-based inhibition of SPRED1 translation, indicating that miR-126 regulates VEGF signaling, in part, through SPRED1.
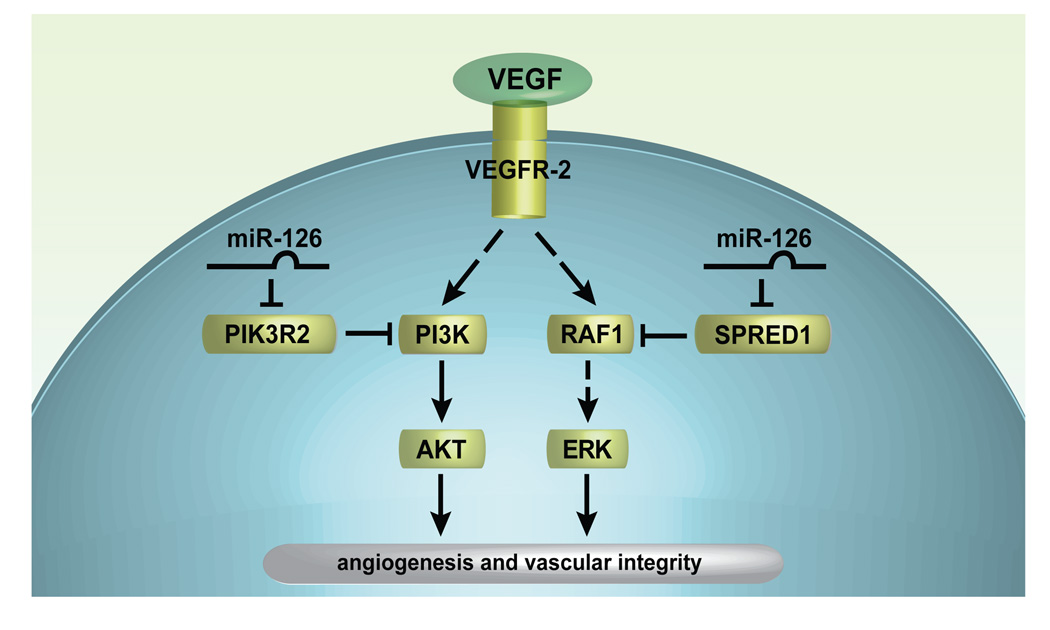
Figure 7. Putative model of miR-126 function in endothelial cells.
miR-126 represses SPRED1 and PIK2R2, which negatively regulate VEGF signaling (and possibly other growth factor signaling pathways) via the MAP kinase and PI3 kinase pathways, respectively. Thus, miR-126 promotes VEGF signaling, angiogenesis and vascular integrity by inhibiting protein production of endogenous VEGF repressors within endothelial cells.
miR-126 regulation of VEGF and other growth factor signaling appears to be reinforced by targeting of PIK3R2 (p85-β), which negatively regulates the activity of PI3 kinase (Ueki et al., 2003). A role for PIK3R2 in the regulation of VEGF signaling had not previously been addressed. Importantly, we found that siRNA-mediated knockdown of PIK3R2 in cells with reduced miR-126 levels was able to rescue the defect in VEGF-dependent AKT phosphorylation, indicating the significance of PIK3R2 as a target. These findings support the emerging theme of microRNAs functioning as “master regulators” of key pathways by their ability to target multiple mRNAs in a coordinate fashion (Silver et al., 2007; Zhao et al., 2007).
Other genes regulated by miR-126 may also contribute to the attenuation of growth factor responsiveness in cells with lower levels of miR-126. For example, the down-regulation of PDGF-A, -B, -C and -D in human cells upon inhibition of miR-126 was striking given the role of PDGFs in promoting angiogenesis (Cao et al., 2002; Guo et al., 2003; Li et al., 2003). Similarly, EphrinB2, which inhibits MAP kinase signaling downstream of VEGF (Kim et al., 2002), was upregulated in both human and zebrafish endothelial cells with reduced miR-126 expression. Finally, RGS5, which represses phosphorylation of ERK (Cho et al., 2003), and Rgs4, a closely related RGS protein that inhibits tubulogenesis by reducing ERK phosphorylation (Albig and Schiemann, 2005), were upregulated in endothelial cells with decreased miR-126 expression. Thus, the alteration of numerous genes involved in angiogenic signaling pathways in response to miR-126 inhibition suggests a global effect of this microRNA on titrating growth factor signals.
Recent studies in the mouse have illustrated that autocrine VEGF signaling in endothelial cells is essential for vascular homeostasis (Lee et al., 2007b). In mice with VEGF deleted from the endothelium, blood vessels were highly abnormal, and hemorrhages occurred in multiple vascular beds. Interestingly, the lumens of several blood vessels appeared to be collapsed in VEGF mutants. This phenotype is similar in many respects to zebrafish with reduced miR-126, and inhibition of VEGF signaling after the establishment of circulation produced similar defects in zebrafish embryos. We propose that increased expression of the miR-126 targets, SPRED1 and PIK3R2, in the endothelium of miR-126 morphants inhibits VEGF signaling (Rousseau et al., 2000). Since SPRED1 also regulates the cytoskeleton (Johne et al., 2008; Miyoshi et al., 2004), the mechanism responsible for vessel collapse in miR-126 morphants may involve disruption of cytoskeletal structure. In support of this hypothesis, we noted that several cytoskeletal genes were dysregulated in cultured endothelial cells with reduced miR-126, and that the arrangement of actin fibers in basal and VEGF-stimulated endothelial cells was defective.
Our findings have important implications not only for vascular development but also tumor biology. We have shown that angiogenesis and vascular integrity can be disrupted through modulation of miR-126 expression. Interestingly, EGFL7 is down-regulated in quiescent endothelial cells, but is up-regulated in the endothelium of proliferating tissue, including tumors (Parker et al., 2004). Since miR-126 is embedded within EGFL7, it is possible that miR-126 increases the sensitivity of these activated endothelial cells to VEGF or other growth factors through repression of SPRED1 and/or PIK3R2 expression. Thus, miR-126 may contribute to angiogenesis in this setting. Considering the importance of miR-126 in the regulation of angiogenesis and vascular integrity, we propose that miR-126 may be an important target for either pro- or anti-angiogenic therapies.
Experimental Procedures
Cell Biology Assays
For a detailed description of cell culture, and endothelial cell biology assays (cell survival, adhesion, migration, tube formation) please see the Supplemental Methods online.
Fluorescence-activated cell sorting
Single-cell suspensions were generated by digesting EBs or mouse embryos with Accutase (Chemicon). Cells were resuspended in PBS containing 1% BSA and labeled with fluorochrome-conjugated primary antibodies. For the separation of Flk1-positive cells from mouse EBs, phycoerytherin (PE)-conjugated anti-mouse Flk1 antibody (BD Pharmingen, Avas 12α, Cat. Number: 555308) was used. For the separation of CD31-positive cells from mouse EBs or embryos, FITC-conjugated anti-mouse CD31 antibody (BD Pharmingen, MEC 13.3, Cat. Number: 553372) was used. Antibodies were used at 5 µg/mL and were incubated with cells for 30 min at 4°C with rotation, and cells were then washed with PBS-1% BSA. Cells were sorted in PBS-0.1% BSA with a FACS Diva flow cytometer and cell sorter (Becton Dickinson). For the sorting of endothelial cells from Tg(flk1:GFP)s843 zebrafish, embryos were digested with trypsin and sorted based on GFP fluorescence.
Transfection/electroporation of plasmids, MOs and microRNA mimics
HeLa cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations. Cells were transfected at 90% confluency in six-well dishes with 2 µg of pGL3 (empty or with 3′ UTR of potential targets inserted), 2 µg of expression construct (empty or with miR-1 or miR-126 pri-cursor sequence), and 0.1 µg of Renilla construct (for normalizing transfection efficiency). Cells were analyzed at 48 h post-transfection.
HUVECs (0.5 × 106) were electroporated using the Amaxa Nucleofector according to the manufacturer’s recommendations with 15 nmol of control MO or MOs that block processing of the miR-126 pri-cursor or translation of SPRED1 (Gene Tools; see below for sequences). For rescue experiments, HUVECs were co-electorporated with 15 nmol of control or miR-126 MOs together with 15 nmol of SPRED1 MO. For luciferase experiments, HUVECs were transfected with 1 µg of pGL3 luciferase constructs and 0.5 µg Renilla construct, together with MOs. Cells were analyzed 72 h post-transfection. For transfection of endothelial cells with miR-126 mimic, Oligofectamine (Invitrogen) was used according to the manufacturers’s recommendations with 300 nM of control or miR-126 mimic (Dharmacon). Cells were analyzed 48 h post-transfection.
For siRNA-mediated knockdown of PIK3R2, endothelial cells were transfected with 300 nM Stealth siRNA (top strand: 5′-UUG UCG AUC UCU CUG UUG UCC GAG G-3′) or a GC-matched control (Invitrogen) using Oligofectamine. For rescue experiments, HUVECs were electroporated with control or miR-126 MO using the Amaxa kit, and 24 h later were transfected with control or PIK3R2 siRNAs. Analysis of protein or RNA was performed after an additional 48 h.
Luciferase assays
Firefly and Renilla luciferase activities were quantified in lysates using the Dual Luciferase Reporter Assay kit (Promega) on a Victor3 1420 multilabel counter (PerkinElmer) according to the manufacturer’s recommendations. Luciferase readings were corrected for background, and Firefly luciferase values were normalized to Renilla to control for transfection efficiency.
Microarrays and quantitative reverse-transcriptase real-time PCR (qRT-PCR)
For detailed methods please see the Supplemental Methods online.
Chromatin immunoprecipitation (ChIP)
ChIP was performed essentially as described (Fish et al, 2005). For detailed methods please see the Supplemental Methods online.
Zebrafish experiments
Tg(flk1:GFP)s843;Tg(gata1:dsRed)sd2 zebrafish embryos (Jin et al., 2007) were injected at the one-cell stage with 1–2 ng of spred1 splice-blocking or translation blocking MOs (see sequences below). For miR-126 MOs, embryos were injected with 4–8 ng of MO. For spred1 mRNA injection, full-length zebrafish spred1 was cloned into pcDNA3.1. mRNA was generated by T7 mMessage mMachine (Ambion), and 100 pg of mRNA was injected into embryos. As a control, zebrafish heart adapter protein 1 (hadp1) mRNA (Accession number: EU380770) was synthesized and injected in parallel. While development was essentially normal with 100 pg of injected hadp1, developmental abnormalities were evident at higher amounts of injected mRNA (J. Wythe and D. Li, unpublished results). Embryo development was assessed at 24–72 hpf. For VEGF receptor inhibitor treatment, 48 hfp embryos were treated with 5 µM Vatalanib (LC Laboratories) in DMSO. Control embryos were treated with the same volume of DMSO alone.
Morpholinos
MOs targeting dre-pri-miR-126 were 5′-TGC ATT ATT ACT CAC GGT ACG AGT TTG AGT C-3′ (dre-miR-126 MO-1) and GCC TAG CGC GTA CCA AAA GTA ATA A (dre-miR-126 MO-2), and those targeting hsa-miR-126 were GCA TTA TTA CTC ACG GTA CGA GTT T (hsa-miR-126 MO). To direct nonsense-mediated decay of zebrafish spred1, a MO was designed to cause exon 2 skipping and the generation of a premature stop codon. The MO was CCT GAG GAC CAG AAA CAG TCT CAC C (zebrafish spred1 MOsb). A second, non-overlapping MO was used to block translation of zebrafish spred1: GTT CTT CGC TCA TGT TTC TCT CAC G (zebrafish spred1 MOATG). To block translation of human SPRED1, the following MO was used: GTC TCC TCG CTC ATC TTT CCC TCA C (human spred1 MOATG).
Western blotting
Western blots were performed with 20–40 µg of protein. The antibodies used were anti-AKT1 (Santa Cruz), phospho-AKT1/2/3 (Ser473) (Cell Signaling), ERK2 (Santa Cruz), - phospho-ERK1/2 (Thr202/Tyr204) (Cell Signaling), SRC (Cell Signaling), phospho-SRC (Tyr416) (Cell Signaling), -EGFL7 (Santa Cruz or Abnova), -SPRED1 (Abgent and kindly provided by Dr. Yoshimura), -PIK3R2 (Abcam), -GAPDH (Santa Cruz), and VCAM1 (Santa Cruz). For phospho-AKT, -ERK and p-SRC western blots, HUVECs were serum-starved in medium containing 0.1% FBS without growth factors overnight and then stimulated with 10 ng/mL of human recombinant VEGF (BD Biosciences), EGF (R&D Systems), bFGF (Invitrogen) or 50 ng/mL TNF-α (R&D Systems) for 10 min.
Statistical analyses
All experiments were repeated a minimum of three times, and the error bars on graphs represent the mean ± the standard error of the mean, unless otherwise stated. Statistical significance was determined by a Student’s T-test or ANOVA, as appropriate, and a p-value of < 0.05 was considered as significant.
Microarray data submission
Microarray data for human and zebrafish arrays have been submitted to the Gene Expression Omnibus (GEO) and have been given a provisional accession number: GSE12039.
Supplementary Material
Acknowledgements
We thank Gary Howard and Bethany Taylor for editorial assistance, members of the Srivastava lab for helpful comments on the manuscript, and Kimberly Cordes for preparation of graphics. We thank the Genomics Core, Flow Cytometry Core and the Microscopy Core at the J. David Gladstone Institute for experimental support. We thank Dr. A. Yoshimura (Kyushu University, Japan) for providing Spred1 antibodies and Michael McManus (University of California, San Francisco) for providing constructs. J.E.F. was a recipient of a Canadian Institute of Health Research Postdoctoral Fellowship. M.M.S. was supported by a Human Frontiers Science Organization Career Development Award. S.U.M. was supported by an American Heart Association Western Affiliates Predoctoral Fellowship. J.D.W. was supported by a postdoctoral NIH Cardiovascular Developmental Biology Fellowship. D.Y.R.S. was supported by grants from the National Institutes of Health and the Packard Foundation. D.S. was supported by grants from the National Institutes of Health, American Heart Association, March of Dimes Birth Defects Foundation and the California Institute of Regenerative Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albig AR, Schiemann WP. Identification and characterization of regulator of G protein signaling 4 (RGS4) as a novel inhibitor of tubulogenesis: RGS4 inhibits mitogen-activated protein kinases and vascular endothelial growth factor signaling. Mol Biol Cell. 2005;16:609–625. doi: 10.1091/mbc.E04-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman EP, Campbell JJ, Druey KM, Scheschonka A, Kehrl JH, Butcher EC. Regulation of chemotactic and proadhesive responses to chemoattractant receptors by RGS (regulator of G-protein signaling) family members. J Biol Chem. 1998;273:28040–28048. doi: 10.1074/jbc.273.43.28040. [DOI] [PubMed] [Google Scholar]
- Bruhl T, Urbich C, Aicher D, Acker-Palmer A, Zeiher AM, Dimmeler S. Homeobox A9 transcriptionally regulates the EphB4 receptor to modulate endothelial cell migration and tube formation. Circ Res. 2004;94:743–751. doi: 10.1161/01.RES.0000120861.27064.09. [DOI] [PubMed] [Google Scholar]
- Cao R, Brakenhielm E, Li X, Pietras K, Widenfalk J, Ostman A, Eriksson U, Cao Y. Angiogenesis stimulated by PDGF-CC, a novel member in the PDGF family, involves activation of PDGFR-alphaalpha and -alphabeta receptors. Faseb J. 2002;16:1575–1583. doi: 10.1096/fj.02-0319com. [DOI] [PubMed] [Google Scholar]
- Chan J, Bayliss PE, Wood JM, Roberts TM. Dissection of angiogenic signaling in zebrafish using a chemical genetic approach. Cancer Cell. 2002;1:257–267. doi: 10.1016/s1535-6108(02)00042-9. [DOI] [PubMed] [Google Scholar]
- Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kozasa T, Bondjers C, Betsholtz C, Kehrl JH. Pericyte-specific expression of Rgs5: implications for PDGF and EDG receptor signaling during vascular maturation. Faseb J. 2003;17:440–442. doi: 10.1096/fj.02-0340fje. [DOI] [PubMed] [Google Scholar]
- De Maziere A, Parker L, Van Dijk S, Ye W, Klumperman J. Egfl7 knockdown causes defects in the extension and junctional arrangements of endothelial cells during zebrafish vasculogenesis. Dev Dyn. 2008;237:580–591. doi: 10.1002/dvdy.21441. [DOI] [PubMed] [Google Scholar]
- Dejana E, Taddei A, Randi AM. Foxs and Ets in the transcriptional regulation of endothelial cell differentiation and angiogenesis. Biochim Biophys Acta. 2007;1775:298–312. doi: 10.1016/j.bbcan.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Guo P, Hu B, Gu W, Xu L, Wang D, Huang HJ, Cavenee WK, Cheng SY. Platelet-derived growth factor-B enhances glioma angiogenesis by stimulating vascular endothelial growth factor expression in tumor endothelia and by promoting pericyte recruitment. Am J Pathol. 2003;162:1083–1093. doi: 10.1016/S0002-9440(10)63905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstrom M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- Jin SW, Herzog W, Santoro MM, Mitchell TS, Frantsve J, Jungblut B, Beis D, Scott IC, D'Amico LA, Ober EA, et al. A transgene-assisted genetic screen identifies essential regulators of vascular development in vertebrate embryos. Dev Biol. 2007;307:29–42. doi: 10.1016/j.ydbio.2007.03.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne C, Matenia D, Li XY, Timm T, Balusamy K, Mandelkow EM. Spred1 and TESK1--Two New Interaction Partners of the Kinase MARKK/TAO1 That Link the Microtubule and Actin Cytoskeleton. Mol Biol Cell. 2008;19:1391–1403. doi: 10.1091/mbc.E07-07-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CA, London NR, Chen H, Park KW, Sauvaget D, Stockton RA, Wythe JD, Suh W, Larrieu-Lahargue F, Mukouyama YS, et al. Robo4 stabilizes the vascular network by inhibiting pathologic angiogenesis and endothelial hyperpermeability. Nature medicine. 2008;14:448–453. doi: 10.1038/nm1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kim I, Ryu YS, Kwak HJ, Ahn SY, Oh JL, Yancopoulos GD, Gale NW, Koh GY. EphB ligand, ephrinB2, suppresses the VEGF- and angiopoietin 1-induced Ras/mitogen-activated protein kinase pathway in venous endothelial cells. Faseb J. 2002;16:1126–1128. doi: 10.1096/fj.01-0805fje. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007a;104:20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Chen TT, Barber CL, Jordan MC, Murdock J, Desai S, Ferrara N, Nagy A, Roos KP, Iruela-Arispe ML. Autocrine VEGF signaling is required for vascular homeostasis. Cell. 2007b;130:691–703. doi: 10.1016/j.cell.2007.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Fredriksson L, Li X, Eriksson U. PDGF-D is a potent transforming and angiogenic growth factor. Oncogene. 2003;22:1501–1510. doi: 10.1038/sj.onc.1206223. [DOI] [PubMed] [Google Scholar]
- Lien S, Lowman HB. Therapeutic anti-VEGF antibodies. Handb Exp Pharmacol. 2008:131–150. doi: 10.1007/978-3-540-73259-4_6. [DOI] [PubMed] [Google Scholar]
- Lu Q, Sun EE, Klein RS, Flanagan JG. Ephrin-B reverse signaling is mediated by a novel PDZ-RGS protein and selectively inhibits G protein-coupled chemoattraction. Cell. 2001;105:69–79. doi: 10.1016/s0092-8674(01)00297-5. [DOI] [PubMed] [Google Scholar]
- Mason JM, Morrison DJ, Basson MA, Licht JD. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006;16:45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Wakioka T, Nishinakamura H, Kamio M, Yang L, Inoue M, Hasegawa M, Yonemitsu Y, Komiya S, Yoshimura A. The Sprouty-related protein, Spred, inhibits cell motility, metastasis, and Rho-mediated actin reorganization. Oncogene. 2004;23:5567–5576. doi: 10.1038/sj.onc.1207759. [DOI] [PubMed] [Google Scholar]
- Myers C, Charboneau A, Boudreau N. Homeobox B3 promotes capillary morphogenesis and angiogenesis. J Cell Biol. 2000;148:343–351. doi: 10.1083/jcb.148.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonami A, Kato R, Taniguchi K, Yoshiga D, Taketomi T, Fukuyama S, Harada M, Sasaki A, Yoshimura A. Spred-1 negatively regulates interleukin-3-mediated ERK/mitogen-activated protein (MAP) kinase activation in hematopoietic cells. J Biol Chem. 2004;279:52543–52551. doi: 10.1074/jbc.M405189200. [DOI] [PubMed] [Google Scholar]
- Park TJ, Boyd K, Curran T. Cardiovascular and craniofacial defects in Crk-null mice. Mol Cell Biol. 2006;26:6272–6282. doi: 10.1128/MCB.00472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker LH, Schmidt M, Jin SW, Gray AM, Beis D, Pham T, Frantz G, Palmieri S, Hillan K, Stainier DY, et al. The endothelial-cell-derived secreted factor Egfl7 regulates vascular tube formation. Nature. 2004;428:754–758. doi: 10.1038/nature02416. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- Rousseau S, Houle F, Huot J. Integrating the VEGF signals leading to actin-based motility in vascular endothelial cells. Trends Cardiovasc Med. 2000;10:321–327. doi: 10.1016/s1050-1738(01)00072-x. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Hagen JW, Okamura K, Perrimon N, Lai EC. Functional screening identifies miR-315 as a potent activator of Wingless signaling. Proc Natl Acad Sci U S A. 2007;104:18151–18156. doi: 10.1073/pnas.0706673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Kohno R, Ayada T, Kato R, Ichiyama K, Morisada T, Oike Y, Yonemitsu Y, Maehara Y, Yoshimura A. Spreds are essential for embryonic lymphangiogenesis by regulating vascular endothelial growth factor receptor 3 signaling. Mol Cell Biol. 2007;27:4541–4550. doi: 10.1128/MCB.01600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki K, Fruman DA, Yballe CM, Fasshauer M, Klein J, Asano T, Cantley LC, Kahn CR. Positive and negative roles of p85 alpha and p85 beta regulatory subunits of phosphoinositide 3-kinase in insulin signaling. J Biol Chem. 2003;278:48453–48466. doi: 10.1074/jbc.M305602200. [DOI] [PubMed] [Google Scholar]
- Wakioka T, Sasaki A, Kato R, Shouda T, Matsumoto A, Miyoshi K, Tsuneoka M, Komiya S, Baron R, Yoshimura A. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412:647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Molecular cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Srivastava D. A developmental view of microRNA function. Trends in biochemical sciences. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.