Abstract
In insulin-resistant status such as obesity, failure of pancreatic islets to increase insulin secretion leads to diabetes. We sought to screen for the islet genes that facilitate islet adaptation to obesity by comparing gene expression profiles between two strains of obesity-prone inbred mice with different propensities for hyperglycemia. C57BL/6J and AKR/J were fed regular rodent chow or high-fat diet, after which islet morphology, secretory function, and gene expression were assessed. AKR/J had lower blood glucose and higher insulin levels compared with C57BL/6J mice on regular rodent chow or high-fat diet. Insulin secretion was 3.2-fold higher in AKR/J than C57BL/6J mice following intraperitoneal glucose injection. Likewise, glucose-stimulated insulin secretion from isolated islets was higher in AKR/J. Additionally, islet mass was 1.4-fold greater in AKR/J compared with C57BL/6J. To elucidate the factors associated with the differences in islet function, we analyzed the gene expression profiles in islets in AKR/J and C57BL/6J mice. Of 14,000 genes examined, 202 were upregulated and 270 were downregulated in islets from diet-induced obese AKR/J mice compared with C57BL/6J mice. Key genes involved in islet signaling and metabolism, e.g., glucagon-like peptide-1 receptor, sterol Co-A desaturase 1 and 2, and fatty acid desaturase 2 were upregulated in obese AKR/J mice. The expression of multiple extracellular matrix proteins was also increased in AKR/J mice, suggesting a role in modulation of islet mass. Functional analyses of differentially regulated genes hold promise for elucidating factors linking obesity to alterations in islet function.
Keywords: C57BL/6J, AKR/J, insulin secretion, islet mass, microarray
the incidence of type 2 diabetes (DM) is increasing rapidly as a consequence of the obesity epidemic (18, 25). Normally, insulin resistance associated with obesity is compensated for by increasing pancreatic islet mass and insulin secretion (16, 30). However, in some patients β-cell adaptation is attenuated, leading to the onset of diabetes and related complications (30). Both genetic and environmental factors are important in determining the susceptibility to type 2 DM and are likely to play critical roles in the attenuation of β-cell adaptation in high-risk individuals (28). Although rare cases of a single gene mutation can cause type 2 DM, the genetic defects that account for the deterioration of islet function in the majority of type 2 DM are complex, polygenic, and still not fully understood (28, 30).
Inbred mice show wide variation in the propensity for obesity and diabetes and serve as valuable models to analyze genetic factors that confer susceptibility to the disease (35, 36, 39). Previous studies indicate that genetic factors have a strong influence on islet function and islet mass and thereby contribute to the disparity in glucose homeostasis between different lines of inbred mice (1, 3, 21). Perifused islets from C57BL/6J (Bl6) mice had significantly lower glucose-stimulated insulin secretion compared with A/J mice (21). The comparison of islet mass and total islet numbers in mice from seven different genetic backgrounds demonstrated that strains are the major determinant of these parameters (3). Therefore, the genes differentially regulated in pancreatic islets between inbred strains of mice may reflect factors that regulate islet size and its response to secretagogues.
In the present study we aimed to screen for the genes that promote islet compensation for insulin resistance by utilizing inbred mice. We used microarray analysis as it allows for the screening of a wide range of genes and has the potential to identify gene clusters that concomitantly regulate islet function. Moreover, the polygenic nature of diabetes is difficult to address with linkage analyses. Therefore, we performed gene expression profiles in pancreatic islets between AKR/J (AKR) and Bl6 mice, both of which are susceptible to diet-induced obesity but show different propensities for diet-induced hyperglycemia.
MATERIALS AND METHODS
Animal studies.
Experiments were performed in accordance with the University of Pennsylvania Institutional Animal Care and Use Committee guidelines with its approvals. We housed 4 wk-old male Bl6 mice and AKR mice (Jackson Laboratories, Bar Harbor, ME) (n = 5/cage) in 12 h light/dark cycle, at ambient temperature of 22°C, and allowed them free access to food and water. Groups of mice were fed regular rodent chow (NC, 4 kcal% fat; no. 5001 from Lab Diet, Richmond, IN) or high-fat diet (HF, 45 kcal% fat; no. D124551i from Research Diets, New Brunswick, NJ) for up to 12 wk.
Body composition.
Bl6 mice and AKR mice NC or HF were anesthetized with pentobarbital sodium (50 mg/kg BW ip), and body composition was measured using dual-emission x-ray absorptiometry (PIXImus DEXA, General Electric, Madison, WI; Ref. 15).
Glucose testing.
Glucose tolerance test was performed after overnight fast by giving 2 gm/kg glucose ip. For insulin tolerance test, mice were fasted for 5 h and given Humalin 0.75 U/kg ip (Eli Lilly, Indianapolis, IN). Tail blood was drawn at various times for glucose measurement with OneTouch Ultra Glucometer (Lifescan; Johnson & Johnson, Milpitas, CA). To assess glucose-stimulated insulin secretion in vivo, mice were given 3 gm/kg glucose ip, and 20 μl of tail blood was obtained at the indicated time for insulin measurement by ELISA from Crystal Chem (Chicago, IL) using mouse insulin standard.
Islet isolation and ex vivo perifusion assay.
Mice were anesthetized with pentobarbital sodium (50 mg/kg ip), and pancreatic islets were isolated using collagenase digestion followed by Ficoll density gradient centrifugation as was described before (10, 20). Then islets were hand picked under a dissecting microscope (SMZ 800; Nikon, Melville, NY). The purity of islet preparation judged by dithizone staining was ∼90%. Around 100 freshly isolated islets were loaded to a perifusion apparatus and perifused for 35 min with the Krebs buffer (pH 7.4) containing 2.2 mM Ca2+, 0.25% bovine serum albumin, and 10 mM HEPES under 5% CO2 atmosphere at 37 °C without glucose. Then perifusion was continued with the Krebs buffer containing increasing concentrations of glucose, ramped from 0 to 30 mM at 0.8 mmol/min. At the end of each experiment, islets were tested for the maximum insulin secretion by adding 30 mM KCl in the perifusate (14). Samples were collected at 1 ml/min for insulin measurement by radioimmunoassay (Linco Research, St. Charles, MO). At the end of experiments, islets in a perifusion apparatus were recovered on the filter and stored at −80°C for DNA and insulin measurement as described before (14). The threshold of glucose-stimulated insulin secretion was obtained as the glucose concentration that resulted in significantly higher level of insulin secretion compared with the baseline in the perifusion assay.
Histology.
Paraffin-embedded secretions were prepared from pancreas fixed with 10% buffered formalin overnight and visualized with 1:1,000 guinea pig anti-insulin antibody (Linco Research) as described before (14). An image of a pancreatic section with the maximum footprint from each mouse was captured by a color video camera attached to a Nikon light microscope. Pancreas area, total tissue area, and β-cell area were measured using IP lab (Scanalytics, Fairfax, VA). Islet area (%) was calculated as (β-cell area/pancreas area). Islet weight was calculated as (weight of prefixed pancreas) × (β-cell area/total tissue area) and divided by body weight to adjust for the difference in body weight between Bl6 and AKR.
RNA extraction and gene expression analyses.
RNA was extracted from freshly isolated islets using RNeasy kit (Qiagen, Valencia, CA), and cDNA was generated by SprintPowerScript for cDNA synthesis (Clontech, Mountain View, CA) using 500 ng of islet RNA as a template. Gene expression was analyzed by ABI Prism 7900HT sequence detection system (Applied Biosystems, Foster City, CA) with commercial primers for the system. The results were expressed using 36B4 gene expression as an internal standard.
Microarray analyses.
Microarray analyses were performed on quadruplicate RNA samples of pancreatic islets from AKR and Bl6 mice placed on HF for 3 mo. Pancreases from two mice were combined to yield one sample of islet RNA. All protocols were conducted as described in the Affymetrix GeneChips Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA) using 5 μg total RNA and GeneChip Mouse Expression Arrays MOE 430v2 (Affymetrix). For data analyses, probe intensity data (cel files) were input into ArrayAssist Lite version 3.4 (Stratagene, La Jolla, CA), and expression values for the probe sets were calculated using GCRMA. Affymetrix “present,” “absent,” or “marginal” flags were also calculated. Subsequently intensity and flag data were imported into GeneSpring GX version 7.3.1 (Agilent Technologies, Palo Alto, CA) and filtered to retain probe sets flagged as present in at least three out of eight samples. Finally, the statistical test Significance Analysis of Microarrays (v2.23b; Stanford University, Palo Alto, CA) was applied using a two-class unpaired analysis, and differentially expressed genes were identified using a fold change cutoff of ≥1.8 (up or down) and a false discovery rate of 0.08%. The gene lists thus obtained (472 genes) were uploaded to DAVID (http://david.abcc.ncifcrf.gov), and cell compartment annotation and functional annotation chart were obtained. The genes present in at least three samples out of eight (22,245 probe sets) were used as the background list. Microarray data were submitted to Gene Expression Omnibus (www.ncbi.nlm.nih.gov/projects/geo) under the accession number GSE10639.
Statistics.
The data obtained from in vivo analyses and islet morphometry are presented as means ± SE. Differences between two groups were assessed with unpaired Student's t-test. Regression lines were obtained using GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego CA). Multiple parameters in Figs. 1 and 3 were analyzed by ANOVA test. P < 0.05 was considered significant.
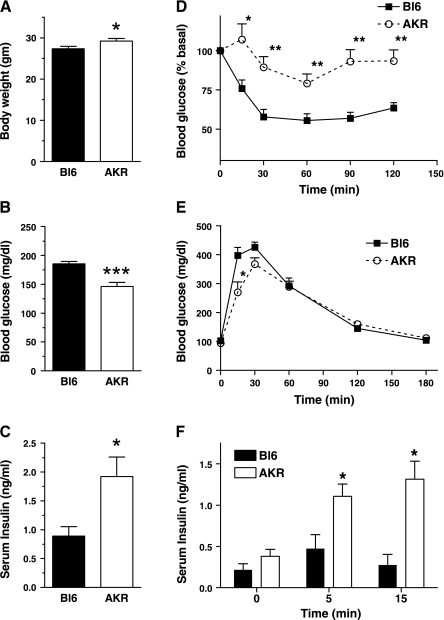
Fig. 1.
Body weight (BW, A), ad libitum blood glucose levels (B), and blood insulin levels (C) were obtained from 3-mo-old male Bl6 and AKR mice on regular rodent chow (NC). D: blood glucose levels in insulin tolerance test by 0.75 U/kg BW insulin intraperitoneal injection (ip) in C57BL/6J (Bl6, solid) and AKR/J (AKR, open) on NC. E: blood glucose levels in glucose tolerance test by 2 gm/kg BW glucose ip. F: insulin levels in glucose tolerance test by 3 gm/kg BW glucose ip in Bl6 (solid) and AKR (clear) on NC. Data are means ± SE, n = 12–16 (A–C), and 4–5 (D–F). *P < 0.05 vs. Bl6, **P < 0.01 vs. Bl6, ***P < 0.0005 vs. Bl6.
RESULTS
Both AKR and Bl6 are prone to obesity, but AKR maintain lower glucose levels with higher insulin levels on HF.
Inbred strains of mice that effectively increase insulin secretion on HF serve as an ideal model to identify genes that promote islet adaptation to insulin resistance. Since AKR are known to develop diet-induced obesity (DIO) like Bl6 but are more glucose tolerant on HF (31, 39), we evaluated their glucose homeostasis and islet function on NC and HF. On NC, AKR were slightly heavier than Bl6 (Fig. 1A, P < 0.05). However, serum glucose levels were significantly lower in AKR (Fig. 1B, P = 0.0001) while insulin levels were higher in AKR (Fig. 1C, P < 0.05). Insulin tolerance testing revealed that AKR are resistant to hypoglycemic effect of insulin compared with Bl6 (Fig. 1D; ANOVA test, P < 0.0001). Despite being insulin resistant, AKR had better glucose tolerance at an early time point, and glucose-stimulated insulin secretion was significantly higher in AKR (Fig. 1, E and F). Higher glucose-stimulated insulin secretion in vivo strongly implies that AKR have better islet function compared with Bl6 and serve as an ideal model to analyze genetic factors that improve islet function.
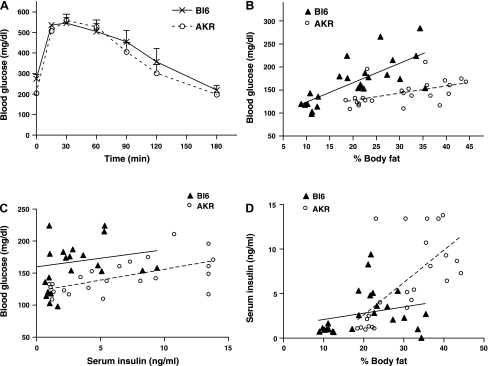
Next, AKR mice and Bl6 mice were placed on NC (4% fat kcal) or HF (45% fat kcal) and monitored for changes in their body weight, blood glucose levels, and serum insulin levels. After 8 wk on the HF, both AKR and Bl6 mice substantially gained weight. AKR weighed 31.3 ± 1.3 g on NC and 46.4 ± 1.3 g on HF (means ± SE, n = 8, P < 0.0001 between NC and HF), while Bl6 weighed 29.7 ± 0.4 g on NC and 35.5 ± 1.5 g on HF (means ± SE, n = 8, P < 0.0001 between NC and HF). The body weight after HF was significantly higher in AKR compared with Bl6 (P < 0.0001), indicating that AKR are more prone to obesity than Bl6. Glucose tolerance test showed that AKR mice have similar glucose tolerance compared with BL6 after HF challenge even though they are more prone to obesity (Fig. 2A). To better delineate differences in response to HF, the relationships between % body fat, glucose levels, and insulin levels from AKR and Bl6 on NC and HF were plotted (Fig. 2, B–D). AKR mice tend to have lower blood glucose levels compared with Bl6 matched for % body fat, indicating that AKR are protected from diet-induced hyperglycemia (Fig. 2B). Moreover, AKR maintained lower blood glucose levels accompanied by higher insulin levels (Fig. 2C). The correlation between serum insulin levels and % body fat indicated that AKR increase insulin levels more effectively when challenged with HF (Fig. 2D).
Fig. 2.
A: blood glucose levels in glucose tolerance test by 1.5 gm/kg BW glucose ip in male Bl6 (×) and AKR (○) on high-fat diet (HF, 45% fat kcal). Data are means ± SE, n = 4. (B–D) Bl6 mice and AKR mice were weaned to NC (4% fat kcal), or HF (45% fat kcal) to compare the response to high-fat feeding between the two species. After 8 wk on the diet, the relationship between blood glucose levels and % body fat (B), and the correlation between blood glucose levels and serum insulin levels (C), and the correlation between serum insulin levels and % body fat (D) were plotted; n = 24 for each strain. Bl6 (▴) and AKR (○).
Pancreatic islets from AKR mice have higher glucose-stimulated insulin secretion and larger islet mass.
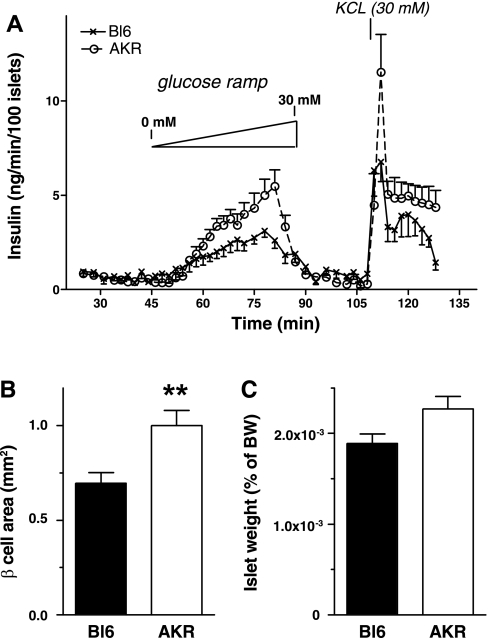
Since AKR showed higher insulin secretion in vivo compared with Bl6, we investigated whether ex vivo insulin secretion and islet mass are different in Bl6 and AKR. Here, islets from AKR and Bl6 on NC were compared, considering that higher insulin secretion in vivo was seen even on NC (Fig. 1). Thus we aimed to address whether islet characteristics differ between the two strains at minimally challenged status. Perifusion of pancreatic islets from AKR and Bl6 revealed that glucose-stimulated insulin secretion was significantly higher in AKR (Fig. 3A, two-way ANOVA, P <0.05). The threshold concentration of glucose was 10.5 mM for AKR islets and 12.2 mM for Bl6 islets. We did not observe a significant difference in DNA content (15.6 ± 2.4 ng/islet in Bl6 vs. 12.4 ± 1.0 ng/islet in AKR) or in insulin content between the two strains (3.4 ± 0.1 ng/ng of DNA in Bl6 vs. 3.6 ± 0.7 ng/ng of DNA in AKR). Morphometric analysis of pancreatic islets showed that β-cell area was significantly larger in AKR than in Bl6 (Fig. 3B). Islet weight adjusted for body weight did not reach statistical significance but also showed a trend of increased mass in AKR compared with Bl6 (P = 0.052).
Fig. 3.
A: ex vivo perifusion compared glucose-stimulated insulin secretion from Bl6 islets (×) and AKR islets (○) on NC. Data are means ± SE, n = 5–6. Islet area (B) and islet weight (C) (% of BW) were obtained in Bl6 and AKR on NC as is described in materials and methods. Data are means ± SE, n = 8. **P < 0.01 vs. Bl6.
Differential expression profile of pancreatic islets from AKR and Bl6.
Since higher serum insulin levels in AKR were associated with elevated insulin secretion in vivo and ex vivo and larger islet mass, we compared islet gene expression profile in DIO AKR and Bl6 (on 45% fat kcal diet) to screen for the factors involved in islet adaptation to obesity. To maximize the differential expression of genes involved in islet adaptation to insulin resistance, the analysis was performed using islets from AKR and Bl6 on HF.
Out of 14,000 distinct genes tested, we observed upregulation of 202 genes and downregulation of 270 genes in AKR islets using a fold change cutoff of ≥1.8 and a false discovery rate of 0.08% (Supplementary Table I).1 We did not observe a significant difference in the expression levels of insulin or pancreatic amylase between the two groups, indicating that the purity of islets preparation was comparable between AKR islets and Bl6 islets. Cell compartment assignment was done based on Gene Ontology and UniProt Knowledgebase Keyword classification through DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov). As is shown in Fig. 4A, 42% of differentially expressed genes belong to extracellular space and 40% are associated with the cell membrane; 4.9% of genes belong to vesicles, cytosol, and endoplasmic reticulum (Fig. 4A). To elucidate functional features of differentially regulated genes, functional Annotation Clustering was performed using DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov). The analyses showed that genes that have signal sequence (Table 1; category; SP_PIR_KEYWORDS, signal) are enriched in both upregulated (P = 0.0022, enrichment compared with background genes) and downregulated genes (P = 3.3E-11, enrichment compared with background genes). In addition, genes involved in extracellular matrix (Table 1; category; GOTERM_CC_ALL, extracellular region) and fatty acid desaturases (Table 1; category; INTERPRO_NAME, fatty acid desaturase) were enriched in upregulated genes in AKR islets (P = 0.0073 and 0.0046, respectively, enrichment compared with background genes). The representative genes from these clusters were validated using real-time PCR in islets from AKR and Bl6 on NC and HF. Stearoyl-CoA desaturase 1 gene (SCD1), stearoyl-CoA desaturase 2 (SCD2), and fatty acid desaturase-2 (FAD) catalyze desaturation of fatty acids and are implied in the regulation of lipid metabolism (5, 33). Asporin and periostin are extracellular proteins that are shown to play a role in cell-cell interactions in other organs (17, 26). These genes were confirmed to be upregulated in AKR compared with Bl6 in islets from both NC- and HF-fed mice (Fig. 4B).
Fig. 4.
A: cell compartment assignment of differentially regulated genes between diet-induced obese (DIO) AKR and Bl6 islets (on 45% fat kcal diet) was performed using DAVID Bioinformatic Resources (http://david.abcc.ncifcrf.gov). B: real-time PCR of islet RNA from Bl6 and AKR on NC and HF. The expression level of 36B4 was used as an internal standard and the value was expressed taking AKR on HF as 100%. White bars, AKR on NC; black bars, Bl6 on HF; gray bars, Bl6 on NC. Data are means ± SE, n = 4. *P < 0.05 AKR vs. Bl6 of the same diet, **P < 0.01 AKR vs. Bl6 of the same diet, #P < 0.05 AKR on HF vs. Bl6 on regular rodent chow.
Table 1.
Gene clusters enriched in AKR islets versus Bl6 islets
| Gene | Gene ID* | Fold Change | AffyID† | |||
|---|---|---|---|---|---|---|
| Signal (category; sp_pir_keywords); upregulated in AKR | ||||||
| Serine (or cysteine) peptidase inhibitor, clade a, member 1d | NM_009243, NM_009244, NM_009245, NM_009246, NM_009247 | 25.5, 19.6, 10.9 | 1420553_x_at, 1418282_x_at, 1451513_x_at | |||
| Glucagon-like peptide 1 receptor | NM_021332 | 21.1 | 1458719_at | |||
| Apolipoprotein F | NM_133997 | 12.9 | 1418239_at | |||
| Inter-alpha trypsin inhibitor, heavy chain 1 | NM_008406 | 10.1 | 1417973_at | |||
| Dehydrogenase/reductase (sdr family) member 8 | NM_053262 | 9.1 | 1421011_at | |||
| Syndecan 4 | NM_011521 | 7.5 | 1448793_a_at | |||
| Astrotactin 1 | NM_007495 | 4.9 | 1418615_at | |||
| Ciliary neurotrophic factor receptor | NM_016673 | 4.2 | 1419429_at | |||
| Sparc-like 1 (mast 9, hevin) | NM_010097 | 3.7 | 1416114_at | |||
| Neuroblastoma, suppression of tumorigenicity 1 | NM_008675 | 3.3 | 1448428_at | |||
| Glial cell line derived neurotrophic factor family receptor alpha 1 | NM_010279 | 3.1 | 1450440_at | |||
| Histocompatibility 2, d region locus 1‡ | NM_010380 | 2.7 | 1450170_x_at | |||
| Interleukin 6 signal transducer | NM_010560 | 2.7 | 1437303_at | |||
| Periostin, osteoblast specific factor | NM_015784 | 2.6 | 1423606_at | |||
| Jagged 2 | NM_010588 | 2.4 | 1426430_at | |||
| Sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 | NM_009262 | 2.5, 1.7 | 1419672_at, | |||
| 1419673_at | ||||||
| Folate receptor 1 (adult) | NM_008034 | 2.4 | 1450995_at | |||
| Epidermal growth factor-containing fibulin-like extracellular matrix protein 2 | NM_021474 | 2.3 | 1417018_at | |||
| Spondin 2, extracellular matrix protein | NM_133903 | 2.2 | 1417860_a_at | |||
| Secretory granule neuroendocrine protein 1, 7b2 protein | NM_009162 | 2.2 | 1423150_at | |||
| Asporin | NM_025711 | 2.2 | 1416652_at | |||
| Apolipoprotein A-II | NM_013474 | 2.1 | 1417950_a_at | |||
| Serine (or cysteine) peptidase inhibitor, clade I member 1 | NM_009250 | 1.9 | 1416702_at | |||
| Protease, serine, 23 | NM_029614 | 1.9 | 1431057_a_at | |||
| Vitronectin | NM_011707 | 1.9 | 1455098_a_at | |||
| Phospholipase A2, group XIIa | NM_023196, NM_183423 | 1.8 | 1452026_a_at | |||
| Signal (category; sp_pir_keywords); downregulated in AKR | ||||||
| Angiogenin, ribonuclease a family, member 1 | NM_007447 | −579.1 | 1438936_s_at | |||
| −11.0 | 1438937_x_at | |||||
| Histocompatibility 2, D region locus 1‡ | NM_010380 | −68.5, −48.4, −31.0, −4.1 | 1425545_x_at | |||
| 1426324_at | ||||||
| 1451683_X_AT, | ||||||
| 1451931_X_AT | ||||||
| Histocompatibility 2, K1, K region | NM_001001892 | −38.8, −21.9, −2.30 | 1425336_x_at, 1427746_x_at, 1451784_x_at | |||
| Kallikrein 1-related peptidase b5 | NM_008456 | −13.0, −14.4 | 1419010_x_at, | |||
| 1449313_at | ||||||
| Galanin | NM_010253 | −10.9 | 1460668_at | |||
| Trefoil factor 2 (spasmolytic protein 1) | NM_009363 | −6.6 | 1422448_at | |||
| Serine (or cysteine) peptidase inhibitor, clade I, member 2 | NM_026460 | −6.1 | 1425152_s_at | |||
| Phospholipase a2, group Ib, pancreas | NM_011107 | −5.4, −5.0 | 1416626_at, | |||
| 1437015_x_at | ||||||
| Left right determination factor 1 | NM_010094 | −5.2 | 1417638_at | |||
| Complement component 8, beta subunit | NM_133882 | −5.0 | 1427472_a_at | |||
| Phospholipase a2, group IIF | NM_012045 | −4.6 | 1434852_at | |||
| Kallikrein 1-related peptidase b22 | NM_010114, NM_010116 | −4.5 | 1425182_x_at | |||
| Fxyd domain-containing ion transport regulator 3 | NM_008557 | −4.5 | 1418374_at | |||
| Islet neogenesis associated protein-related protein | NM_013893 | −4.5 | 1424009_at | |||
| Polymeric immunoglobulin receptor | NM_011082 | −4.4 | 1450060_at | |||
| Kallikrein 1 | NM_010639 | −4.3 | 1415837_at | |||
| Insulin-like growth factor 1 | NM_010512, NM_184052 | −4.3 | 1452014_a_at | |||
| Hydroxysteroid (17-beta) dehydrogenase 13 | NM_198030 | −4.2 | 1460606_at | |||
| Fxyd domain-containing ion transport regulator 6 | NM_022004 | −4.0 | 1417343_at | |||
| Chemokine (c-c motif) ligand 28 | NM_020279 | −4.03 | 1455577_at | |||
| Serine peptidase inhibitor, kazal type 3 | NM_009258 | −4.0 | 1415938_at | |||
| Interleukin 11 receptor, alpha chain 2 | NM_010549,NM_010550 | −3.9 | 1417505_s_at | |||
| Kallikrein 1 | NM_010639 | −3.9 | 1415837_at | |||
| Amylase 1, salivary | NM_007446 | −3.6 | 1417765_a_at | |||
| Complement component 1, q subcomponent, c chain | NM_007574 | −3.5 | 1449401_at | |||
| Matrilin 2 | NM_016762 | −3.4 | 1419442_at | |||
| Glycoprotein 2 (zymogen granule membrane) | NM_025989 | −3.4 | 1449452_a_at | |||
| Pancreatic lipase-related protein 2 | NM_011128 | −3.26, −2.96 | 1437438_x_at, 1448186_at | |||
| Osteoglycin | NM_008760 | −3.25, −2.17 | 1419662_at, 1419663_at | |||
| Complement component 1, q subcomponent, beta polypeptide | NM_009777 | −3.0 | 1417063_at | |||
| Deleted in malignant brain tumors 1 | NM_007769 | −3.7 | 1418287_a_at | |||
| CD200 antigen | NM_010818 | −3.0 | 1448788_at | |||
| Contactin associated protein-like 2 | NM_025771, NM_001004357 | −3.0 | 1422798_at | |||
| Stromal cell derived factor 4 | NM_011341 | −2.9 | 1440340_at | |||
| Histocompatibility 2, class II antigen a, alpha | NM_010378 | −2.8 | 1435290_x_at | |||
| Protein tyrosine phosphatase, receptor type, t | NM_021464 | −2.8 | 1439725_at | |||
| Transmembrane emp24 protein transport domain containing 6 | NM_025458 | −2.7 | 1416490_at | |||
| Contactin associated protein-like 2 | NM_025771, NM_001004357 | −2.7 | 1422798_at | |||
| Prominin 1 | NM_008935 | −2.6 | 1419700_a_at | |||
| MHC (a.ca/j(h-2k-f) class I antigen | NM_019909, NM_001001892 NM_010380 | −2.6 | 1424948_x_at | |||
| Cathepsin S | NM_021281 | −2.5 | 1448591_at | |||
| Platelet-derived growth factor receptor, alpha polypeptide | NM_011058 | −2.5 | 1421917_at | |||
| Cathepsin H | NM_007801 | −2.4 | 1418365_at | |||
| Activated leukocyte cell adhesion molecule | NM_009655 | −2.4, | 1426300_at, | |||
| −2.1, | 1426301_at, | |||||
| −2.0 | 1437466_at | |||||
| Macrophage expressed gene 1 | XM_890648, XM_129176 | −2.4 | 1427076_at | |||
| Delta/notch-like EGF-related receptor | NM_152915 | −2.3, | 1423671_at, | |||
| −2.3 | 1456379_x_at | |||||
| Regenerating islet-derived 1 | NM_009042 | −2.1 | 1415905_at | |||
| Signal sequence receptor, alpha | NM_025965 | −2.1 | 1441327_a_at | |||
| Torsin family 3, member a | NM_023141 | −2.0 | 1428660_s_at | |||
| Protein tyrosine phosphatase, receptor type, j | NM_008982 | −2.0 | 1455030_at | |||
| Protein tyrosine phosphatase, receptor type, d | NM_001014288 NM_011211 | −1.8 | 1445767_at | |||
| HTRA serine peptidase 1 | NM_019564 | −1.8 | 1416749_at | |||
| Fibrinogen-like protein 2 | NM_008013 | −1.8 | 1421855_at | |||
| Gamma-glutamyl hydrolase | NM_010281 | −1.8 | 1419595_a_at | |||
| Extracellular region (category; goterm_cc_all); upregulated in AKR | ||||||
| Sparc-like 1 (mast 9, hevin) | NM_010097 | 3.7 | 1416114_at | |||
| Periostin, osteoblast specific factor | NM_015784 | 2.6 | 1423606_at | |||
| Sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 | NM_009262 | 2.5, 1.7 | 1419672_at, 1419673_at | |||
| Epidermal growth factor-containing fibulin-like extracellular matrix protein 2 | NM_021474 | 2.3 | 1417018_at | |||
| Spondin 2, extracellular matrix protein | NM_133903 | 2.2 | 1417860_a_at | |||
| Asporin | NM_025711 | 2.2 | 1416652_at | |||
| Vitronectin | NM_011707 | 1.9 | 1455098_a_at | |||
| Fatty acid desaturase (category; interpro_name); upregulated in AKR | ||||||
| Stearoyl-coenzyme a desaturase 2 | NM_009128 | 3.6, 2.1 | 1415822_at, 1415824_at | |||
| Stearoyl-coenzyme a desaturase 1 | NM_009127 | 2.3 | 1415964_at | |||
| Fatty acid desaturase 2 | NM_019699 | 2.1 | 1449325_at | |||
NCBA accession Refseq mRNA (http://www.ncbi.nlm.nih.gov).
AffymetrixID (www.affymetrix.com).
Histocompatibility 2, d region locus 1 (NM_010380) is probed by 5 separate Affymetrix probe sets, of which one is upregulated in AKR islets while the rest are downregulated (29). Histocompatibility 2, d region.
DISCUSSION
To investigate factors involved in islet adaptation to insulin resistance, the expression profiles of islet genes were compared between inbred mice of high and low competence for islet compensation in response to HF. By using a cutoff of ≥1.8-fold change and a false discovery rate of 0.08%, 202 genes were found to be upregulated and 270 genes to be downregulated in hyperinsulinemic AKR islets. These genes are promising in the identification of new factors that aid islet adaptation to obesity and prevent development of diabetes.
In agreement with previous reports, AKR maintained lower glucose levels despite being more insulin resistant (31, 39). Although the expression of insulin genes in pancreatic islets was comparable between AKR and Bl6 islets, following observation supports the notion that elevated insulin secretion confers improved glucose homeostasis in AKR. First, AKR had higher glucose-stimulated insulin secretion in vivo (Fig. 1F). Second, AKR increased serum insulin levels effectively and maintained lower blood glucose levels on HF (Fig. 2, C and D). In addition, ex vivo insulin secretion and islet mass were increased in AKR compared with Bl6 (Fig. 3, A and B). Therefore, we used AKR islet as a model of pancreatic islets that show efficient adaptation to insulin resistance.
Previously, several quantitative trait locus analyses addressed candidate genes that confer vulnerability to hyperglycemia in inbred mice (37, 38). One of the genes identified was nicotinamide nucleotide transhydrogenese (NNT), a mitochondrial proton pump gene, which is functionally missing in Bl6 mice and proposed to impair glucose-stimulated insulin secretion (9). Aston-Mourney et al. (2) further demonstrated that NNT activity correlates with first-phase insulin secretion in five mouse strains. However, whether NNT plays any role in the development of diabetes in humans awaits further study. So far, candidate loci in human type 2 DM do not include NNT. A recent study also pointed out that NNT might not be solely responsible for lower insulin secretion in Bl6 compared with DBA/2 (2). Also, the mechanisms of the regulation of insulin secretion by NNT are not fully understood. Therefore, our study that identified multiple genes and gene clusters has the potential to elucidate additional genes that affect islet functions independently or in concert with NNT.
We used freshly isolated islets for gene expression profiling. Islet isolation has been shown to alter the expression of multiple genes including those involved in inflammation, hypoxia, stress, and apoptosis (24). However, the functional cluster analysis of our microarray did not show enrichment of genes in these categories. Since islets from both AKR and Bl6 were isolated by the same protocol, the effects of isolation on gene expression was likely cancelled when the comparison was made between the two.
The functional cluster analysis and cell compartment assignment indicated that the most prominent difference between AKR islets and Bl6 islets is in the category of peptides with signal sequences, which includes secreted proteins and receptors that reside in the cell membrane. Thus, response to external stimuli may yield improved islet function and mass in AKR. For example, a receptor for glucagon-like peptide-1 (GLP-1), an incretin hormone known to increase insulin secretion and islet mass, was upregulated in AKR islets (6). Furthermore, extracellular matrix (ECM) genes were among the enriched genes in AKR islets. Together, these findings indicate that the interaction between β-cells and the surrounding matrix may contribute significantly to islet adaptation to insulin resistance. Interestingly, the two independent microarray analyses of rat models of diabetes, Goto-Kawasaki rats and Zucker Diabetes Fatty rats, also detected the major changes in the expression of ECM genes (11, 42). The importance of ECM has been shown for the maintenance of insulin secretion in cultured islets and for the proper development of islets (4, 12, 13, 23). As such, further studies that address the role of islet architecture in islet adaptation to insulin resistance are warranted.
The elevation of circulating fatty acid (FA) is proposed to link obesity with changes in islet functions ranging from adaptive hypersecretion to islet dysfunction (27). Although there are conflicting data indicating both positive and detrimental effects of FA on islet function and size, the significant impact of lipid metabolism on islet function is well recognized (41). In the current study, three fatty acid desaturases were upregulated in AKR islets, while acetyl-coenzyme A dehydrogenase and CDP-diacylglycerol synthase 1 were downregulated. SCD-1 and -2 are the rate-limiting enzymes in monounsaturated fatty acid synthesis, and they negatively affect fatty acid oxidation and increase lipogenesis in liver (5). However, the deficiency of SCD-1 promotes the development of diabetes in leptin-deficient mice due to islet dysfunction, indicating a protective role for SCD in insulin secretion (8). Ultimately, comparison of lipid metabolism in AKR and Bl6 islet should address whether differential regulation of these genes has functional consequence. However, modification of lipid metabolism is one plausible mechanism that results in better islet function in AKR islets.
Insulin-like growth factor I (IGF-I) was downregulated in AKR islets in our microarray analysis (Table 1). The studies using various islet-specific knockout mice have shown that the insulin signaling plays a critical role in islet growth and islet compensation to insulin resistance (19). However, in contrast to insulin receptor and its signaling molecules that positively regulate islet mass, endogenous IGF-I in islets may inhibit islet growth. Pancreatic-specific inactivation of IGF-I gene results in islet enlargement and confers resistance to high fat-induced diabetes (22). Thus downregulation of IGF-I seen in AKR islet may aid adaptation to HF in AKR islets.
In addition to genes mentioned above, the list of differentially expressed gene contains noble candidate genes that may modulate islet functions. For example Rab3C, a GTP-binding protein, was upregulated 58.9 times in AKR islets compared with Bl6 in microarray analysis and was one of the most prominently differentiated genes (Supplementary Table Ia). Rab3C is known to be colocalized to secretory vesicles and play a role in exocytosis (32). In PC12 cells, Rab3C inhibited Ca2+-triggered exocytosis (32). Further studies may reveal that Rab3C regulates insulin release from β-cells. Dipeptidyl aminopeptidase 6 (DPP6) is another differentially expressed gene that might be a noble regulator of insulin secretion (Supplementary Table Ia). DPP6 is the single transmembrane protein that is associated with a voltage-gated potassium channel subtype 4 (Kv4) and alters its biophysical properties (34). Since Kv channels have a functional role in the process of glucose-stimulated insulin secretion, DPP6 may regulate insulin secretion by modifying the property of a Kv4 channel (7, 40).
As presented above, our analysis revealed many new candidate genes for the islet adaptation to obesity that were previously not connected with insulin secretion nor islet function (Supplementary Table I). On the other hand, limited number of genes previously implicated in islet growth and islet compensation are found in the list (30). GLP-I receptor, IGF-I, SCD1, and peroxisome proliferator-activated receptor γ are examples of such genes (6, 8, 22, 30). However, transcription factors associated with islet growth such as pancreatic and duodenal homeobox 1 (PDX-1), mafA, and NeuroD were not detected in the present analysis (Supplementary Table I). This may be in part due to stringent selection criteria used in the study (a cutoff of ≥1.8-fold change and a false discovery rate of 0.08%).
In summary, cDNA microarray analyses compared gene expression profiles in pancreatic islets from two strains of mice that both develop DIO but show differences in islet adaptation to obesity. The differentially expressed genes identified may serve as molecular markers of islet adaptation to DIO. Future studies should test whether the candidate genes are functionally linked to different propensities for islet dysfunction in the two strains.
GRANTS
The study was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK-071536 and Institute for Diabetes Obesity and Metabolism at Penn (IDOM) pilot and feasibility grant to Y. Imai, DERC Mouse Metabolic Phenotyping and Islet Biology Cores (P30-DK-19525), the Penn Genome Frontiers Institute, and a grant with the Pennsylvania Department of Health to J. W. Tobias.
DISCLAIMER
The Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Supplementary Material
Acknowledgments
We thank Edem Abotsi and Dr. Yong Qi for technical support and David Faleck for critical reading of the manuscript. The technical supports for the study were provided by Penn Microarray Facility, Morphology Core at Center for Molecular Studies in Digestive and Liver Disease, and Radioimmunoassay and Biomarker Core in Diabetes and Endocrinology Research Center (DERC) at University of Pennsylvania School of Medicine.
Address for reprint requests and other correspondence: Y. Imai, Dept. of Medicine, Div. of Endocrinology, Diabetes and Metabolism, Univ. of Pennsylvania School of Medicine, Philadelphia, PA (e-mail: imai@mail.med.upenn.edu).
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Andrikopoulos S, Massa CM, Aston-Mourney K, Funkat A, Fam BC, Hull RL, Kahn SE, Proietto J. Differential effect of inbred mouse strain (C57BL/6, DBA/2, 129T2) on insulin secretory function in response to a high fat diet. J Endocrinol 187: 45–53, 2005. [DOI] [PubMed] [Google Scholar]
- 2.Aston-Mourney K, Wong N, Kebede M, Zraika S, Balmer L, McMahon JM, Fam BC, Favaloro J, Proietto J, Morahan G, Andrikopoulos S. Increased nicotinamide nucleotide transhydrogenase levels predispose to insulin hypersecretion in a mouse strain susceptible to diabetes. Diabetologia 50: 2476–2485, 2007. [DOI] [PubMed] [Google Scholar]
- 3.Bock T, Pakkenberg B, Buschard K. Genetic background determines the size and structure of the endocrine pancreas. Diabetes 54: 133–137, 2005. [DOI] [PubMed] [Google Scholar]
- 4.Bosco D, Meda P, Halban PA, Rouiller DG. Importance of cell-matrix interactions in rat islet beta-cell secretion in vitro: role of alpha6beta1 integrin. Diabetes 49: 233–243, 2000. [DOI] [PubMed] [Google Scholar]
- 5.Dobrzyn A, Ntambi JM. Stearoyl-CoA desaturase as a new drug target for obesity treatment. Obes Rev 6: 169–174, 2005. [DOI] [PubMed] [Google Scholar]
- 6.Drucker DJ The biology of incretin hormones. Cell Metab 3: 153–165, 2006. [DOI] [PubMed] [Google Scholar]
- 7.Dukes ID, Philipson LH. K+ channels: generating excitement in pancreatic beta-cells. Diabetes 45: 845–853, 1996. [DOI] [PubMed] [Google Scholar]
- 8.Flowers JB, Rabaglia ME, Schueler KL, Flowers MT, Lan H, Keller MP, Ntambi JM, Attie AD. Loss of stearoyl-CoA desaturase-1 improves insulin sensitivity in lean mice but worsens diabetes in leptin-deficient obese mice. Diabetes 56: 1228–1239, 2007. [DOI] [PubMed] [Google Scholar]
- 9.Freeman H, Shimomura K, Horner E, Cox RD, Ashcroft FM. Nicotinamide nucleotide transhydrogenase: a key role in insulin secretion. Cell Metab 3: 35–45, 2006. [DOI] [PubMed] [Google Scholar]
- 10.Gotoh M, Maki T, Kiyoizumi T, Satomi S, Monaco AP. An improved method for isolation of mouse pancreatic islets. Transplantation 40: 437–438, 1985. [DOI] [PubMed] [Google Scholar]
- 11.Homo-Delarche F, Calderari S, Irminger JC, Gangnerau MN, Coulaud J, Rickenbach K, Dolz M, Halban P, Portha B, Serradas P. Islet inflammation and fibrosis in a spontaneous model of type 2 diabetes, the GK rat. Diabetes 55: 1625–1633, 2006. [DOI] [PubMed] [Google Scholar]
- 12.Hulinsky I, Cooney S, Harrington J, Silink M. In vitro growth of neonatal rat islet cells is stimulated by adhesion to matrix. Horm Metab Res 27: 209–215, 1995. [DOI] [PubMed] [Google Scholar]
- 13.Hulinsky I, Harrington J, Cooney S, Silink M. Insulin secretion and DNA synthesis of cultured islets of Langerhans are influenced by the matrix. Pancreas 11: 309–314, 1995. [DOI] [PubMed] [Google Scholar]
- 14.Imai Y, Patel HR, Hawkins EJ, Doliba NM, Matschinsky FM, Ahima RS. Insulin secretion is increased in pancreatic islets of neuropeptide Y-deficient mice. Endocrinology 148: 5716–5723, 2007. [DOI] [PubMed] [Google Scholar]
- 15.Imai Y, Varela GM, Jackson MB, Graham MJ, Crooke RM, Ahima RS. Reduction of hepatosteatosis and lipid levels by an adipose differentiation-related protein antisense oligonucleotide. Gastroenterology 132: 1947–1954, 2007. [DOI] [PubMed] [Google Scholar]
- 16.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444: 840–846, 2006. [DOI] [PubMed] [Google Scholar]
- 17.Katsuragi N, Morishita R, Nakamura N, Ochiai T, Taniyama Y, Hasegawa Y, Kawashima K, Kaneda Y, Ogihara T, Sugimura K. Periostin as a novel factor responsible for ventricular dilation. Circulation 110: 1806–1813, 2004. [DOI] [PubMed] [Google Scholar]
- 18.Kopelman PG Obesity as a medical problem. Nature 404: 635–643, 2000. [DOI] [PubMed] [Google Scholar]
- 19.Kulkarni RN Receptors for insulin and insulin-like growth factor-1 and insulin receptor substrate-1 mediate pathways that regulate islet function. Biochem Soc Transact 30: 317–322, 2002. [DOI] [PubMed] [Google Scholar]
- 20.Lacy PE, Kostianovsky M. Method for the isolation of intact islets of Langerhans from the rat pancreas. Diabetes 16: 35–39, 1967. [DOI] [PubMed] [Google Scholar]
- 21.Lee SK, Opara EC, Surwit RS, Feinglos MN, Akwari OE. Defective glucose-stimulated insulin release from perifused islets of C57BL/6J mice. Pancreas 11: 206–211, 1995. [DOI] [PubMed] [Google Scholar]
- 22.Lu Y, Herrera PL, Guo Y, Sun D, Tang Z, LeRoith D, Liu JL. Pancreatic-specific inactivation of IGF-I gene causes enlarged pancreatic islets and significant resistance to diabetes. Diabetes 53: 3131–3141, 2004. [DOI] [PubMed] [Google Scholar]
- 23.Maldonado TS, Crisera CA, Kadison AS, Alkasab SL, Longaker MT, Gittes GK. Basement membrane exposure defines a critical window of competence for pancreatic duct differentiation from undifferentiated pancreatic precursor cells. Pancreas 21: 93–96, 2000. [DOI] [PubMed] [Google Scholar]
- 24.Marselli L, Thorne J, Ahn YB, Omer A, Sgroi DC, Libermann T, Otu HH, Sharma A, Bonner-Weir S, Weir GC. Gene expression of purified beta-cell tissue obtained from human pancreas with laser capture microdissection. J Clin Endocrinol Metab 93: 1046–1053, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mokdad AH, Ford ES, Bowman BA, Dietz WH, Vinicor F, Bales VS, Marks JS. Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA 289: 76–79, 2003. [DOI] [PubMed] [Google Scholar]
- 26.Nakajima M, Kizawa H, Saitoh M, Kou I, Miyazono K, Ikegawa S. Mechanisms for asporin function and regulation in articular cartilage. J Biol Chem 282: 32185–32192, 2007. [DOI] [PubMed] [Google Scholar]
- 27.Nolan CJ, Madiraju MS, Delghingaro-Augusto V, Peyot ML, Prentki M. Fatty acid signaling in the beta-cell and insulin secretion. Diabetes 55, Suppl 2: S16–S23, 2006. [DOI] [PubMed] [Google Scholar]
- 28.Permutt MA, Wasson J, Cox N. Genetic epidemiology of diabetes. J Clin Invest 115: 1431–1419, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pullen JK, Horton RM, Cai ZL, Pease LR. Structural diversity of the classical H-2 genes: K, D, and L. J Immunol 148: 953–967, 1992. [PubMed] [Google Scholar]
- 30.Prentki M, Nolan CJ. Islet beta cell failure in type 2 diabetes. J Clin Invest 116: 1802–1812, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossmeisl M, Rim JS, Koza RA, Kozak LP. Variation in type 2 diabetes-related traits in mouse strains susceptible to diet-induced obesity. Diabetes 52: 1958–1966, 2003. [DOI] [PubMed] [Google Scholar]
- 32.Schluter OM, Khvotchev M, Jahn R, Sudhof TC. Localization versus function of Rab3 proteins. Evidence for a common regulatory role in controlling fusion. J Biol Chem 277: 40919–40929, 2002. [DOI] [PubMed] [Google Scholar]
- 33.Sjogren P, Sierra-Johnson J, Gertow K, Rosell M, Vessby B, de Faire U, Hamsten A, Hellenius ML, Fisher RM. Fatty acid desaturases in human adipose tissue: relationships between gene expression, desaturation indexes and insulin resistance. Diabetologia 51: 328–335, 2008. [DOI] [PubMed] [Google Scholar]
- 34.Soh H, Goldstein SA. I SA channel complexes include four subunits each of DPP6 and Kv4.2. J Biol Chem 283: 15072–15077, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surwit RS, Feinglos MN, Rodin J, Sutherland A, Petro AE, Opara EC, Kuhn CM, Rebuffe-Scrive M. Differential effects of fat and sucrose on the development of obesity and diabetes in C57BL/6J and A/J mice. Metabolism 44: 645–651, 1995. [DOI] [PubMed] [Google Scholar]
- 36.Surwit RS, Kuhn CM, Cochrane C, McCubbin JA, Feinglos MN. Diet-induced type II diabetes in C57BL/6J mice. Diabetes 37: 1163–1167, 1988. [DOI] [PubMed] [Google Scholar]
- 37.Surwit RS, Seldin MF, Kuhn CM, Cochrane C, Feinglos MN. Control of expression of insulin resistance and hyperglycemia by different genetic factors in diabetic C57BL/6J mice. Diabetes 40: 82–87, 1991. [DOI] [PubMed] [Google Scholar]
- 38.Toye AA, Lippiat JD, Proks P, Shimomura K, Bentley L, Hugill A, Mijat V, Goldsworthy M, Moir L, Haynes A, Quarterman J, Freeman HC, Ashcroft FM, Cox RD. A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia 48: 675–686, 2005. [DOI] [PubMed] [Google Scholar]
- 39.West DB, Boozer CN, Moody DL, Atkinson RL. Dietary obesity in nine inbred mouse strains. Am J Physiol Regul Integr Comp Physiol 262: R1025–R1032, 1992. [DOI] [PubMed] [Google Scholar]
- 40.Yan L, Figueroa DJ, Austin CP, Liu Y, Bugianesi RM, Slaughter RS, Kaczorowski GJ, Kohler MG. Expression of voltage-gated potassium channels in human and rhesus pancreatic islets. Diabetes 53: 597–607, 2004. [DOI] [PubMed] [Google Scholar]
- 41.Yaney GC, Corkey BE. Fatty acid metabolism and insulin secretion in pancreatic beta cells. Diabetologia 46: 1297–1312, 2003. [DOI] [PubMed] [Google Scholar]
- 42.Zhou YP, Madjidi A, Wilson ME, Nothhelfer DA, Johnson JH, Palma JF, Schweitzer A, Burant C, Blume JE, Johnson JD. Matrix metalloproteinases contribute to insulin insufficiency in Zucker diabetic fatty rats. Diabetes 54: 2612–2619, 2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.