Abstract
Tumor protein D52 is expressed at relatively high levels in cells within the gastrointestinal tract that undergo classical exocytosis and is overexpressed in several cancers. Current evidence supports a role for D52 in the regulation of vesicular trafficking. D52 function(s) are regulated by calcium-dependent phosphorylation; however, the intracellular mechanisms that mediate this process are not well characterized. The goal of this study was to identify the calcium-dependent phosphorylation site(s) in D52 and to characterize the protein kinase(s) that mediate this phosphorylation. Using mass spectrometry and site-directed mutagenesis, we identified a single amino acid residue, S136, that undergoes increased phosphorylation upon elevation of intracellular Ca2+ concentration. A phosphospecific antibody (pS136) was produced and used to characterize D52 kinase activity in gastric mucosal, colonic T84, and HEK293 cells. By using D52 as a substrate, a protein kinase with a molecular weight (Mr) of ∼50 kDa was identified with “in gel” assays. This kinase comigrated with rat brain calcium/calmodulin-dependent protein kinase (CAMK2)α cross-reacted with pan-specific CAMK2 antibodies as well as with anti-active CAMK2 (pT286/287) antibody when activated. Carbachol-stimulated phosphorylation of S136 was inhibited by the CAMK2 inhibitor KN93 (IC50 38 μM) and by the calmodulin antagonist W7 (IC50 3.3 nM). A previously uncharacterized CAMK2 isoform, CAMK2δ6, which has the same domain structure and Mr as CAM2α, was identified in gastric mucosa by RT-PCR. The cloned, expressed protein comigrated with D52 kinase and colocalized with D52 protein in T84 and HEK293 cells. These findings support a role for CAMK2δ6 in the mediation of D52 phosphorylation.
Keywords: mouse gastric glands, T84 cells, HEK293 cells, protein phosphorylation
d52 (tpd52)-like proteins are members of a highly conserved gene family that includes at least three, possibly four orthologous groups: D52, D53, D54, and NYD-SP25 (predicted) (3). The founding family member is hD52, which was identified as an overexpressed gene product in breast cancers (4) and in tumor-derived cell lines from colon, kidney, lung, breast, and pancreas (6). hD52 is also overexpressed in prostate and ovarian cancers (see Ref. 3 and citations therein). Native D52 protein was initially characterized as a cholinergically regulated, 28 kDa calcium-sensitive phosphoprotein in rabbit gastric mucosal and in rat pancreatic acinar cells (where it was named CSPP28 and CRHSP-28, respectively) (22, 37). In the gastrointestinal (GI) tract, D52 is prominently expressed in granule-rich cells such as acinar cells in the exocrine pancreas, gastric chief cells, and intestinal goblet and Paneth cells (23). D52 is also expressed in mature secretory plasma B cells (45).
Although the biological functions of D52 have not been clearly established, studies with permeabilized pancreatic acinar cells support a role for this protein in the regulation of vesicular membrane trafficking (42). D52 is localized on apically directed vesicles in pancreatic acinar cells along with the early endosomal protein EEA-1 and the trans-Golgi marker TGN38 (44). In addition, D52 binds to two proteins that have been implicated in the regulation of endocytosis: annexin VI (43, 45) and MAL2 (50). The intracellular signaling pathways that regulate D52 functions are not clearly established. Indirect evidence supports a role for calcium/calmodulin-dependent protein kinase (CAMK2) (27, 37). However, neither the CAMK2 inhibitors, KN93 and KN62, nor the less specific calmodulin antagonists, W7 and trifluoperazine (TFP), have been found to inhibit calcium-dependent D52 phosphorylation in colonic T84 cells (27).
The goal of the present study was to identify the specific phosphorylation site(s) in D52 that are regulated by activation of calcium-dependent signaling pathways and to define the protein kinase(s) that mediate this process. Evidence presented herein identifies a highly conserved serine residue (S136) in D52 as the in vivo target for cholinergic, calcium-dependent stimulation and a previously uncharacterized ∼50-kDa CAMK2 isoform as the mediator of this process.
METHODS
Antibodies and inhibitors.
Antibodies were obtained from the following sources: anti-hemagglutinin (HA.11, clone 16B12), Babco/Covance, Madison, WI; anti-CAMKIV (rabbit polyclonal), Cell Signaling Technology, Danvers, MA; anti-casein kinase 2 (CK2), Upstate Biotechnology/Millipore; anti-active CAMK2, which recognizes autophosphorylated CAMK2 isoforms [Thr-286 (α) or Thr-287 (β, γ, δ)], Promega, Madison, WI; anti-CAMK2, Stressgen, Ann Arbor, MI and Upstate Biotechnology/Millipore, Billerica, MA. Anti-CAMK2, rabbit polyclonal antibody G-301 was a gift from Dr. James Goldenring, Vanderbilt University; rabbit polyclonal anti-green fluorescent protein (GFP) was a gift from Dr. Nevin Lambert, Medical College of Georgia (MCG); cyanine (Cy)-5-tagged donkey anti-mouse IgG, Jackson Immunoresearch Laboratories, Westgrove, PA; Alexa 488 phalloidin and Oregon green phalloidin, which specifically recognize F-actin, and donkey anti-rabbit Alexa 555 secondary antibody were from Molecular Probes/Invitrogen, Eugene, OR.
A monoclonal antibody directed against His-tagged D52 (clone 3C10) was produced by the Monoclonal Antibody facility at the University of Georgia. Specificity and titer of the antibody were confirmed by Western blotting using previously described techniques (15). Phosphorylation state-specific polyclonal antibodies were raised in rabbits and affinity purified by 21st Century Biochemicals.
With the exception of wortmannin and Rp-8-Br-PET-cGMPs, which were from Sigma Chemicals, St. Louis, MO and Biomol International, Plymouth Meeting, PA, respectively, protein kinase inhibitors were from Calbiochem/EMD Biosciences.
Gastric mucosal cell isolation, cell lines, and DNA transfection.
Glands and mucosal cells were isolated from rabbit gastric mucosa and pepsinogen-secreting chief cells cultured as previously described for parietal cells (7, 8, 11). Gastric glands from mouse were isolated by using a modified protocol (9) in which gastric mucosal scrapings were briefly incubated with pronase (0.25 mg/ml, 5 min) prior to collagenase digestion (0.5 mg/ml, 30 min). All cell isolation protocols were reviewed and approved by the IACUC at MCG.
Colonic T84 and human embryonic kidney (HEK) 293 cell lines were obtained from American Type Culture Collection and cultured according to their recommendations. These cell lines were used for transfection experiments because native gastric cells have low transfection efficiencies (36). Cells were transfected with Lipofectamine 2000 (Invitrogen) at a ratio of 1:5 (DNA: reagent, 2 μg DNA/35 mm dish) or Effectene (Qiagen) at a ratio of 0.5:10 (DNA: reagent, 0.5 μg/35 mm dish). siIMPORTER (Invitrogen) was used for small interfering RNA (siRNA)/plasmid cotransfection experiments.
Localization of fluorescently tagged proteins in tissues and cells.
Rabbit tissues and cultured cells were fixed and processed for fluorescent analyses as previously described (12, 14). Antibody dilutions are indicated in the figure legends. Images were acquired with a confocal microscope (Zeiss LSM 510 Meta running Meta 3.2 software) with controls for “crossover” fluorescence and nonspecific binding as previously described (12, 14).
RNA isolation, plasmid constructs, and site-directed mutagenesis.
Total RNA was isolated from gastric glands by use of a Perfect Pure RNA kit (5′-Prime). First-strand cDNA was synthesized with a SuperScript First-Strand Synthesis System for RT-PCR with random hexamers (Invitrogen). PCR screening was performed by using Platinum pfx DNA Polymerase per manufacturer's instructions (Invitrogen). Full-length PCR products were cloned by using standard molecular biological techniques in conjunction with the pGEM-T Easy Vector System II (Promega) after A-tailing with GoTaq DNA polymerase. Positive clones expressed in JM109 bacteria were identified and purified (Qiagen mini-prep). For protein expression analyses, hemagglutinin (HA)-tagged constructs were generated by using pGEM-T Easy Vectors as PCR templates followed by subcloning into pcDNA3.1. Enhanced green fluorescent protein (EGFP)-tagged D52 was produced as previously described (36).
His-tagged D52 protein was produced by PCR amplification of rabbit gastric mucosal cell cDNA and bacterial expression in a PET15b expression vector (Novagen, Madison, WI) followed by sequential purification on His-bind nickel chelate resin and Mono Q anion exchange as previously described (37). A cDNA construct for HA-tagged D52 was produced by using the PET15b vector as a template and standard PCR conditions with the following primers: forward, 5′-CCC GAT ATC ATG GGC TAC CCA TAC GAT GTT CCA GAT TAC GCT GAC CGC GGC GAG CAA-3′; reverse complement, 5′-CGC CTC GAG TCA CAG GCC CTC CTG TGT CT-3′. Primers used for other cDNA constructs are included in the figure legends.
Mutations in D52 were introduced by using pcDNA 3.1 plasmids containing HA-tagged D52 DNA inserts with a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) (10, 12). All plasmid cDNA constructs were confirmed by sequencing in the Molecular Biology Core at MCG.
siRNA analyses.
Synthetic siRNA constructs for CK2 (4 pooled SMARTselected siRNA duplexes with “UU” overhangs plus 5′-phosphate on the antisense strand) and negative controls (4 pooled nonspecific similarly engineered siRNA duplexes) were from Dharmacon/Upstate/Millipore. Constructs were cotransfected along with pcDNA 3.1 plasmids containing cDNA inserts encoding for HA-tagged D52 into HEK293 cells by use of the siIMPORTER reagent. Three days after transfection, cells were stimulated with carbachol (100 μM, 2 min) and lysed. Lysates were subjected to Western blotting to confirm CK2 knockdown (anti-CK2 antibody, 1:500) and to quantify the level of phosphorylated D52 (affinity purified anti-phospho D52, 1:100–1:1,000) and total D52 (3C10 or anti-HA antibody) (10).
Phosphorylation site analyses based on metabolic 32P labeling.
For phosphopeptide analyses, His-tagged D52 was phosphorylated with recombinant CAMK2α catalytic subunit (NEN BioLabs) by use of [γ32P]ATP (37). Native D52 was immunoprecipitated following calcium-dependent phosphorylation of metabolically 32P-labeled rabbit gastric mucosal cells as described in the next section. After resolution on SDS-PAGE gels, D52 was subjected to “in gel” tryptic digestion and phosphopeptides were resolved by use of a Pharmacia SMART system as previously described (37). Immunoprecipitation was also used to isolate D52 for phosphorylation site-directed mutagenesis studies.
To immunoprecipitate D52, cells were lysed in a nondenaturing lysis buffer [50 mM Tris·HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% NP40 supplemented with freshly prepared protease and phosphatase inhibitors including 1 mM NaH2PO4, 10 mM NaF, 0.2 mM AEBSF, 0.1 μM calyculin A, and one Roche protease inhibitor tablet (Complete, EDTA-free/7 ml)]. After centrifugation (10 min, 10,000 g), supernatants were incubated with the 3C10 antibody (1:100 dilution) to isolate native rabbit protein or with anti-HA antibody (1:500 dilution) to isolate HA-tagged D52 from cell lines. Immunoprecipitates were collected by incubation with Dynabeads (Invitrogen) or Protein G plus/Protein A-agarose beads (Calbiochem) per manufacturers' instructions. All immunoprecipitation steps were performed at 4°C.
Phosphorylation site analyses by mass spectrometry.
For mass spectrometry (MS), native D52 protein was isolated from rabbit gastric glands by preparative 2D gel electrophoresis as previously described (37) except that preparative isoelectric focusing gel strips (pH 3.5–5, Amersham/GE) replaced tube gels in the first dimension. D52 was partially enriched prior to 2D gel electrophoresis as follows: Glands were lysed by sonication in 20 mM Tris, pH 7.8, 0.25 M sucrose, 2 mM EDTA, 2 mM sodium pyrophosphate, 15 mM 2-mercaptoethanol, 0.2 mM AEBSF, and 1 EDTA-free Roche protease inhibitor tablet/7 ml and then centrifuged (150,000 g, 30 min). Supernatants were heated (90°C, 7 min), and precipitated proteins were recovered by centrifugation (2,000 g, 10 min), dissolved in 3% SDS/10% β-mercaptoethanol, reprecipitated with acetone at room temperature, washed with deionized water, and solubilized in rehydration buffer [8 M urea, 2% CHAPS, 10 mM DTT, 0.5% IPG buffer, pH 3.5–5 (Amersham/GE)].
In gel tryptic digestion and MS analyses were performed by TwentyFirst Century Biochemicals. Nanospray and liquid chromatography (LC)/MS experiments were conducted with a QStar XL (MDS/Sciex) mass spectrometer equipped with a nanospray source or interfaced with a LC Packings Ultimate micropump equipped with a 150-μm ID column packed in-house with Magic C-18 reversed phase packing material (Michrom). Data were analyzed by manually reconstructing MS and MS/MS data and comparing to theoretical MS and MS/MS data via Bioanalyst software.
Partial purification of D52 kinase by column chromatography.
D52 kinase in cell lysates was partially purified by anion exchange chromatography, calmodulin-Sepharose affinity chromatography, and gel filtration. Lysates were prepared by sonicating cells in a buffer supplemented with protease and phosphatase inhibitors (10 mM Tris·HCl, pH 7.5, 1 mM DTT, 1 mM EDTA, 10 mM β-glycerophosphate, 1 μM calyculin A, 1 μM microcystin LR, 0.1 mM AEBSF, and 1 Roche inhibitor tablet/7 ml). Lysate supernatants (14,000 g for 10 min or 100,000 g for 20 min, 4°C) and Triton X-100-solubilized particulate fractions (extracted with lysis buffer containing 0.2% Triton for 30 min, 4°C) were assayed for D52 kinase activity before and after column fractionations.
Anion exchange chromatography was performed by use of a Mono Q HR 5/5 column (Pharamacia) equilibrated with 25 mM HEPES, pH 7.5, 1 mM DTT, 0.5 mM EDTA. Bound proteins were eluted (1 ml fractions) with a linear NaCl gradient (30 ml of 0–1 M NaCl or 0–0.4 M NaCl) into tubes on ice containing 100 μl of 50% glycerol. Column fractions were assayed for D52 kinase activity immediately after collection. Peaks were pooled, concentrated, and, in some experiments, further fractionated on a gel filtration column (Superose 6 HR 10/30, Pharmacia) by using a buffer composed of 25 mM HEPES, pH 7.5, 1 mM DTT, 0.5 mM EDTA, 0.15 M NaCl and/or by batch-wise elution using calmodulin-Sepharose 4B beads (Pharmacia/GE) per manufacturer's instructions.
Protein kinase assays.
Assays for calcium/calmodulin-dependent D52 kinase and CAMK2 activities in cellular extracts and column fractions were performed by using both unlabeled and radiolabeled ATP. In the latter case, cellular extracts (5–10 μg protein) or column fractions (20 μl) were preincubated (5 min, 30°C) in 50 mM HEPES, pH 7.5, 1 mM DTT, 10 mM MgCl2, 4 mM EGTA, 0.2 mM [γ32P]ATP (specific activity 6,000 counts·min−1·pmol−1), in the presence and absence of 200 nM calmodulin, 7 mM CaCl2. Reactions were initiated by the addition of either recombinant His-tagged D52 (1 μg protein) or the CAMK2 substrate peptide, autocamtide-2 (0.1 mM). 32P labeling of His-tagged D52 was quantified after fractionation on 12% SDS-PAGE minigels with a Phosphoimager (Molecular Dynamics) or by Cerenkov counting (37). Autocamtide-2 phosphorylation was quantified via a filter-based protocol in which assay tube aliquots were spotted onto P81 filters (31). The same incubation conditions were used for assays with unlabeled ATP except that 1–10 mM ATP was used in place of [γ32P]ATP and changes in D52 phosphorylation were quantified by Western blotting using affinity-purified pS136 antibody (1:1,000–1:2,500 dilution) with enhanced chemiluminescent (ECL) detection. CAMK2 activation was similarly assessed by use of anti-active CAMK2 (T286/287) antibody (1:1,000 dilution). CK2 was assayed in T84 cell lysates with a CK2 assay kit (Upstate Biotechnology) per manufacturer's instructions.
For phosphorylation with recombinant proteins, CK2α [Stressgen; 300 ng, specific activity (SA) 107 nmol PO4·min−1·mg−1] and CAMK2δ (Invitrogen; 2 ng, SA 15,400 nmol PO4·min−1·mg−1) were incubated with 1 μg His-tagged D52 protein, 10 mM MgCl2, 100 μM [γ32P]ATP (10,000 cpm/pmol) in assay buffers composed of 50 mM Tris·HCl, pH 7.5, 100 mM NaCl, and 1 mM DTT or 50 mM HEPES, pH 7.5, and 1 mM DTT (50 μl final volume), respectively. Prior to addition of D52 substrate, CAMK2 was preincubated with 0.7 mM CaCl2, 0.4 mM EGTA, 0.2 μM calmodulin for 10 min at 30°C. 32P incorporation into D52 was quantified as for autocamtide-2. Site-specific phosphorylation was analyzed by Western blotting as described above.
In gel kinase assays were performed as previously described (31) except that 2 mg of His-tagged D52 replaced myelin basic protein as a substrate in the gels and calcium/calmodulin were included in the assay buffer, which was the same as that used for in vitro phosphorylation analyses. Controls for protein kinase autophosphorylation were similarly analyzed by using gels in which the D52 protein was omitted.
Western blotting, protein assays, and data analysis.
Western blotting, ECL detection, and quantitation were performed as previously described (10, 12, 13). Protein in cell and tissue extracts was quantified with the Bio-Rad DC assay kit or with a QUANT-IT Protein assay kit (Invitrogen) per manufacturers' instructions. GraphPad Prism, Mac version 4.0c was used for nonlinear curve fitting, calculation of Kd and Ki values, and statistical analyses. For inhibitors, curves were analyzed by using the one-site competition equation. Where applicable, values in figures are expressed as means ± SE, and N represents the number of independent experiments.
RESULTS
Immunolocalization of D52 in cells in the GI tract, kidney, and brain.
The few studies that have analyzed the tissue and cellular distributions of D52 have focused mainly on the GI tract where relatively high levels of expression have been detected in pancreas, gastric mucosa, and intestine (23, 37). Within these tissues, D52 is predominately immunolocalized to granule-rich cells including pepsinogen-secreting chief cells in the gastric mucosa, pancreatic acinar cells, salivary gland cells, and intestinal goblet, crypt, and Paneth cells (23). Varying levels of expression have been detected by Western and Northern blotting in several other tissues including kidney, liver, spleen, and brain (23, 37).
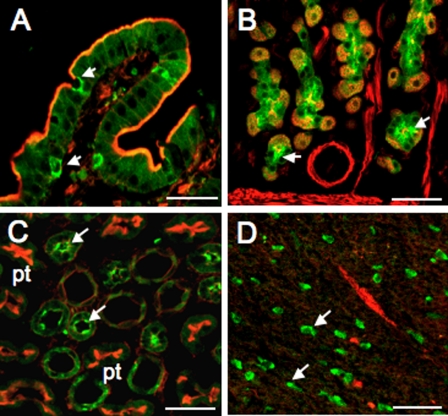
To determine whether D52 is also expressed in granule-rich cells outside the GI tract, we immunolocalized D52 in kidney and brain using GI tissues and cultured gastric chief cells to confirm the specificity of the 3C10 antibody. As expected, on the basis of previous analyses (23), high levels of D52 expression were detected in goblet/Paneth cells in intestine (Fig. 1A) as well as in chief cells in the gastric mucosa (Fig. 1B). In gastric chief cells in primary culture, D52 was also immunolocalized to granules known to contain the enzyme, pepsinogen (not shown). In kidney, D52 expression was pronounced in cells in the distal tubule but was undetectable in cells within the proximal tubule (Fig. 1C). In brain, D52 was immunolocalized predominately within a population of granule-rich cells in the hippocampus (Fig. 1D). No D52 expression was detected in cells within the cerebral cortex or in heart or skeletal muscle (not shown). These data confirm that D52 is widely expressed in granule-rich cells within epithelial tissues and in brain. The finding that a subpopulation of cells within the hippocampal region expresses high levels of D52 is particularly intriguing and suggests that these cells possess highly specialized functions that may involve classical exocytotic activity.
Fig. 1.
Immunolocalization of D52 in intestine, stomach, kidney, and brain. Tissues were fixed, permeabilized, and stained as described in methods. After overnight incubation with monoclonal D52 antibody (3C10, 1:50 dilution), D52 was immunolocalized with donkey anti-mouse Cy-5-tagged IgG secondary antibody, 1:100 dilution) and F-actin was localized with Alexa 488 phalloidin (1:40 dilution). A: goblet and Paneth cells (arrows) in small intestine. Strong apical staining was also detected in intestinal crypt cells (not shown). B: chief cells in gastric mucosa (arrows). D52 was more clearly immunolocalized to pepsinogen-containing granules in chief cells in primary culture (not shown). C: distal tubule cells in kidney. Note that proximal tubule cells (pt) stain strongly for F-actin but do not cross-react with the 3C10 antibody. D: unidentified granular cells in brain (hippocampal region). Arrows point to examples of strongly staining cells. To improve visualization, atypical green pseudocoloring was used to localize the D52 monoclonal antibody/Cy5-tagged secondary antibody signal and red pseudocoloring was used to localize the Alexa 488 phalloidin signal. Bars, 100 μm.
Analysis of D52 kinase activity in rabbit gastric mucosal cell extracts.
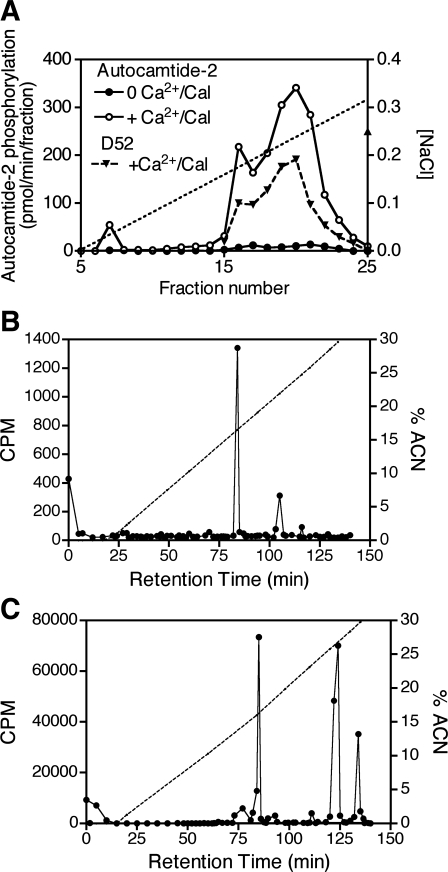
Calcium-dependent phosphorylation of D52 has been demonstrated in gastric mucosal cell extracts (37). To characterize further the protein kinase(s) involved, extracts were prepared from rabbit gastric mucosal cells, fractionated on a Mono Q anion exchange column, and assayed for their ability to phosphorylate recombinant His-tagged D52 protein by the transfer of 32P from [γ32P]ATP. A broad peak of calcium/calmodulin-dependent D52 kinase activity, eluting between ∼0.18 and 0.3 M NaCl, was identified (Fig. 2A). This activity profile was essentially the same when the specific CAMK2 substrate, autocamtide-2, was used in place of D52 (Fig. 2A). These findings supported the hypothesis that CAMK2 or a related kinase phosphorylates D52; however, phosphopeptide mapping of D52 after in vivo and in vitro phosphorylation was inconclusive because the elution patterns of tryptic digests were dissimilar (Fig. 2, B and C), suggesting that either 1) CAMK2 phosphorylates additional sites in vitro or 2) an enzyme other than CAMK2 mediates calcium-dependent phosphorylation of D52 in vivo.
Fig. 2.
Characterization of D52 kinase activity in rabbit gastric mucosal cells and analysis of tryptic digests of the D52 protein after in vivo/in vitro phosphorylation. A: rabbit gastric mucosal cell extracts were fractionated on a Mono Q column then assayed for kinase activity by using either His-tagged D52 protein or autocamtide-2 as a substrate as described in methods. 32P incorporation into His-tagged D52 was quantified with a Phosphoimager after resolution on SDS-PAGE gels. Phosphorylation of autocamtide-2 was quantified with a scintillation counter. Phosphoimager units were scaled to fall within the range of autocamtide-2 values. B: elution profile of tryptic digests of native D52 immunoprecipitated from metabolically 32P-labeled rabbit gastric cells (∼10 × 106 cells, 2 mCi carrier-free 32P/ml) following incubation with ionomycin (3 μM, 5 min) to elevate intracellular Ca2+ concentration. C: elution profile of tryptic digests of recombinant His-tagged D52 after in vitro phosphorylation with recombinant CAMK2α with [γ32P]ATP as the phosphate donor. ACN, acetonitrile.
Isolation and mass spectrometric analysis of phosphorylated D52.
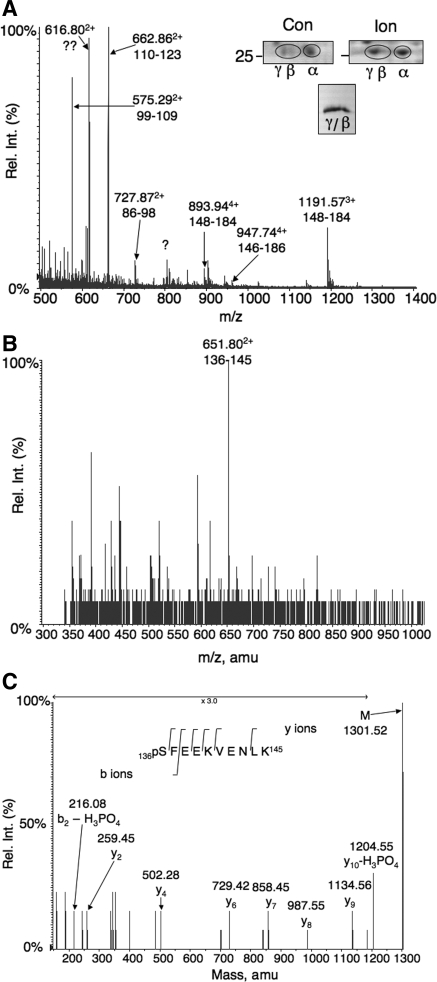
To avoid uncertainties associated with in vitro analyses, we adopted a MS-based approach to identify in vivo, calcium-dependent phosphorylation sites. Native D52 was phosphorylated in rabbit gastric mucosal cells by incubation with the calcium ionophore ionomycin, isolated on preparative 2D gels, and subjected to mass spectrometric analysis. Three major isoforms of D52 (α, β, and γ) were identified by parallel Western blotting. Ionomycin increased the concentration of the most acidic (γ) isoform (Fig. 3A, inset, top). The β and γ isoforms were isolated from preparative 2D gels, concentrated on a single minigel (Fig. 3A, inset, bottom), and used for mass spectrophotometric analyses (Fig. 3, A–C). Figure 3A shows a nanospray mass spectrum of the digested D52 sample. Multiple ions corresponding to D52 tryptic peptides were detected. Mass reconstruction of LC/MS experiments showed 67% sequence coverage of the native D52 protein with unequivocal identification based on multiple tandem MS assignments. Additionally, several potential phosphopeptides corresponding to residues 130–145 and 136–145 were detected. A peak at 52.1 min [651.802+ mass-to-charge ratio (m/z)] that corresponded to residues 136–145 (Fig. 3B) was analyzed by tandem MS. Collision-induced dissociation of this ion produced fragmentation data identifying serine 136 as the modified amino acid. Of particular importance is the detection of the neutral loss of phosphoric acid (−98 Da) and the subsequent loss of the resultant dehydroalanine (−71 Da). These two ions are labeled y10-H3PO4 (phosphoric acid loss) and y9 (dehydroalanine loss) in Fig. 3C. Double phosphorylation of this residue was not detected.
Fig. 3.
Isolation of phosphorylated D52 from rabbit gastric mucosal cells and mass spectrometry (MS) analyses. A, inset, top: regions of preparative 2D gels stained with modified Coomassie blue showing D52 migration patterns in extracts from control (Con) and ionomycin (Ion)-stimulated rabbit gastric glands. Note acidic shift from α to β/γ isoforms after ionomycin stimulation. Inset, bottom: Pooled β/γ spots from 11 preparative 2D gels after concentration on a single SDS-PAGE minigel. The identity of D52 was confirmed by Western blot using 3C10 antibody, which specifically cross-reacts with rabbit D52 (not shown). Graph: liquid chromatography (LC)/MS analysis of tryptic digests of pooled β/γ spots identifying 67% of the protein. B: mass spectrum at 52.1 min from an LC/MS analysis of a tryptic D52 digest identifying an ion at mass-to-charge ratio (m/z) 651.802+ corresponding to residues 136–145. This value matches the theoretical mass of this peptide with a single phosphorylation site [1,221.6 Da, which is increased to 1,301.6 Da by phosphorylation]. With the addition of 2 hydrogens, m/z = (1,301.6 + 2/2) = 651.8 Da. C: mass reconstruction of m/z 651.802+ identifying serine 136 as the phosphorylated form. The theoretical m/z values of the y ions identified in the figure are as follows: y2, 260; y4, 503; y6, 730; y7, 859; y8, 988; y9, 1,135; y10, 1,204. Rel. Int., relative intensity.
In agreement with previous analyses of V8 digests by classical Edman degradation (37), mass spectrometric analysis indicated that the NH2 terminus was blocked. Further analysis of tryptic digests by capillary LC/MS detected acetylation in residues 1–10, m/z 608.792+. The presence of an acidic aspartate residue in the penultimate position in D52 supports NH2-terminal acetylation (38). Although previously thought be a widespread occurrence, NH2-terminal acetylation is now thought to be present in less than 30% of proteins (30). Interestingly, there is some evidence that this type of acetylation can facilitate protein-protein interactions to assist in the recruitment of proteins to vesicular compartments (21).
The main unidentified region in D52 fell within residues no. 47-69. The highly acidic nature of residues in this region, which was previously sequenced by Edman degradation (37), presumably hindered the mass spectrometric analysis. Of potential regulatory importance, this region also contains a histidine residue and has the predicted capacity to undergo several posttranslational modifications including methylation.
Effects of site-directed mutagenesis of S136 on calcium-dependent D52 phosphorylation.
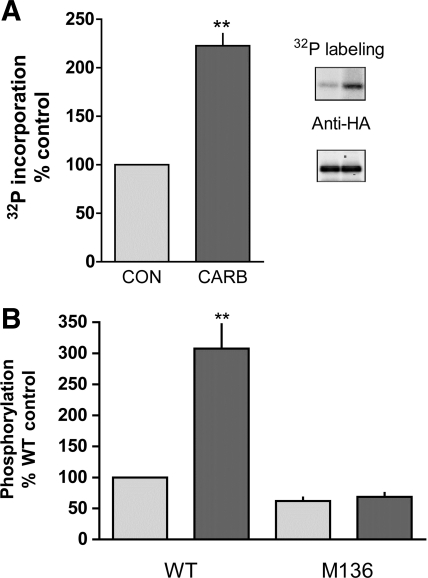
To confirm that S136 is the physiologically relevant target of calcium-dependent D52 regulation, T84 cells were used because D52 is expressed in this cell line and is known to be phosphorylated following cholinergic stimulation (17, 27). The ability of the cholinergic agonist carbachol to elicit a similar response in transiently transfected T84 cells expressing HA-tagged D52 was analyzed by immunoprecipitating the HA-tagged protein from metabolically 32P-labeled cells. As shown in Fig. 4A, carbachol (100 μM, 2 min) induced a ∼2.5-fold increased in 32P incorporation into immunoprecipitated HA-D52. Ionomycin also induced a significant increase in 32P incorporation into HA-tagged D52 (Fig. 4B). Importantly, this response was completely abolished in cells expressing HA-tagged D52 containing an S→A136 mutation (Fig. 4B). Thus these data confirmed the mass spectrometric analyses and provide strong evidence that S136 is the sole site that is regulated by agonist-dependent elevation of intracellular Ca2+ concentration. Because the S136 mutation did not completely abolish basal phosphorylation (Fig. 4B), D52 is likely phosphorylated on site(s) in addition to S136.
Fig. 4.
Comparison of effects of carbachol (Carb) and ionomycin on 32P incorporation into T84 cells expressing either hemagglutinin (HA)-tagged D52 or HA-tagged D52 in which S136 was mutated to alanine. Mutations were introduced by use of pcDNA 3.1 plasmids containing HA-tagged D52 DNA inserts with the following primers: forward, 5′-CCC CAA CCT TCA AAG CCT TTG AAG AGA AAG-3′; reverse complement, 5′-CTT TCT CTT CAA AGG CTT TGA AGG TTG GGG-3′. Cells were transfected with Lipofectamine 2000 and metabolically labeled 3 days later with 400 μCi/ml carrier-free 32P for 6 h in phosphate-free DMEM. A: quantitation of carbachol-stimulated (100 μM, 2 min) 32P incorporation into immunoprecipitated HA-tagged D52 (data normalized on the basis of ECL signals obtained with anti-HA antibody). N = 3, **P < 0.001. Inset: representative experiment showing 32P labeling of immunoprecipitated HA-tagged D52 and Western analysis of the same blot after probing with anti-HA antibody. Control, left lane; carbachol, right lane. B: in T84 cells transfected with D52 (WT) or mutant D52 (M136, Ser136 mutated to Ala), mutation of S136 to alanine was found to inhibit ionomycin-stimulated (3 μM, 5 min) 32P incorporation into D52 and to reduce basal phosphorylation levels. **P < 0.001, N = 3.
Multiple sequence alignments established that S136 and the surrounding amino acids are highly conserved in several mammalian species as well as in quail (Table 1). Phosphorylation site analyses predicted, with high probability, that S136 is phosphorylated. However, analyses with three independent databases (Scansite, ELM, and NetPhosK 1.0) failed to identify a strong consensus site for any protein kinase. Depending on the database, CK2, CAMK2, and glycogen synthase kinase (GSK3) were identified, with low probabilities, as potential candidates (Table 1). Data in Table 1 also indicate that there is a high probability that S32 is phosphorylated, possibly by CK2. If so, we reasoned that this might explain why D52 is a good substrate for CK2 in vitro (27) since both S32 and S136 are predicted substrates for this kinase. S32 may also be basally phosphorylated in vivo, which would explain the residual phosphorylation that was detected in S136 mutational analyses (Fig. 4B).
Table 1.
Conserved D52 phosphorylation sites with the highest phosphorylation potentials and kinase predictions for these sites
|
Phosphorylation Potential |
Kinase Predictions
|
||||||
|---|---|---|---|---|---|---|---|
| Species | Position | Sequence | NetPhos | Disphos | NetPhosK 1.0 | Scansite | ELM |
| Human | 32 | TETLSEEEQ | 0.997 | 0.921 | CK2/CK1/CAMK2 | CK2 | CK2 |
| Mouse | 32 | PEALTEEEQ | 0.790 | 0.436 | CK2/CK1/cdc2/GSK3/CAMK2 | CK2 | CK2 |
| Rabbit | 32 | TETLSEEEQ | 0.997 | 0.933* | CK2/CK1/CAMK2 | CK2 | CK2 |
| Quail | 77 | SETLSEEEQ | 0.995 | 0.859* | CK2/CK1/CAMK2/GSK3 | CK2 | CK2 |
| Human | 136 | PTFKSFEEK | 0.978 | 0.698 | CK2/CAMK2/GSK3 | CAMK2 | CK2 |
| Mouse | 136 | PTFKSFEEK | 0.978 | 0.699 | CK2/CAMK2/GSK3 | CAMK2 | CK2 |
| Rabbit | 136 | PTFKSFEEK | 0.978 | 0.701* | CK2/CAMK2/GSK3 | CAMK2 | CK2 |
| Quail | 204 | PTFKSFEEK | 0.978 | 0.678* | CK2/CAMK2/GSK3/cdc2 | CAMK2 | CK2 |
Predicted phosphorylation sites are highlighted in bold.
Homo sapiens, regulation used in search. Phosphorylation potentials analyzed using default settings: Quail 0.819, 0.610; rabbit 0.912, 0.636.
Our in vitro analyses of gastric cell extracts (Fig. 2 and Ref. 37) as well as a previous study with T84 cells (27) favored CAMK2 as a candidate mediator. However, several lines of evidence did not support this conclusion. These included the following observations: 1) The CAMK2 inhibitor, KN62, did not inhibit D52 phosphorylation at expected concentrations in either the T84 cell line or in isolated gastric glands from rabbit (Ref. 27 and C. S. Chew, unpublished observations). 2) The calmodulin antagonists, W7 and TFP, did not inhibit carbachol-stimulated D52 phosphorylation in T84 cells (27). 3) CK2 catalyzed ∼10 times more phosphate incorporation into serine residues within D52 than did CAMK2 (27). These findings, coupled with the observation that the pattern of D52 phosphorylation by CAMK2 in vitro is not the same as that induced by calcium-dependent phosphorylation in vivo (Fig. 2, B and C), prompted us to develop a phosphorylation site-specific antibody to provide a more direct, sensitive, and powerful approach for analyzing agonist-dependent D52 phosphorylation.
Development and characterization of a phosphospecific antibody directed against S136.
Two phosphospecific peptides were synthesized and used to produce a polyclonal antibody. A third, nonphosphorylated peptide with a similar sequence was used to affinity purify the phosphospecific antibody (Fig. 5, legend). To confirm antibody specificity, responses to carbachol and ionomycin were analyzed in both T84 and HEK293 cell lines expressing HA-tagged D52, and the competitive effects of phosphorylated and nonphosphorylated peptides were assessed. These analyses demonstrated that stimulation of HEK293 cells with either carbachol or ionomycin was correlated with increased binding of affinity-purified pS136 antibody to a 28-kDa protein (Supplemental Fig. S1; the online version of this article contains supplemental data). The 28-kDa protein also cross-reacted with anti-HA antibody, confirming that the pS136 antibody recognized authentic HA-tagged D52 protein (data not shown). Preincubation of affinity-purified pS136 antibody with phosphopeptide antigen (10 μg) suppressed antibody binding on Western blots of extracts from unstimulated cells and from cells stimulated with either carbachol or ionomycin. In contrast, 10 μg of nonphosphorylated peptide did not suppress these responses nor did it reduce the basal signal (Supplemental Fig. S1). In agreement with the S136 mutational analyses (Fig. 4B), these latter data also suggest that S136 is weakly phosphorylated in unstimulated cells.
Fig. 5.
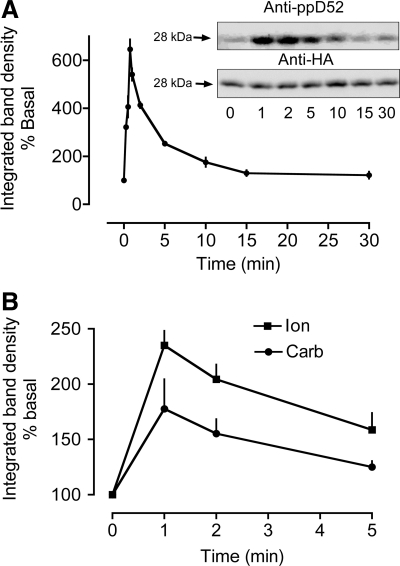
Time courses for calcium-dependent D52 S136 phosphorylation in HEK 293 cells and mouse gastric glands. S136 phosphorylation was quantified by using affinity-purified pS136 antibody that was produced by injecting rabbits with 2 phosphopeptides: Ac-CNSPTFK(pS)FEEKVEN-amide and Ac-NSPTF K(pS) FEEKVENC-amide. The antibody was affinity purified by passing immunized rabbit serum through an affinity column coupled to a nonphosphopeptide (Ac-CNSPTFKSFEEKVEN-amide) and then passing flowthrough fractions from this column through a second affinity column coupled to the 2 phosphopeptides. Phosphospecific antibodies were eluted from the latter column with low pH glycine. A: time course for carbachol stimulation of HEK293 cells (100 μM Carb, N = 3–8) expressing HA-D52. Cells were transfected with Effectene as described in Fig. 4. Inset: a representative experiment depicting Western blots of samples from HEK293 cells probed for S136 phosphorylation (anti-ppD52) and for the HA tag. The HA signals were used to correct for differences in HA-D52 expression levels. After normalization, S136 phosphorylation was expressed as % of basal. B: time courses for carbachol and ionomycin stimulation of mouse gastric glands. N = 3, 30 mice. 100 μM Carb, 3 μM Ion.
Further analyses confirmed that findings with the pS136 antibody mirrored those obtained with metabolic 32P labeling. Thus carbachol stimulation (100 μM, 2 min) induced a 207.5 ± 9.8% in the pS136 antibody binding signal in both T84 cells and in HEK293 cells (N = 4 and N = 2, respectively), a response that was quantitatively similar to that obtained with metabolic 32P labeling/immunoprecipitation of HA-tagged D52 (Fig. 4A). Because all tested approaches supported the specificity of the pS136 antibody, this antibody was used exclusively in subsequent experiments.
Time courses of carbachol-induced phosphorylation in HEK293 cells and in mouse gastric glands.
The temporal response to carbachol was assessed in HEK293 cells expressing HA-tagged D52 as well as in mouse gastric glands. In both models, carbachol induced a rapid, transient increase in S136 phosphorylation (Fig. 5). With HEK293 cells, S136 phosphorylation peaked within 1 min of stimulation and fell to near basal levels within 15 min (Fig. 5A). A similar response occurred in mouse gastric glands (Fig. 5B). In this case, however, it was necessary to use pS136 antibody serum because the affinity-purified antibody was not sufficiently sensitive to detect S136 phosphorylation in native D52 protein. Temporal limitations for gland fixation also precluded measurements of responses prior to 1 min of stimulation (Fig. 5B).
Effects of various protein kinase inhibitors on S136 phosphorylation.
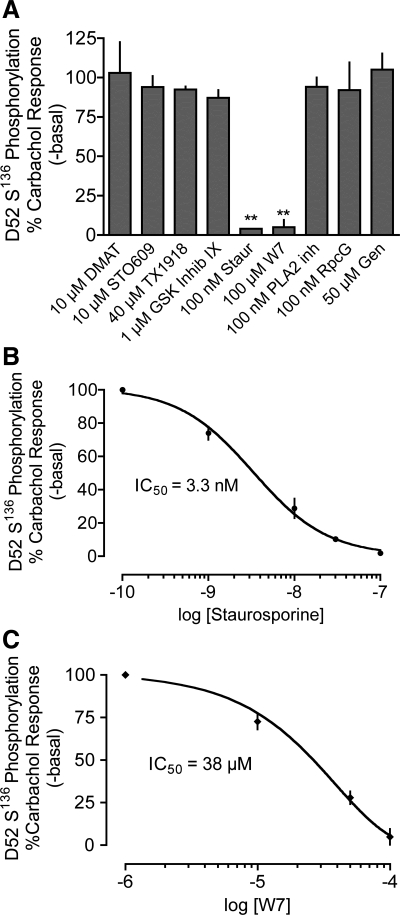
A wide range of protein kinase inhibitors was tested to determine whether CK2, GSK3, or an unidentified calcium-dependent, calmodulin-independent protein kinase might modulate calcium-dependent S136 phosphorylation. A portion of these data is depicted in Fig. 6A. The complete data set and inhibitor selectivities are presented in Supplemental Table S1. Of the inhibitors tested, only the calmodulin-dependent protein kinase inhibitor W-7 and the pan kinase inhibitor staurosporine suppressed carbachol-stimulated S136 phosphorylation with appropriate IC50 values (38 μM and 3 nM, respectively; Fig. 6, B and C). These findings led us to tentatively eliminate members of the recently identified α-helical kinase family because these kinases are highly resistant to staurosporine inhibition (40). We also tentatively eliminated the CAMK family members CAMK1 and CAMK4 because the ineffective CAMKK inhibitor STO-609 blocks CAMKK-dependent activation of CAMK1 and CAMK4 but does not inhibit CAMK2 activity (47, 49). Finally, we eliminated CK2 as a candidate for the following reasons: 1) Two different CK2 inhibitors, TBB and the more potent inhibitor DMAT (35), failed to inhibit carbachol-stimulated S136 phosphorylation (Fig. 6 and Supplemental Table S1). 2) siRNA knockdown of CK2 (>70%, not shown) did not significantly alter carbachol-stimulated S136 phosphorylation. 3) Recombinant CK2 catalyzed the incorporation of 32P from [γ32P]ATP into recombinant D52 protein in vitro but did not target S136 (Supplemental Fig. S2). In contrast, recombinant CAMK2 robustly phosphorylated S136 as assessed by Western blot analyses using the pS136 antibody (Supplemental Fig. S2).
Fig. 6.
Effects of selected protein kinase inhibitors on carbachol-stimulated D52 S136 phosphorylation in HEK293 cells expressing HA-tagged D52. S136 phosphorylation was analyzed as described in Fig. 5. A: cells were incubated with predicted maximal concentrations of the indicated inhibitors then stimulated with 10 μM carbachol for 1 min. Only staurosporine and W7 significantly inhibited S136 phosphorylation. **P < 0.001, N = 3–6 experiments. B: dose-dependent effects of staurosporine on carbachol-stimulated S136 phosphorylation. The apparent IC50 was 3.3 nM, N = 4. C: dose-dependent effects of W7 on carbachol-stimulated S136 phosphorylation. The apparent IC50 was 38 μM, N = 4.
Partial purification and characterization of D52 kinase in HEK293 cells and mouse gastric glands.
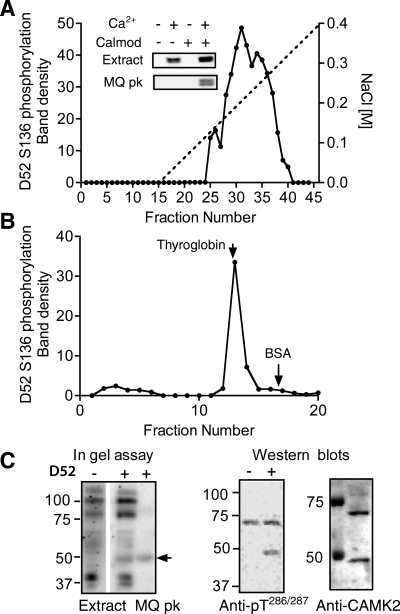
Because calcium-dependent D52 phosphorylation was sensitive to inhibition by W7 and recombinant CAMK2 appeared to phosphorylate S136 specifically (Supplemental Fig. S2), we explored the possibility that a KN62-resistant CAMK2 isoform was involved in mediating S136 phosphorylation. We reasoned that the most direct approach would be to use an anti-active CAMK2 antibody (directed against pT286/287) to determine whether CAMK2 is activated by carbachol and ionomycin within a time frame appropriate for mediating S136 phosphorylation; however, this approach was not productive because several proteins in crude cellular extracts from both HEK293 cells and mouse gastric glands cross-reacted nonspecifically with the antibody and no specific Ca2+-dependent changes were detected. To circumvent this problem, HEK 293 cell and mouse gastric gland extracts were fractionated on ion exchange and gel filtration columns as well as on calmodulin-Sepharose affinity columns. As shown in Fig. 7A, a broad Ca2+-calmodulin-dependent peak of D52 kinase was eluted from a Mono Q ion exchange column (between 0.18 and 0.3 M NaCl). These results were similar to those obtained with rabbit gastric gland extracts via a 32P-labeling approach (Fig. 2A). In both analyses, the phosphorylation of S136 in column fractions was dependent on the inclusion of calcium and calmodulin; however, in crude cellular extracts, S136 phosphorylation was calmodulin independent (Fig. 7A, inset), presumably because the endogenous calmodulin levels in these unfractionated extracts were sufficient to fully activate the enzyme upon addition of calcium. Calmodulin binding to D52 kinase was confirmed on the basis of calcium-dependent binding of D52 kinase to calmodulin-Sepharose and elution by chelation of calcium with EGTA (data not shown).
Fig. 7.
Characterization of D52 kinase in HEK293 cells. A: D52 kinase activity profile obtained after fractionation of HEK293 cell extract (100,000 g supernatant) on a Mono Q anion exchange column. Kinase activity in column fractions was analyzed by incubation of aliquots with recombinant His-tagged D52 protein in the presence of calcium/calmodulin/ATP followed by Western blotting with affinity-purified pS136 antibody. Inset: S136 phosphorylation assayed +/− calcium/calmodulin (Calmod) in the supernatant and in Mono Q peak fractions (nos. 30–36). B: D52 kinase activity profile of Mono Q peak after fractionation on a Superose 6 HR 10/30 size exclusion column. Arrows indicate elution positions of 2 external standards: thyroglobin (Mr 660,000) and BSA (Mr 68,000). C: migration patterns of the monomeric form of D52 kinase on SDS-PAGE gels. Left: “in gel” assay comparing phosphorylation patterns of the unfractionated extract and Mono Q peak in the absence (−) and presence (+) of His-tagged D52 protein in the gel. Arrow indicates the position of D52 phosphorylating activity that was detected when D52 included in the 10% SDS PAGE gel. Center: Western blot of Mono Q peak assayed +/− calcium/calmodulin (Calmod) and probed with anti-active CAMK2 antibody (anti-pT286/287). Samples were resolved on 8% SDS PAGE gels prior to Western blotting. Right: Western blot of the same Mono Q peak probed with anti-CAMK2 antibody (Stressgen). The identity of the ∼70-kDa protein detected on this blot and the anti-pT286/287 blot is unknown.
Gel filtration analyses of pooled D52 kinase peaks from Mono Q fractionations identified a single peak of D52 S136 phosphorylating activity with an apparent molecular mass (Mr) of ∼600,000 kDa (Fig. 7B). As only CAMK2 isoforms are known assemble into large multimeric complexes (24, 25), these findings provided strong support for the hypothesis that D52 kinase is a CAMK2 isoform.
Mr analyses of the monomeric form of D52 kinase.
In gel assays were used to determine the apparent Mr of the monomeric form of the enzyme. With crude cellular extracts, multiple phosphorylated bands were detected when D52 substrate was omitted from the gels (Fig. 7C, in gel assay, left), presumably resulting from autophosphorylation and/or phosphorylation of comigrating proteins. Inclusion of the D52 protein in the gels led to the appearance of a single additional band that migrated with an apparent Mr of ∼50 kDa (Fig. 7C, in gel assay, right, Extract). A similarly sized protein was present in D52 kinase activity peaks obtained from Mono Q (MQ) column fractionations (Fig. 7C, in gel assay, right, MQ peak). This ∼50 kDa D52 kinase specifically cross-reacted with anti-active CAMK2 in the presence of calcium/calmodulin and migrated at a position slightly below the 50-kDa Mr standard with an apparent Mr of 46 kDa on 8% SDS PAGE gels (Fig. 7C, Western blots). In contrast, another unidentified protein of ∼70 kDa cross-reacted indiscriminately with anti-active CAMK2 antibody in the presence and absence of calcium/calmodulin (Fig. 7C, Western blot, anti-pT286/287). Interestingly, the anti-CAMK2 antibody, which detects both active and inactive forms, also cross-reacted with a protein with a similar Mr (Fig. 7C, Western blot, anti-CAMK2).
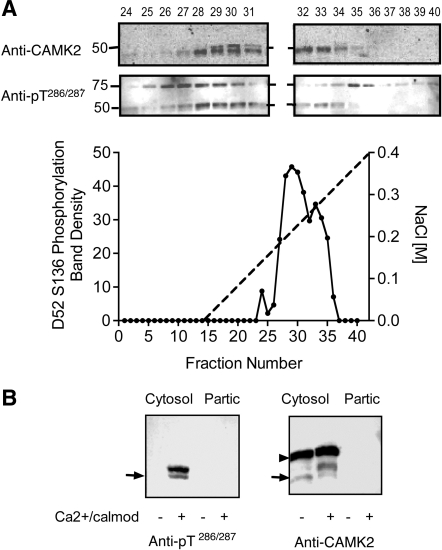
Fractionation of mouse gastric gland extracts on a Mono Q column yielded a similar activity profile (Fig. 8A). In this case, however, two proteins with apparent Mr values of 46 and 48 kDa were detected with both the anti-CAMK2 and the anti-active CAMK2 antibodies (Fig. 8A). A similar profile was observed with rabbit gastric gland extracts (not shown). Further analyses of mouse extracts indicated that the 48-kDa protein is a hyperphosphorylated form of the 46-kDa isoform (Fig. 8B). It is unclear why a phosphorylation-dependent Mr band shift occurs in rabbit and mouse gastric cells but not in HEK293 cells. One possibility is that minor differences in amino acid sequence affect the migration pattern. There is precedence for such a species-dependent phenomenon in that the highly conserved Lasp1 protein undergoes a band shift in response to cAMP elevation in rabbit gastric mucosal cells, but not in mouse (26).
Fig. 8.
Characterization of D52 kinase in mouse gastric glands. A: D52 kinase activity profile in mouse gastric gland extracts after fractionation on a Mono Q column. Cells were extracted and D52 kinase activity was assayed as described in Fig. 7. Inset shows patterns of cross-reactivity of column fractions with anti-CAMK2 and anti-pT286/287 following stimulation with calcium/calmodulin. The identity of the ∼75-kDa protein(s) that cross-reacted with the anti-CAMK2 antibody is unknown. B: D52 kinase activity in mouse gastric gland extract before column fractionation. Glands were sonicated in lysis buffer and centrifuged (100,000 g, Cytosol) as described in methods. Pellets were extracted with 0.1% Triton X-100 (Partic) then assayed along with the cytosolic extract. Left: Western blot probed with anti-active CAMK2 (anti-pT286/287). Right: Western blot probed with anti-CAMK2. Arrow indicates position of 50 kDa Mr standard. Although no CAMK2 kinase activity was detected in the particulate fraction in this experiment, minor activity (15–20% of the total) was detected in others (not shown). Note the prominent Mr band shift in CAMK2 signal following activation with calcium/calmodulin and absence of detectable pT286/287 signal when these activators were omitted. This band shift was not detected in HEK293 cell extracts (Fig. 7). The identity of the ∼60-kDa protein that cross-reacted with anti-CAMK2 antibody (arrowhead) but not the anti-active CAMK2 antibody is unknown; however, since this protein did not cross-react with anti-active CAMK2 antibody, it was presumed not to be a CAMK2 isoform.
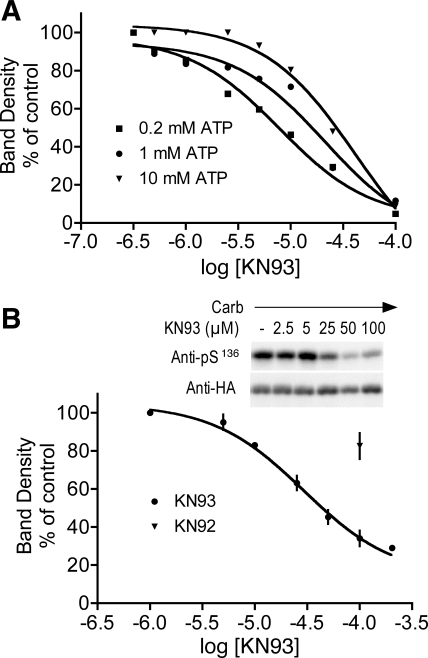
Effects of the CAMK2 inhibitor, KN93, on D52 kinase activity.
Because the column fractionation analyses strongly supported a role for CAMK2 in mediating D52 phosphorylation, more stringent approaches were used to define the effects of CAMK2 inhibitors with S136 phosphorylation as an end point. Effects of KN93 were first assessed in vitro by using D52 kinase activity peaks obtained from Mono Q column fractionations. In these experiments, KN93 inhibited calcium/calmodulin-dependent S136 phosphorylation and, as previously observed with KN62 (46), the IC50 for KN93 inhibition was dependent on the prevailing concentration of ATP (Fig. 9A). With 0.2 mM ATP, the apparent IC50 was 7.8 μM and the calculated Ki was 0.37 μM. With 1 mM ATP, the IC50 was 21.1 μM, and Ki was 1.0 μM; with 10 mM ATP, the IC50 increased to 41.8 μM and Ki to 1.99 μM. Similar results were obtained by using D52 kinase activity peaks from mouse gland extracts after fractionation on a Mono Q column (not shown). In intact cells, KN93 dose dependently inhibited S136 phosphorylation with an apparent IC50 of ∼30 μM (Fig. 9B). KN92, the less active analog of KN93, also inhibited D52 phosphorylation, but less potently (17.7 ± 2.2% inhibition at 100 μM, Fig. 9B). Taken together, these results provided further support for a role for CAMK2 in the regulation of calcium-dependent D52 S136 phosphorylation.
Fig. 9.
Comparison of effects of the CAMK2 inhibitor KN93 on S136 phosphorylation in intact cells and in vitro. A: in vitro analyses. D52 kinase was partially purified from HEK293 cells by fractionation on a Mono Q column. Peak activity fractions were identified, pooled as described in Fig. 7, and then assayed in the presence ATP (0.2, 1, and 10 mM) and varying concentration of KN93. The calmodulin concentration was kept constant at 200 nM. Changes in D52 S136 phosphorylation were quantified by Western blotting with affinity-purified pS136 antibody. Values are expressed as % of band densities obtained in the absence of KN93. For Ki calculations, the Kd for calmodulin binding was assumed to be 10 nM (18). In similar assays with fractionated mouse gastric gland extracts, the apparent IC50 with 0.2 mM ATP was 5.4 μM and the Ki was 0.26 μM (data not shown). B: cellular analyses. HEK293 cells expressing HA-tagged D52 were preincubated with varying concentrations of KN93 or with 100 μM KN92 or DMSO vehicle (where appropriate) for 1 h then stimulated with carbachol (10 μM, 1 min). Extracts were analyzed by Western blotting using affinity purified pS136 antibody. Corrections for variations in the levels of HA-D52 expression were performed as described in Fig. 5.
Analysis of CAMK2δ isoforms expressed in gastric glands.
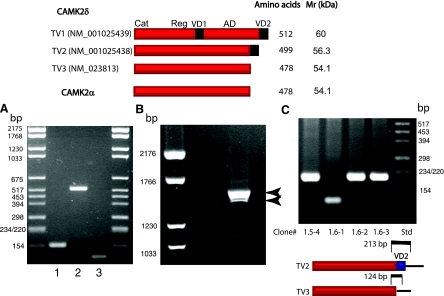
The CAMK2 family contains four distinct genes that encode for four major isoforms, α, β, δ, and γ, each of which has a number of alternatively spliced variants (25). The α isoforms appear to be expressed exclusively in brain, and RT-PCR analyses have not detected CAMK2α in stomach, intestine, or pancreas [Freeman TC, Dixon AK, Campbell EA, Tait EM, Richardson J, Rice KM, Maslen GL, Metcalfe GL, Streuli LH, and Bentley DR, Mouse Genome Informatics (MGI) Direct Data Submission 1998, accession ID. 1199209]. The β isoforms also have a fairly restricted tissue distribution (24, 48), which, on the basis of expressed sequence tag (EST) counts (from NCBI Unigene EST profile viewer), does not include stomach, pancreas, or colon. In contrast, δ and γ isoforms are widely expressed in tissues including stomach, intestine, and kidney (29). Because the predicted Mr values of CAMK2 γ isoforms are higher than the δ isoforms (48), we used a PCR-based approach to define δ isoform expression using total RNA isolated from mouse gastric glands.
In mouse, three transcript variants encode for CAMK2δ isoforms. Transcript variant (TV)1 (NM_001025439) encodes for the largest protein (512 amino acids). TV2 (NM_001025438) encodes for CAMK2δ2, a protein of intermediate size (499 amino acids) that is 100% identical to human CAMK2δ2 (alternatively named CAMK2δC) (25, 48). Mouse TV3 (NM_023813) encodes for the smallest protein (478 amino acids). This protein is 100% identical to a human CAMK2δ isoform that has been variously named CAMK2δ6 and CAMK2δG (25).
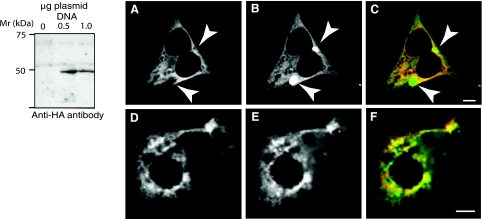
The strategy used to discriminate between the three TVs is outlined in Fig. 10. As shown, mRNAs encoding for TV2 and 3, but not TV1, were identified in mouse gastric mucosa. To confirm that TV3 encodes for a protein that comigrates with D52 kinase, pcDNA3 plasmids containing cDNA inserts encoding for HA-tagged CAMK2δ6 were transfected into HEK293 cells and cellular extracts were analyzed by Western blotting with anti-HA antibody. The expressed protein migrated as a ∼46-kDa protein on an 8% SDS PAGE gel (Fig. 11, left). Thus the migration pattern for CAMK2δ6 is similar to that for D52 kinase, providing further support to our hypothesis that the two proteins are one and the same.
Fig. 10.
Identification of CAMK2δ transcript variant (TV) 2 and TV3, but not TV1, in mouse gastric mucosa. A: an initial screen eliminated TV1. cDNA derived from mouse gastric mucosa was used as a template for RT-PCR-based analyses. Three sets of primers that bridged both variable domains were used: set 1, forward: 5′-CGC CAT CTT GAC AAC TAT GC; reverse 5′-CAG TGA CTT TGA TGA TCT CC; set 2, forward, same as set 1; reverse, 5′-TTA GTT GAT GGG TAC TGT GGG; set 3, forward, 5′-GCA GAA TGT TCA CTT TCA CCG TTC; reverse, 5′-GAT GTT TTG CCA CAA AGA GGT GCC (see diagram). These primers were predicted to generate the following products: set 1, 260 bp (TV1), 158 bp (TV2 and 3); set 2, 630 bp (TV1), 528 bp (TV2 and 3); set 3, 112 bp (TV2) with no product for TV1 and 3 (see diagram at top). As shown, PCR products for TV2, and possibly TV3, but not TV1 were obtained. B: the second screen identified TV2 and TV3. A single set of PCR primers that spanned the entire open reading frame of TV2 and 3 was used (forward: 5′-GCC ATG GCT TCG ACC ACC; reverse: 5′-GGA CGT CCC AGG ATC ACC C). Predicted sizes of PCR products were 1593 and 1505 bp for TV2 and TV3, respectively. As shown, 2 products within this size range were produced. C: a tertiary screen provided further confirmation that TV3 is expressed in mouse gastric mucosa. TV3 was of particular interest because it encodes for CAMK2δ6/G, the only known CAMK2 isoform that has the same domain structure and predicted Mr as CAMK2α. PCR products from B were gel purified, PCR cloned into the pGEM-T vector, and bacterially transformed. Positive clones were screened with a single set of primers directed against a conserved region in both transcripts that spanned VD2 (forward: 5′-GGG AAG TGG CAG AAT GTT C; reverse, 5′-GAA GAG GAG AGG ACG TCC C; see diagram on right). As shown in the figure, PCR products of predicted sizes for both TV2 and TV3 were obtained. The identity of these products was confirmed by DNA sequencing (not shown).
Fig. 11.
Western blot analysis of CAMK2δ6 and subcellular localization of CAMK2δ6 and D52 proteins in HEK293 and T84 cells. A: Western blotting demonstrated that the expressed protein product of TV3 (CAMK2δ6/G) has a migration pattern that is similar to D52 kinase and CAMK2α. cDNA encoding for HA-tagged CAMK2δ6/G was generated by PCR using the pGEM-T Easy-CAMK2δ-V3 construct (Fig. 10C) as a template then subcloned into pcDNA 3.1. PCR primers were as follows: forward with EcoRV cut site, 5′-CCC GAT ATC ATG GGC TAC CCA TAC GAT GTT CCA GAT TAC GCT GCT TCG ACC ACC ACC; reverse with XhoI cut site, 5′-CGC CTC GAG TTA GTT GAT GGG TAC TGT GGG. pcDNA 3.1 plasmids with and without the cDNA insert were transfected into HEK293 cells as described in methods. The expressed protein was identified by Western blotting using anti-HA antibody. B: CAMK2δ6 and D52 proteins display an overlapping subcellular distribution when coexpressed in HEK293 and T84 cells. Both cell lines were transfected with Effectene by using 0.5 μg plasmid DNA for each construct as described in methods. HA-tagged CAMK2δ6 was immunolocalized with monoclonal anti-HA antibody (1:1,000 dilution) in conjunction with donkey anti-mouse Cy-5-tagged secondary antibody (1:100 dilution) (A and D) and pseudocolored in red in the merged images (C and F). enhanced green fluorescent protein (GFP; EGFP)-tagged D52 was analyzed directly (B) or after staining with a polyclonal anti-GFP antibody and donkey anti-rabbit Alexa 555 secondary antibody (1:100 dilution) (E) and pseudocolored in green, rather than blue, for better visualization in merged images (C and F). The absence of signal crossover was confirmed in all analyses. Arrows in A–C indicate accumulation of EGFP-tagged D52 within subcellular compartments from which CAMK2δ6 is excluded. This accumulation may, at least partially, reflect the “trapping” of the overexpressed EGFP-tagged protein within the Golgi apparatus. Images were acquired with a Zeiss LSM 510 confocal microscope using a ×40, 1.3-NA oil immersion objective. Bars, 5 μm.
Finally, we reasoned that if D52 is, indeed, an in vivo substrate for CAMK2δ6, then one would expect that the subcellular locations of these two proteins to overlap, at least partially. To assess this possibility, HEK293 cells were cotransfected with plasmids containing cDNA inserts encoding for HA-tagged CAMK2δ6 and EGFP-tagged D52. As shown in Fig. 11, A–F, these analyses detected extensive overlap in the subcellular localizations of the two proteins.
DISCUSSION
In this study, we identify the calcium-responsive phosphorylation site in the tumor-promoting protein, D52, at S136 and provide compelling evidence that D52 is directly regulated by a previously uncharacterized CAMK2 isoform, CAMK2δ6, which bears a striking structural similarity to CAMK2α. The highly conserved nature of the amino acid residues surrounding S136 in mammalian as well as in avian species suggests that the phosphorylation of this residue plays a central role in regulating calcium-dependent functions of the D52 protein. On the basis of our temporal analyses, we believe exocytosis is a strong functional candidate. Thus the rapid, biphasic calcium-dependent phosphorylation of S136 closely mimics cholinergically induced calcium transients in gastric chief and pancreatic acinar cells (8, 33), which, in turn, is correlated with the exocytotic “burst” of enzyme release in the cells. It will be interesting to determine, in future experiments, whether D52 phosphorylation alters interactions between D52 and previously identified candidate endocytosis-associated binding proteins (43, 45, 50), thereby possibly enhancing the exocytic response.
Further support for the importance of S136 in agonist-dependent regulation is the finding, in a global phosphorylation site-specific study in HeLa cells, that the same serine residue in TPD54 isoform 2 (TPD52L2) (in this case, S146 within the conserved sequence: NSATFKS*FEDRV) is progressively dephosphorylated following exposure of cells to epidermal growth factor (34). Given that the TPD53 isoform (TPD52L1) contains the same sequence string as D52 (NSPTFKSFEEKV), it is likely that the corresponding S154 within this isoform is also regulated by changes in phosphorylation. It will also be interesting to determine whether the testes-specific isoform, D55 (TPD52L3), in which this region is less well conserved (KSATLRSFEGLN) (5), is regulated by agonist-dependent phosphorylation/dephosphorylation.
We applied multiple criteria to confirm that the cholinergically activated D52 kinase is a CAMK2 isoform. In brief, the pattern of D52 phosphorylation by gastric mucosal extracts fractionated on Mono Q columns was the same as that for the CAMK2-specific substrate, autocamtide-2. Moreover, a similar elution profile was obtained by using extracts from HEK293 cells. Significantly, D52 kinase activity eluted from gel filtration columns as part of a large multimeric complex and the monomeric form of D52 kinase cross-reacted with anti-CAMK2 antibodies as well as with antiactive (phosphoT286/287) CAMK2 antibody upon exposure to calcium/calmodulin. Of the known multifunctional calcium/calmodulin-dependent protein kinase family members, only CAMK2 isoforms exist within multimeric complexes and only CAMK2 isoforms undergo autophosphorylation on T286/287 within their multimeric domains (reviewed in Refs. 24 and 25). In cells, we found that the calmodulin antagonist W7 dose dependently inhibits carbachol-dependent S136 phosphorylation and confirmed that recombinant CAMK2 rapidly and specifically phosphorylates S136 whereas recombinant CK2 does not. These latter findings are in general agreement with previous peptide mapping experiments (27) in which the pattern of CAMK2-dependent 32P labeling, but not CK2-dependent labeling, was similar to that elicited by carbachol stimulation of T84 cells. The apparent conflict between findings in this study and the earlier study in which carbachol-stimulated D52 phosphorylation was not inhibited by W7 or TFP (27) might be explained by our use of a lower concentration of carbachol (10 vs. 100 μM carbachol).
An unexpected observation was that the monomeric form of D52 kinase isolated from rabbit and mouse gastric mucosal cells, as well as from HEK293 cells, migrated on 8% SDS PAGE gels with an apparent Mr of 46 kDa. Moreover, although this protein was clearly categorized as a CAMK2 isoform on the basis of criteria described above, in striking contrast to findings with excitable cells, the isoform was highly resistant to inhibition by the CAMK2 inhibitor, KN93. Why was this the case? One potential explanation is that the ∼10 times lower concentrations of KN93 that inhibit a range of responses in excitable cells are suppressing cellular functions other than CAMK2, such as voltage-dependent calcium entry (1, 2, 39, 41). Because voltage-sensitive calcium channels are not present in gastric mucosal and other nonexcitable epithelial cell types, KN93-dependent suppression of calcium entry presumably does not occur (although there may be other types of nonspecific effects). As previously reported for KN62 (46), we found that the IC50 for the inhibition of CAMK2 by KN93 in vitro is dependent on the concentration of ATP. Thus, assuming the intracellular concentration of ATP is ∼2 mM, there is good agreement between the in vivo and in vitro IC50 values that we obtained for KN93 inhibition of D52 S136 phosphorylation.
On the basis of our PCR and protein expression analyses, we have tentatively identified the ∼50 kDa CAMK2 isoform as CAMK2δ6/G. Importantly, the coexpression analyses indicate that this CAMK2 isoform can colocalize with D52 in cellular subcompartments as would be predicted if these two proteins directly interact as an enzyme-substrate complex. Our findings are the first to support a role for this previously uncharacterized CAMK2 isoform in mediating calcium-dependent signaling events in epithelial cells. The predicted Mr of known δ and γ isoforms range from 54.1 to 60.0 kDa and 56 to 62.2 kDa, respectively (48). In rabbit gastric mucosal cells, the respective Mr values of the δ and γ isoforms have been reported to be 54 and 56.5 kDa, based on cross-reactivity with antibodies raised against these isoforms (19). However, a ∼50 KDa calmodulin binding protein that comigrates with CAMK2α immunoreactivity was identified in an earlier study of microsomal membranes isolated from rabbit gastric cells (20), and multifunctional CAMKs of similar Mrs have been found in several GI tissues as well as in spleen, lung, and adipose tissue (16, 28, 32). Interestingly, amino acid sequence alignments indicate that the domain structure of CAMK2δ6 has the same configuration as that for CAMK2α. These include a catalytic domain, a conserved linker region, and association domains. Mouse CAMK2δ6 (Uniprot accession no. Q6PHZ2-2) also shares 94% homology and 87% identity with mouse CAMK2α (accession no. P11798). Unlike all other CAMK2δ isoforms, there are no variable domains in CAMK2δ6 (25). The predicted Mr for both the δ6 and α isoforms is 54 kDa (48); however, CAMK2α migrates on SDS PAGE gels with an apparent Mr of ∼50 kDa and, as found here, comigrates with D52 kinase. Because CAMK2δ6 lacks both the internal and COOH-terminal variable domains that are variously present in other CAMK2δ isoforms, it will not cross-react with antibodies raised against these regions. This lack of cross-reactivity presumably explains the discrepancy between earlier studies of gastric cells in which a 50-kDa CAMK2 isoform was not detected (19) and those in which the isoform was detected (20). In the latter case, the no. 301 CAMK2 antibody was used. The no. 301 antibody was raised against a peptide encompassing residues 281–302 in CAMK2α (28), and these residues fall within the regulatory domain, which is present in all CAMK2 isoforms. Identical residues are present in mouse CAMK2δ, but this homology is not as stringently conserved in CAMK2γ isoforms (C. S. Chew, unpublished observations).
In summary, we present the first evidence to support a role for CAMK2δ6 in mediating calcium-dependent signaling events that occur within the gastric mucosa, including the phosphorylation of D52 on S136. Further analyses will be required to confirm unequivocally that CAMK2δ6 mediates the in vivo functions of D52 and to define the specific physiological functions and cellular/subcellular localization of this heretofore uncharacterized CAMK2 isoform.
GRANTS
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK-31900.
Supplementary Material
Acknowledgments
We acknowledge Dr. John Parente for contributions in the initial stages of this study. We thank Drs. Nevin Lambert and Deborah Lewis (MCG) for providing HEK 293 cells and anti-GFP antibody. We also thank Dr. Katz Miyake and Darren Baker for outstanding technical assistance in the Imaging Core at MCG.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Anderson ME, Braun AP, Wu Y, Lu T, Wu Y, Schulman H, Sung RJ. KN-93, an inhibitor of multifunctional Ca++/calmodulin-dependent protein kinase, decreases early afterdepolarizations in rabbit heart. J Pharmacol Exp Ther 287: 996–1006, 1998. [PubMed] [Google Scholar]
- 2.Bhatt HS, Conner BP, Prasanna G, Yorio T, Easom RA. Dependence of insulin secretion from permeabilized pancreatic beta-cells on the activation of Ca2+/calmodulin-dependent protein kinase II: a re-evaluation of inhibitor studies. Biochem Pharmacol 60: 1655–1663, 2000. [DOI] [PubMed] [Google Scholar]
- 3.Boutros R, Fanayan S, Shehata M, Byrne JA. The tumor protein D52 family: many pieces, many puzzles. Biochem Biophys Res Commun 325: 1115–1121, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Byrne JA, Tomasetto C, Garnier JM, Rouyer N, Mattei MG, Belloq JP, Rio MC, Basset P. A screening method to identify genes commonly overexpressed in carcinomas and the identification of a novel complementary DNA sequences. Cancer Res 55: 2896–2903, 1995. [PubMed] [Google Scholar]
- 5.Cao Q, Chen J, Zhu L, Liu Y, Zhou Z, Sha J, Wang S, Li J. A testis-specific and testis developmentally regulated tumor protein D52 (TPD52)-like protein TPD52L3/hD55 interacts with TPD52 family proteins. Biochem Biophys Res Commun 344: 798–806, 2006. [DOI] [PubMed] [Google Scholar]
- 6.Chen SL, Maroulakou IG, Green JE, Romano-Spica V, Modi W, Lautenberger J, Bhat NK. Isolation and characterization of a novel gene expressed in multiple cancers. Oncogene 12: 741–751, 1996. [PubMed] [Google Scholar]
- 7.Chew CS cAMP technologies, functional correlates in gastric parietal cells. Methods Enzymol 191: 640–661, 1990. [DOI] [PubMed] [Google Scholar]
- 8.Chew CS, Brown MR. Release of intracellular Ca2+ and elevation of inositol trisphosphate by secretagogues in parietal and chief cells isolated from rabbit gastric mucosa. Biochim Biophys Acta 888: 116–125, 1986. [DOI] [PubMed] [Google Scholar]
- 9.Chew CS, Chen X, Bollag RJ, Isales CM, Ding KH, Zhang H. Targeted disruption of the Lasp1 gene is linked to increases in histamine-stimulated gastric HCl secretion. Am J Physiol Gastrointest Liver Physiol 295: G37–G44, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chew CS, Chen X, Parente JA Jr, Tarrer S, Okamoto C, Qin HY. Lasp-1 binds to nonmuscle F-actin in vitro and is localized within multiple sites of dynamic actin assembly in vivo. J Cell Sci 115: 4787–4799, 2002. [DOI] [PubMed] [Google Scholar]
- 11.Chew CS, Ljungstrom M, Smolka A, Brown MR. Primary culture of secretagogue-responsive parietal cells from rabbit gastric mucosa. Am J Physiol Gastrointest Liver Physiol 256: G254–G263, 1989. [DOI] [PubMed] [Google Scholar]
- 12.Chew CS, Okamoto CT, Chen X, Qin HY. IQGAPs are differentially expressed and regulated in polarized gastric epithelial cells. Am J Physiol Gastrointest Liver Physiol 288: G376–G387, 2005. [DOI] [PubMed] [Google Scholar]
- 13.Chew CS, Okamoto CT, Chen X, Thomas R. Drebrin E2 is differentially expressed and phosphorylated in parietal cells in the gastric mucosa. Am J Physiol Gastrointest Liver Physiol 289: G320–G331, 2005. [DOI] [PubMed] [Google Scholar]
- 14.Chew CS, Parente JA Jr, Chen X, Chaponnier C, Cameron RS. The LIM and SH3 domain-containing protein, lasp-1, may link the cAMP signaling pathway with dynamic membrane restructuring activities in ion transporting epithelia. J Cell Sci 113: 2035–2045, 2000. [DOI] [PubMed] [Google Scholar]
- 15.Chew CS, Parente JA Jr, Zhou C, Baranco E, Chen X. Lasp-1 is a regulated phosphoprotein within the cAMP signaling pathway in the gastric parietal cell. Am J Physiol Cell Physiol 275: C56–C67, 1998. [DOI] [PubMed] [Google Scholar]
- 16.Cohen ME, Reinlib L, Watson AJ, Gorelick F, Rys-Sikora K, Tse M, Rood RP, Czernik AJ, Sharp GW, Donowitz M. Rabbit ileal villus cell brush border Na+/H+ exchange is regulated by Ca2+/calmodulin-dependent protein kinase II, a brush border membrane protein. Proc Natl Acad Sci USA 87: 8990–8994, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohn JA, Dougherty NC, King WF Jr. Histamine stimulates calcium-mediated protein phosphorylation in a colonic epithelial cell line. Biochem Biophys Res Commun 165: 810–816, 1989. [DOI] [PubMed] [Google Scholar]
- 18.Edman CF, Schulman H. Identification and characterization of delta B-CaM kinase and delta C-CaM kinase from rat heart, two new multifunctional Ca2+/calmodulin-dependent protein kinase isoforms. Biochim Biophys Acta 1221: 89–101, 1994. [DOI] [PubMed] [Google Scholar]
- 19.Fahrmann M, Jacob P, Seidler U, Osterhoff M, Mohlig M, Pfeiffer A. Ca2+/calmodulin-dependent protein kinase II isoenzymes γ and δ are both present in H+/K+-ATPase-containing rabbit gastric tubulovesicles. Eur J Biochem 266: 1036–1042, 1999. [DOI] [PubMed] [Google Scholar]
- 20.Funasaka M, Fox LM, Tang LH, Modlin IM, Goldenring JR. The major calmodulin-binding protein in rabbit parietal cells is Ca2+/calmodulin-dependent protein kinase II. Biochem Int 27: 1101–1109, 1992. [PubMed] [Google Scholar]
- 21.Graham TR Membrane targeting: getting Arl to the Golgi. Curr Biol 14: R483–R485, 2004. [DOI] [PubMed] [Google Scholar]
- 22.Groblewski GE, Wishart MJ, Yoshida M, Williams JA. Purification and identification of a 28-kDa calcium-regulated heat-stable protein. A novel secretagogue-regulated phosphoprotein in exocrine pancreas. J Biol Chem 271: 31502–31507, 1996. [DOI] [PubMed] [Google Scholar]
- 23.Groblewski GE, Yoshida M, Yao H, Williams JA, Ernst SA. Immunolocalization of CRHSP28 in exocrine digestive glands and gastrointestinal tissues of the rat. Am J Physiol Gastrointest Liver Physiol 276: G219–G226, 1999. [DOI] [PubMed] [Google Scholar]
- 24.Hook SS, Means AR. Ca2+/CaM-dependent kinases: from activation to function. Annu Rev Pharmacol Toxicol 41: 471–505, 2001. [DOI] [PubMed] [Google Scholar]
- 25.Hudmon A, Schulman H. Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem J 364: 593–611, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain RN, Brunkan CS, Chew CS, Samuelson LC. Gene expression profiling of gastrin target genes in parietal cells. Physiol Genomics 24: 124–132, 2006. [DOI] [PubMed] [Google Scholar]
- 27.Kaspar KM, Thomas DDH, Taft WB, Takeshita E, Weng N, Groblewski GE. CaM kinase II regulation of CRHSP-28 phosphorylation in cultured mucosal T84 cells. Am J Physiol Gastrointest Liver Physiol 285: G1300–G1309, 2003. [DOI] [PubMed] [Google Scholar]
- 28.Matovcik LM, Haimowitz B, Goldenring JR, Czernik AJ, Gorelick FS. Distribution of calcium/calmodulin-dependent protein kinase II in rat ileal enterocytes. Am J Physiol Cell Physiol 264: C1029–C1036, 1993. [DOI] [PubMed] [Google Scholar]
- 29.Mayer P, Mohlig M, Seidler U, Rochlitz H, Fahrmann M, Schatz H, Hidaka H, Pfeiffer A. Characterization of gamma- and delta-subunits of Ca2+/calmodulin-dependent protein kinase II in rat gastric mucosal cell populations. Biochem J 297: 157–162, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meinnel T, Peynot P, Giglione C. Processed N-termini of mature proteins in higher eukaryotes and their major contribution to dynamic proteomics. Biochimie 87: 701–712, 2005. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura K, Zhou CJ, Parente J, Chew CS. Parietal cell MAP kinases: multiple activation pathways. Am J Physiol Gastrointest Liver Physiol 271: G640–G649, 1996. [DOI] [PubMed] [Google Scholar]
- 32.Nishimura H, Fukunaga K, Okamura H, Miyamoto E. Purification of the multifunctional calmodulin-dependent protein kinases from lung and liver, and the comparison to the brain enzyme. Jpn J Pharmacol 46: 173–182, 1988. [DOI] [PubMed] [Google Scholar]
- 33.Ochs DL, Korenbrot JI, Williams JA. Relation between free cytosolic calcium and amylase release by pancreatic acini. Am J Physiol Gastrointest Liver Physiol 249: G389–G398, 1985. [DOI] [PubMed] [Google Scholar]
- 34.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648, 2006. [DOI] [PubMed] [Google Scholar]
- 35.Pagano MA, Meggio F, Ruzzene M, Andrzejewska M, Kazimierczuk Z, Pinna LA. 2-Dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole: a novel powerful and selective inhibitor of protein kinase CK2. Biochem Biophys Res Commun 321: 1040–1044, 2004. [DOI] [PubMed] [Google Scholar]
- 36.Parente JA Jr, Chen X, Zhou C, Petropoulos AC, Chew CS. Isolation, cloning, and characterization of a new mammalian coronin family member, coroninse, which is regulated within the protein kinase C signaling pathway. J Biol Chem 274: 3017–3025, 1999. [DOI] [PubMed] [Google Scholar]
- 37.Parente JA Jr, Goldenring JR, Petropoulos AC, Hellman U, Chew CS. Purification, cloning, and expression of a novel, endogenous, calcium-sensitive, 28-kDa phosphoprotein. J Biol Chem 271: 20096–20101, 1996. [DOI] [PubMed] [Google Scholar]
- 38.Polevoda B, Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol 325: 595–622, 2003. [DOI] [PubMed] [Google Scholar]
- 39.Rezazadeh S, Claydon TW, Fedida D. KN-93 (2-[N-(2-hydroxyethyl)]-N-(4-methoxybenzenesulfonyl)]amino-N-(4-chlorocinnamyl)-N-methylbenzylamine), a calcium/calmodulin-dependent protein kinase II inhibitor, is a direct extracellular blocker of voltage-gated potassium channels. J Pharmacol Exp Ther 317: 292–299, 2006. [DOI] [PubMed] [Google Scholar]
- 40.Ryazanova LV, Dorovkov MV, Ansari A, Ryazanov AG. Characterization of the protein kinase activity of TRPM7/ChaK1, a protein kinase fused to the transient receptor potential ion channel. J Biol Chem 279: 3708–3716, 2004. [DOI] [PubMed] [Google Scholar]
- 41.Sihra TS, Pearson HA. Ca/calmodulin-dependent kinase II inhibitor KN62 attenuates glutamate release by inhibiting voltage-dependent Ca2+-channels. Neuropharmacology 34: 731–741, 1995. [DOI] [PubMed] [Google Scholar]
- 42.Thomas DD, Taft WB, Kaspar KM, Groblewski GE. CRHSP-28 regulates Ca2+-stimulated secretion in permeabilized acinar cells. J Biol Chem 276: 28866–28872, 2001. [DOI] [PubMed] [Google Scholar]
- 43.Thomas DDH, Kaspar KM, Taft WB, Weng N, Rodenkirch LA, Groblewski GE. Identification of annexin VI as a Ca2+-sensitive CRHSP-28-binding protein in pancreatic acinar cells. J Biol Chem 277: 35496–35502, 2002. [DOI] [PubMed] [Google Scholar]
- 44.Thomas DDH, Weng N, Groblewski GE. Secretagogue-induced translocation of CRHSP-28 within an early apical endosomal compartment in acinar cells. Am J Physiol Gastrointest Liver Physiol 287: G253–G263, 2004. [DOI] [PubMed] [Google Scholar]
- 45.Tiacci E, Orvietani PL, Bigerna B, Pucciarini A, Corthals GL, Pettirossi V, Martelli MP, Liso A, Benedetti R, Pacini R, Bolli N, Pileri S, Pulford K, Gambacorta M, Carbone A, Pasquarello C, Scherl A, Robertson H, Sciurpi MT, Alunni-Bistocchi G, Binaglia L, Byrne JA, Falini B. Tumor protein D52 (TPD52): a novel B-cell/plasma-cell molecule with unique expression pattern and Ca2+-dependent association with annexin VI. Blood 105: 2812–2820, 2005. [DOI] [PubMed] [Google Scholar]
- 46.Tokumitsu H, Chijiwa T, Hagiwara M, Mizutani A, Terasawa M, Hidaka H. KN-62, 1-[N,O-bis(5-isoquinolinesulfonyl)-N-methyl-l-tyrosyl]-4-phenylpiperazine, a specific inhibitor of Ca2+/calmodulin-dependent protein kinase II. J Biol Chem 265: 4315–4320, 1990. [PubMed] [Google Scholar]
- 47.Tokumitsu H, Inuzuka H, Ishikawa Y, Ikeda M, Saji I, Kobayashi R. STO-609, a specific inhibitor of the Ca2+/calmodulin-dependent protein kinase kinase. J Biol Chem 277: 15813–15818, 2002. [DOI] [PubMed] [Google Scholar]
- 48.Tombes RM, Faison MO, Turbeville JM. Organization and evolution of multifunctional Ca2+/CaM-dependent protein kinase genes. Gene 322: 17–31, 2003. [DOI] [PubMed] [Google Scholar]
- 49.Wayman GA, Kaech S, Grant WF, Davare M, Impey S, Tokumitsu H, Nozaki N, Banker G, Soderling TR. Regulation of axonal extension and growth cone motility by calmodulin-dependent protein kinase I. J Neurosci 24: 3786–3794, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson SHD, Bailey AM, Nourse CR, Mattei MG, Byrne JA. Identification of MAL2, a novel member of the MAL proteolipid family, through interactions with TPD52-like proteins in the yeast two-hybrid system. Genomics 76: 81–88, 2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.