Abstract
B cell chronic lymphocytic leukemia (B-CLL) is the most common human leukemia. Deregulation of the T cell leukemia/lymphoma 1 (TCL1) oncogene in mouse B cells causes a CD5-positive leukemia similar to aggressive human B-CLLs. To examine the mechanisms by which Tcl1 protein exerts oncogenic activity in B cells, we investigated the effect of Tcl1 expression on NF-κB and activator protein 1 (AP-1) activity. We found that Tcl1 physically interacts with c-Jun, JunB, and c-Fos and inhibits AP-1 transcriptional activity. Additionally, Tcl1 activates NF-κB by physically interacting with p300/CREB binding protein. We then sequenced the TCL1 gene in 600 B-CLL samples and found 2 heterozygous mutations: T38I and R52H. Importantly, both mutants showed gain of function as AP-1 inhibitors. The results indicate that Tcl1 overexpression causes B-CLL by directly enhancing NF-κB activity and inhibiting AP-1.
The lymphocytes of B cell chronic lymphocytic leukemia (B-CLL) are mostly resting cells with mature appearance and the B220+CD5+ phenotype (1, 2). The T cell leukemia/lymphoma 1 (TCL1) oncogene was discovered as a target of chromosomal translocations and inversions at 14q31.2 in T cell prolymphocytic leukemias (3). We have shown that transgenic mice overexpressing TCL1 in B cells develop the aggressive form of B-CLL (4) and that aggressive human B-CLLs overexpress Tcl1 (5). These results indicate that deregulation of TCL1 is critically important in the pathogenesis of the aggressive form of B-CLL. Previously, we demonstrated that Tcl1 is a coactivator of the Akt oncoprotein, a critical antiapoptotic molecule in T cells (6). More recently, it has been reported that transgenic mice expressing constitutively active myristylated Akt in T cells develop T cell leukemias (7). These results suggest that Akt may be responsible for Tcl1-mediated lymphomagenesis in T cells. Akt could be robustly activated in mouse B cells by homozygous deletion of Pten (8). Surprisingly, these mice did not develop B cell malignancies (8), suggesting that Tcl1 deregulation in B cells causes B-CLL by mechanisms other than Akt activation. Recent studies of transgenic mouse models demonstrated the importance of the NF-κB pathway in B-CLL (reviewed in ref. 9). For example, transgenic expression of a proliferation-inducing TNF ligand (APRIL), a member of the TNF superfamily involved in NF-κB activation, resulted in significant expansions of B220+CD5+ cells (10). Because studies of animal models suggested a role for the NF-κB pathway in the pathogenesis of B-CLL (9), we examined the possibility that Tcl1 might be involved in NF-κB activation.
Results
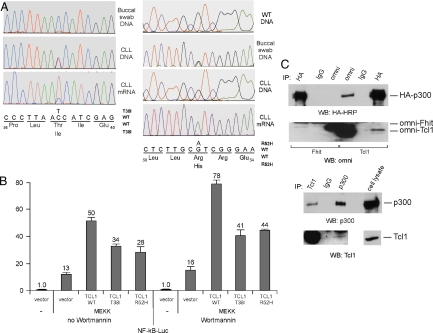
As tools to address this question, B-CLL-specific gain-of-function Tcl1 mutants would be useful. Thus, we have sequenced the TCL1 gene in 600 B-CLL samples. Sequencing analysis of all coding TCL1 exons resulted in the identification of 2 heterozygous mutations resulting in amino acid substitutions, T38I and R52H (Fig. 1A). The normal buccal swab DNA of the first patient did not show the T38I mutation (Fig. 1A). The R52H mutation was also present in the matched normal buccal swab DNA (Fig. 1A Right), suggesting a constitutional variation. Interestingly, RT-PCR results showed that the T38I mutant TCL1 mRNA was the major expressed allele in the B-CLL of origin, accounting for ≈80% of the TCL1 mRNA, and the R52H allele was the only allele expressed (Fig. 1A).
Fig. 1.
Tcl1 activates NF-κB-dependent transcription. (A) Chromatograms of sequences surrounding T38I, E40D, R52H mutations obtained from sequencing of buccal swab constitutional DNA, B-CLL DNA, and results of RT-PCR (for T38I mutant) using RNA from B-CLL cells. (B) Tcl1 activates NF-κB. NIH 3T3 cells were cotransfected with 50 ng of pNF-kB-Luc reporter and 50 ng of pRL-TK Renilla reporter constructs. In addition, 1.5 μg of CMV5-empty vector, or a combination of 0.75 μg of CMV5-empty vector and 0.75 μg of CMV5-Tcl1 WT, or CMV5-Tcl1 T38I constructs were used. Five nanograms of pFC-MEKK was added where indicated. Cells were treated with 200 nmol/L of Wortmannin overnight, where indicated. The normalized promoter activity of pNF-kB-Luc in NIH 3T3 cells transfected with CMV5-empty vector was set as 1. (C) Tcl1 interacts with p300. (Upper) Some 293 cells were cotransfected with p300-HA and Omni-Fhit or p300-HA and Omni-Tcl1 constructs. After lysis, immunoprecipitations were carried out with anti-HA, IgG, or anti-omni antibodies. Western blot analysis was carried out as indicated. (Lower) Daudi cells were lysed and immunoprecipitations were carried out with anti-Tcl1 antibody, IgG, or anti-p300 antibody. Unlabeled higher band in the Tcl1 panel represents IgG. Western blot analysis was carried out as indicated.
To determine whether Tcl1 expression affects the transactivating activity of NF-κB we used a commercial system based on the ability of mitogen-activated protein kinase kinase 1 (MEKK1) to activate an NF-κB reporter construct, pNF-kB-Luc expressing luciferase under the control of an NF-κB-responsive element. NIH 3T3 cells were transfected with the constructs indicated in Fig. 1B. Fig. 1B shows that Tcl1 activated NF-κB activity ≈4-fold (50 versus 13), whereas the 2 mutants activated activity 2- to 3-fold. Because we previously reported that Tcl1 is a coactivator of Akt (6), it could be argued that this NF-κB activation is caused by Akt activation by Tcl1. To eliminate this possibility we performed the same experiment in the presence of wortmannin, a PI3-kinase inhibitor (wortmannin completely inhibits Akt activity). Fig. 1B shows that wortmannin did not affect the ability of Tcl1 to activate NF-κB; in the presence of wortmannin Tcl1 expression activated NF-κB >4-fold (78 versus 16), whereas the expression of Tcl1 mutants resulted in 2.5- to 3-fold activation. In addition, WT Tcl1and T38I mutant did not show any difference in coimmunoprecipitation experiments with Akt (data not shown). These data suggest that Tcl1 activates NF-κB by a mechanism independent of Akt. To elucidate molecular mechanisms of this activation we carried out coimmunoprecipitations between Tcl1 and NF-κB1, NF-κB2, RelA, RelB, and c-Rel by using cotransfections in 293 cells. We did not find evidence of physical interactions between Tcl1 and members of the NF-κB family (data not shown).
The transcriptional activator CREB binding protein/p300 is a ubiquitous nuclear transcription factor involved in transactivation mediated by several signaling pathways, including the NF-κB pathway (11, 12). Because p300 is a coactivator of NF-κB (12, 13) we investigated whether Tcl1 interacts with p300. First, we carried out coimmunoprecipitation experiments, cotransfecting tagged Tcl1 and p300 constructs into 293 cells. Fig. 1C Upper shows that p300 was coimmunoprecipitated with Tcl1, whereas Tcl1 was detected in p300 immune complexes. No coimmunoprecipitation was detected between p300 and Fhit, used as a negative control (Fig. 1C Upper). To prove that the interaction we detected is not the result of overexpression of the 2 proteins, we carried out coimmunoprecipitation experiments in Daudi Burkitt lymphoma cells showing moderate levels of Tcl1 expression (3). Fig. 1C Lower shows that p300 was detected in Tcl1 immune complexes, whereas Tcl1 was coimmunoprecipitated with p300. Thus, we concluded that Tcl1 induces NF-κB-dependent transcription by interacting with p300, perhaps changing its conformation and enhancing its ability to function as an NF-κB coactivator.
Our results indicate that Tcl1 mutants activate NF-κB-dependent transcription to a lesser extent than WT Tcl1 (≈3-fold versus ≈4-fold). Because activation of NF-κB seems to be important in the pathogenesis of B-CLL, the data suggest that the Tcl1 mutants do not exhibit gain of function in the activation of NF-κB. In addition, the Tcl1 T38I mutant protein was similar to WT Tcl1 in coimmunoprecipitation experiments with p300 (data not shown).
Because Tcl1 physically interacts with p300, and p300 plays an important role in transcription activated by several other signal transduction pathways (11), we investigated whether in any of these pathways the Tcl1 mutants exhibit gain of function. One of the most well-studied pathways involving p300 is activator protein 1 (AP-1)-dependent transcription (11). The best-known AP-1 complex contains c-Jun and c-Fos, although other Jun proteins (JunB and JunD) and Fos proteins (Fra-1, Fra-2 and FosB) have been reported (14). Numerous reports indicate that AP-1 induces apoptosis by transactivating proapoptotic genes (15–17). For example, induction of c-Jun and c-Fos was reported in lymphoid cells after growth factor deprivation and this induction was associated with apoptosis (18). In another example, mouse knockout studies showed that JunB deficiency caused development of B cell leukemias (19). Thus, we investigated whether Tcl1 can inhibit AP1-dependent transcription.
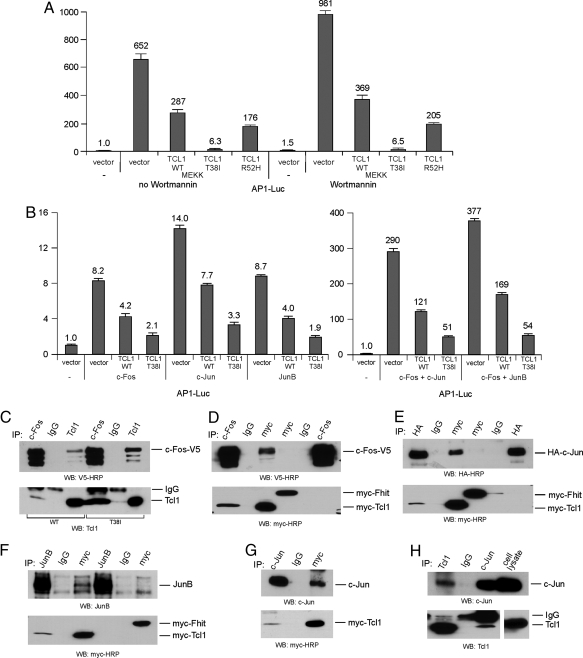
To assess the activity of AP-1 we used a commercial system based on the ability of MEKK1 to activate an AP-1 reporter construct, pAP-1-Luc, expressing luciferase under the control of an AP-1-responsive element. The 293 cells were transfected with the constructs indicated in Fig. 2. First, we investigated whether Tcl1 WT and mutants inhibit the activity of endogenous AP-1 in 293 cells. The 293 cells were transfected with MEKK1 to activate AP-1. Fig. 2A shows that AP-1 activity was induced 652-fold by MEKK1. Tcl1 expression inhibited AP-1 dependent transactivation ≈2.5-fold, whereas Tcl1 T38I caused a dramatic ≈100-fold inhibition (652 versus 6.3). The R52H mutant also showed a more potent effect compared with WT Tcl1 (176 versus 287, compared with 652). Similar results were obtained with cells treated with wortmannin (Fig. 2A): Tcl1 expression inhibited AP-1-dependent transactivation ≈2.5-fold, whereas the T38I mutant caused ≈150-fold inhibition (981 versus 6.5). These results indicate that inhibition of AP-1 by Tcl1 is Akt independent. To determine whether Tcl1 inhibits individual components of the AP-1 complex, we carried out similar experiments using WT Tcl1 and the T38I mutant. We activated AP-1 by overexpression of single AP-1 components rather than by using MEKK1. Fig. 2B Left shows that Tcl1 inhibits separately c-Fos, c-Jun, and Jun-B, whereas Tcl1 T38I mutant inhibited c-Fos, c-Jun, and Jun-B ≈2-fold more effectively. Similar results were obtained with c-Jun/c-Fos and JunB/c-Fos heterodimers (Fig. 2B Right). In all of these cases Tcl1 T38I mutant inhibited more potently than WT Tcl1. These results (Fig. 2 A and B) strongly indicate that Tcl1 mutants show gain-of-function effect in AP-1 inhibition.
Fig. 2.
Tcl1 inhibits AP-1 activity. (A) Some 293 cells were cotransfected with 500 ng of pAP-1-Luc reporter and 50 ng of pRL-TK Renilla reporter constructs. In addition, 1.5 μg of CMV5-empty vector, CMV5-Tcl1 WT, or mutant constructs and 2.5 ng of pFC-MEKK (where indicated) were used. Cells were treated with 200 nmol/L of Wortmannin overnight, where indicated. The normalized promoter activity of pAP-1-Luc in HEK293 cells transfected with CMV5-empty vector was set as 1. (B) Same as in A, except instead of pFC-MEKK construct, 5 ng of c-Fos-V5, c-Jun, JunB, or combinations of 5 ng of c-Fos-V5 and 5 ng of c-Jun or JunB were added as indicated. (C) The 293 cells were cotransfected with c-Fos-V5 and CMV5-Tcl1 WT or c-Fos-V5 and CMV5-Tcl1 T38I constructs. After lysis, immunoprecipitations were carried out with anti-c-Fos, IgG, or anti-Tcl1 antibodies. Western blot analysis was carried out as indicated. (D–F) The 293 cells were cotransfected with myc-Tcl1 T38I or myc-Fhit with c-Fos-V5 (D), c-Jun-HA (E), or JunB (F) as indicated. After lysis, immunoprecipitations were carried out with anti-myc, IgG, and anti-c-Fos (D), anti-HA (E), and anti-JunB (F) antibodies as indicated. Western blot analysis was carried out with the indicated antibodies. (G) The 293 cells were transfected with myc-Tcl1 and treated with 50 ng/mL PMA and 1 μg/mL ionomycin to increase endogenous c-Jun expression, 2 h before lysis. Immunoprecipitations were carried out with anti-c-Jun, IgG, or anti-myc antibodies. (H) Daudi cells were treated with 50 ng/mL PMA and 1 μg/mL ionomycin 2 h before lysis. Immunoprecipitations were carried out with anti-Tcl1, IgG, or anti-c-Jun antibodies.
To elucidate the mechanism of this inhibition we carried out a series of coimmunoprecipitation experiments (Fig. 2 C–H). Fig. 2 C–F shows results of these experiments using transiently expressed proteins. T38I mutant protein showed much robust coimmunoprecitation with c-Fos than WT Tcl1 (Fig. 2C), suggesting a relation with its more potent inhibition of AP-1 compared with WT Tcl1. The specificity of this interaction is shown in Fig. 2D; Tcl1 was coimmunoprecipitated with c-Fos in both directions, whereas no positive coimmunoprecipitates were detected between Fhit (used as a negative control) and c-Fos (Fig. 2D). Similarly, Tcl1 but not Fhit was coimmunoprecipitated with c-Jun (Fig. 2E), and Tcl1 but not Fhit was coimmunoprecipitated with JunB (Fig. 2F). Fig. 2G shows that endogenous c-Jun coimmunoprecipitated with transfected Tcl1 in 293 cells, whereas Tcl1 was detected in immune complexes of endogenous c-Jun. Physical interaction of endogenous Tcl1 and c-Jun in Daudi cells is shown in Fig. 2H: Tcl1 was present in immune complexes of endogenous c-Jun, and c-Jun was coimmunoprecipitated with Tcl1. In these experiments (Fig. 2 G and H), because c-Jun is expressed at very low levels, cells were pretreated with phorbol 12-myristate 13-acetate (PMA) and ionomycin (20). Such treatment significantly induced c-Jun expression in 293 and Daudi cells.
Based on the results described in Fig. 2, we conclude that Tcl1 physically interacts with AP-1 components and functions as an AP-1 inhibitor. The fact that both Tcl1 mutants identified in B-CLL patients show gain-of-function properties in this pathway suggests the ability of Tcl1 to inhibit AP-1-dependent transcription is critical in the pathogenesis of B-CLL.
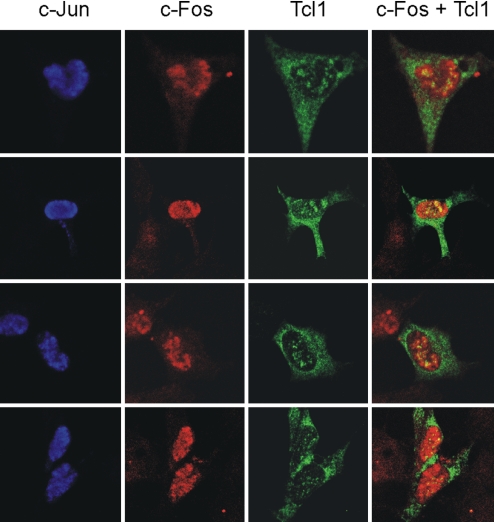
Previously, we had shown that Tcl1 localizes in both nucleus and cytoplasm (6). However, c-Jun and c-Fos are mostly nuclear proteins (16). To determine intracellular localization of Tcl1–AP-1 complexes we carried out immunofluorescence experiments in 293 cells. Fig. 3 shows intracellular location of Tcl1, c-Jun, and c-Fos in 4 different fields. As expected, c-Jun (blue) and c-Fos (red) were colocalized in the nucleus. Tcl1 (green), however, was localized in the nucleus and the cytoplasm. Fig. 3 Right shows that Tcl1–AP-1 complexes (yellow) localized in distinct compartments within the nucleus. These data serve as additional evidence that Tcl1 inhibits AP-1 function by direct association.
Fig. 3.
Intracellular localization of c-Jun, c-Fos, and Tcl1. Some 293 cells were cotransfected with c-Jun-HA, c-Fos-V5, and Omni-Tcl1 constructs. Sixteen hours later, cells were fixed, permeabilized, and immunostained with rat anti-HA, mouse anti-c-Fos, and rabbit anti-omni antibodies. Secondary goat anti-rat Alexa Fluor 647, goat anti-mouse Alexa Fluor 546, and goat anti-rabbit Alexa Fluor 488 antibodies were used to visualize intercellular location of c-Jun (blue), c-Fos (red), and Tcl1 (green). Colocalization of c-Fos and Tcl1 is shown in yellow. (Magnification: 63×.)
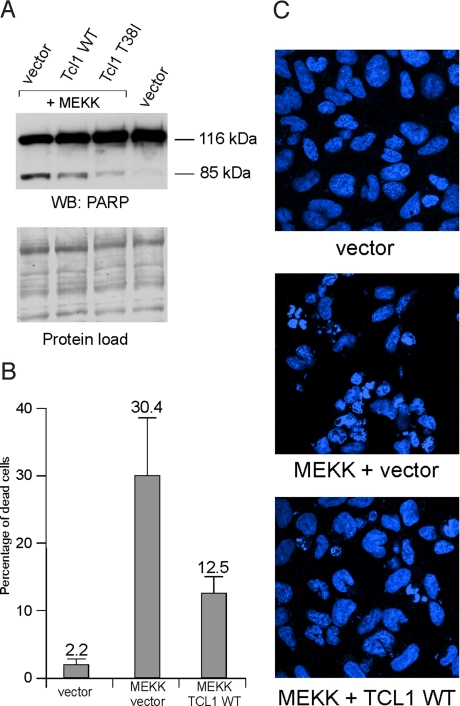
Because we proved that Tcl1 induces NF-κB-dependent transcription and represses AP-1-dependent transcription by participating directly in transcriptional complexes (Figs. 1 and 2), it is reasonable to speculate that these actions of Tcl1 will result in cell death inhibition. To show that this is indeed the case, we took advantage of previously published observations that overexpression of MEKK1 induces apoptosis in 293 cells by c-jun N-terminal kinase (JNK) and AP-1 activation (21, 22). In these experiments we used the construct expressing the kinase domain of MEKK1 that we used to induce AP-1 in Fig. 2. Fig. 4 shows that Tcl1 indeed inhibits AP-1-mediated apoptosis in 293 cells. The 116-kDa intact form of poly(ADP-ribose) polymerase 1 (PARP1) is present in both apoptotic and nonapoptotic cells, whereas the 85-kDa cleaved PARP1 isoform is present only in apoptotic cells. Expression of MEKK1 resulted in the appearance of cleaved 85-kDa PARP1 (Fig. 4A). Tcl1 expression caused decreased intensity of the 85-kDa band, whereas expression of Tcl1 T38I mutant resulted in a further decrease in expression of 85-kDa PARP1 (Fig. 4A). This finding indicates that Tcl1 inhibits MEKK1-induced apoptosis in 293 cells, whereas expression of the Tcl1 T38I mutant results in even stronger inhibition. To evaluate the number of apoptotic cells, we assessed the number of fragmented nuclei in 293 cells 20 h after transfection. Fig. 4 B and C shows that MEKK1 transfection resulted in ≈30% apoptosis in 293 cells, whereasTcl1 expression resulted in a decrease of apoptosis to ≈12.5%. These results suggest that Tcl1 inhibits apoptosis caused by AP-1 activation.
Fig. 4.
Tcl1 inhibits MEKK1-mediated cell death. (A) Some 293 cells were transfected with 1.5 μg of pCMV5-empty vector, 0.5 μg of pFC-MEKK and 1 μg of pCMV5-empty, pCMV5-Tcl1 WT, or pCMV5-Tcl1 T38I constructs. Western blot analysis was carried out as described in Methods. (B and C) Some 293 cells were transfected with 1.5 μg of pCMV5-empty vector, or0.5 μg of pFC-MEKK and 1 μg of pCMV5-empty or pCMV5-Tcl1 WT constructs. Sixteen hours later, cells were fixed, permeabilized, and stained with Hoechst 33342. (B) Percentage of apoptotic cells. For each transfection at least 20 fields were selected for counting the percentage of dead cells (indicated by fragmented nucleus). (C) Results of the same experiment were visualized by using confocal microscopy. (Magnification: 63×.)
Discussion
Studies of animal models conclusively demonstrated that deregulation of TCL1 is a causal event in the development of mature T cell leukemia (T-PLL/T-CLL) and B-CLL (4, 23). Because Tcl1 protein does not contain domains or motifs pointing to its function, investigation of molecular mechanisms involved in Tcl1-induced oncogenesis represents a significant challenge. In 2000 we and others demonstrated that Tcl1 is a coactivator of Akt (6, 24). Successive publications defined molecular details of this function of Tcl1 (25–27) but no progress was made in uncovering other pathways in which Tcl1 might be involved. Because transgenic mice expressing Akt in T cells develop T cell leukemias (7), it is likely that Akt is responsible for Tcl1-mediated lymphomagenesis in T cells. However, transgenic mice expressing activated Akt in B cells do not develop leukemias (Y.P. and C.M.C., unpublished results) and Pten knockout mice expressing constitutively active Akt did not develop B cell malignancies (8).
Recent studies of animal models have demonstrated the importance of the NF-κB pathway in B-CLL development (9), which prompted us to investigate the direct involvement of Tcl1 in transcriptional regulation. Identification of Tcl1 mutants in B-CLL provided an important tool for this study. Although we found that Tcl1 activates NF-κB-dependent transcription, the Tcl1 mutants showed slightly decreased activity compared with WT Tcl1. The Tcl1 mutants still activated NF-κB-dependent transcription 2.5- to 3-fold, suggesting that, although this activation might be important in the pathogenesis of B-CLL, we needed to investigate other pathways to find differences that might explain the selection for expression of these mutant alleles in the original B-CLLs. Our findings that Tcl1 functions as an AP-1 inhibitor provides important insights concerning molecular mechanisms involved in B-CLL development. The importance of these results is greatly enhanced by the fact that the somatic T38I mutant showed gain-of-function properties. The R52H mutation was present in constitutional DNA of the same patient and also led to gain of function in AP-1 inhibition. It is possible that this change represents a rare polymorphism causing genetic predisposition to B-CLL. The physical interaction between Tcl1 and transcription factors such as p300 and AP-1 components provides a novel molecular mechanism of Tcl1 function and proved that this function of Tcl1 is independent of Akt. Because Tcl1 binds to multiple proteins of different structure and function (such as Akt, p300, c-Jun, and c-Fos) it is also possible that it might have other functions, such as transporter, for example.
AP-1 and its components, such as c-Jun, c-Fos, and JunB, are responsible for transactivation of multiple genes (15). AP-1 activation in different cellular contexts can induce proliferation or apoptosis (15, 16). Specifically, inactivation of JunB is widely implicated in leukemogenesis through its proapoptotic function (19). c-Jun and c-Fos are induced in lymphoid cells after growth factor withdrawal, and this induction results in apoptosis (18). However, AP-1 components are known to induce proliferation in a number of solid tumors (15, 16). Because most B-CLL cells do not proliferate (1, 2), function of Tcl1 in these cells may involve inhibition of AP-1-mediated apoptosis and increase of cell survival through activation of the NF-κB pathway. Our findings suggest that novel drugs that inhibit NF-κB or activate AP-1 could be useful in treatment of the aggressive form of B-CLL.
Methods
B-CLL Samples, Genomic Sequencing, and RT-PCR.
A total of 600 B-CLL samples were obtained after informed consent from patients diagnosed with B-CLL from the CLL Research Consortium. Research was performed with the approval of the Institutional Review Board of Ohio State University. Briefly, blood was obtained from CLL patients, and lymphocytes were isolated through Ficoll/Hypaque gradient centrifugation (Amersham) and processed for RNA extraction by using the standard TRIzol method. Oligonucleotides used in genomic DNA PCR and sequencing were: TCL1_149F, 5′-CATGCTGCCCGGATATAAAG-3′; TCL1_539R, 5′-TGCCTGGAGAACTCCTATTCAT-3′; TCL1_31F, 5′- GAAGTGAGCTTCAGGGAACAGT-3′; and TCL1_880R, 5′- ACAGCCACTGTGGACTAAGAGG-3′. Oligonucleotides used in RT-PCR and sequencing were: TCL1_D5, 5′- CCTGTGGGCCTGGGAGAAGT-3′ and TCL1_R5, 5′-TCCTCCACGCCGTCAATCTT-3′.
DNA Constructs.
Full-length human TCL1 and FHIT ORFs were cloned into a pcDNA4-HisMaxC vector (Omni-Tcl1, Omni-Fhit, respectively) (Invitrogen) using standard protocols. The full-length human TCL1 ORF was also cloned into a pCMV5 vector (28) to obtain pCMV5-TCL1 WT construct. pCMV5-TCL1 T38I and pCMV5-TCL1 R52H constructs were created by using a standard PCR-based mutagenesis kit from Stratagene. WT and mutant TCL1 and FHIT ORFs were cloned into the pCMV-2xMyc vector, modified from the pCMV-Myc vector (BD Biosciences) with an added Myc tag, creating Myc tags at both 5′ and 3′ termini. The resulting constructs were named 2xMyc-Tcl1 WT, 2xMyc-Tcl1 T38I, and 2xMyc-Fhit. Mammalian expression constructs for c-Jun and JunB (in pCMV-SPORT6 vector) were purchased from ATCC. c-Jun-HA was constructed by inserting the c-Jun ORF into the pCMV-HA vector (BD Biosciences). The c-Fos-V5 construct was purchased from Invitrogen. The p300-HA construct was purchased from Upstate Biotechnology. The Akt-HA construct has been described (6). The Dual-luciferase Reporter Assay System and Renilla luciferase reporter vector pRL-TK were purchased from Promega. The AP-1 reporter construct, pAP1-Luc, NF-κB reporter construct, pNF-kB-Luc and the construct encoding the kinase domain of MEKK1 under control of the CMV promoter, pFC-MEKK, were purchased from Stratagene.
Cell Culture, Transfection, Western Blot Analysis, and Immunoprecipitation Experiments.
NIH 3T3 and 293 cells were grown in RPMI medium 1640 with 10% FBS and 100 μg/L gentamicin at 37 °C. FuGene 6 transfection reagent and protease inhibitor mixture tablets were obtained from Roche. Transfections, except luciferase assay experiments (see below), cell lysate preparations, and Western blot analysis were carried out as described (6). Immunoblots were developed by using Pierce ECL Western blot analysis substrate or SuperSignal West Femto Maximum Sensitivity Substrate from Thermo Scientific. Antibodies used were: anti-Tcl1 (sc-32331 for Western blot analysis and immunoprecipitation with p300; sc-11156 and sc-11155 for immunoprecipitation with c-Jun), anti-Omni (sc-7270 for immunoprecipitation and Western blot analysis; sc-499 for immunofluorescence), anti-p300 (sc-32244), anti-Myc (9E10), anti-Myc-HRP (9E10), anti-c-Jun (sc-1694 for immunoprecipitation), anti-JunB (sc-8051 for immunoprecipitation; sc-46 for Western blot analysis), anti-c-Fos (sc-447 for immunoprecipitation and immunofluorescence) (Santa Cruz Biotechnology), anti-c-Jun (610326 for Western blot analysis, BD Biosciences), anti-HA (HA.11) (Covance), anti-V5-HRP (Invitrogen), rat anti-HA (for immunofluorescence), and anti-HA-HRP (Roche).
Immunofluorescence.
HEK293 cells were grown on human fibronectin Cellware 2-well culture slides (BD Biosciences). Immunofluorescence experiments were carried out as described with a Zeiss LCM 510 confocal microscope (29). Secondary antibodies used for immunofluorescence were as follows: goat anti-mouse Alexa Fluor 546 (red), goat anti-rat Alexa Fluor 647 (far-red), and goat anti-rabbit Alexa Fluor 488 (green), all purchased from Invitrogen.
Luciferase Assay.
NIH 3T3 or 293 cells were transfected with the indicated constructs. Firefly and renilla luciferase activities were assayed with the dual luciferase assay system (Promega), and firefly luciferase activity was normalized to renilla luciferase activity, as suggested by the manufacturer. All experiments were carried out in triplicate and repeated 3 times with consistent results.
Cell Death Analysis.
Apoptosis was assessed by scoring the number of cells displaying fragmented nuclei, stained with 10 μg/mL of Hoechst 33342 (Invitrogen). An alternative method of apoptosis detection was also used. HEK 293 cells were transfected with either 1.5 μg of pCMV5-empty vector or 0.5 μg of pFC-MEKK with 1 μg of pCMV5-empty or pCMV5-Tcl1 WT or pCMV5-Tcl1 T38I constructs. Twenty-four hours later both dead and live cells were collected and lysed. These lysates were probed with anti-PARP1 antibody (556362; BD Biosciences). The 116-kDa intact form of PARP1 was present in both nonapoptotic and apoptotic cells. The 85-kDa PARP1 cleavage fragment was present only in apoptotic cells.
Acknowledgments.
This work was supported by an American Cancer Society Research Scholar grant, a grant from the Chronic Lymphocytic Leukemia Global Research Foundation (to Y.P.), and National Institutes of Health Grant PO1-CA81534 (to the Chronic Lymphocytic Leukemia Research Consortium, L.R., T.K., and C.M.C.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Sgambati M, Linet M, Devesa S. Chronic lymphocytic leukemia, epidemiological, familial, and genetic aspects. In: Cheson B, editor. Chronic Lymphocytic Leukemias. 2nd Ed. New York: Dekker; 2001. pp. 33–62. [Google Scholar]
- 2.Bullrich F, Croce C. Molecular biology of chronic lymphocytic leukemia. In: Cheson B, editor. Chronic Lymphocytic Leukemias. 2nd Ed. New York: Dekker; 2001. pp. 9–32. [Google Scholar]
- 3.Virgilio L, et al. Identification of the TCL1 gene involved in T cell malignancies. Proc Natl Acad Sci USA. 1994;91:12530–12534. doi: 10.1073/pnas.91.26.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bichi R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA. 2002;99:6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pekarsky Y, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 6.Pekarsky Y, et al. Tcl1 enhances Akt kinase activity and mediates its nuclear translocation. Proc Natl Acad Sci USA. 2000;97:3028–3033. doi: 10.1073/pnas.040557697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malstrom S, Tili E, Kappes D, Ceci JD, Tsichlis PN. Tumor induction by an Lck-MyrAkt transgene is delayed by mechanisms controlling the size of the thymus. Proc Natl Acad Sci USA. 2001;98:14967–14972. doi: 10.1073/pnas.231467698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki A, et al. Critical roles of Pten in B cell homeostasis and immunoglobulin class switch recombination. J Exp Med. 2003;197:657–667. doi: 10.1084/jem.20021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pekarsky Y, Zanesi N, Aqeilan RI, Croce CM. Animal models for chronic lymphocytic leukemia. J Cell Biochem. 2007;100:1109–1118. doi: 10.1002/jcb.21147. [DOI] [PubMed] [Google Scholar]
- 10.Planelles L, et al. APRIL promotes B-1 cell-associated neoplasm. Cancer Cell. 2004;6:399–408. doi: 10.1016/j.ccr.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh AK, Varga J. The transcriptional coactivator and acetyltransferase p300 in fibroblast biology and fibrosis. J Cell Physiol. 2007;213:663–671. doi: 10.1002/jcp.21162. [DOI] [PubMed] [Google Scholar]
- 12.Butscher WG, Haggerty CM, Chaudhry S, Gardner K. Targeting of p300 to the interleukin-2 promoter via CREB-Rel cross-talk during mitogen and oncogenic molecular signaling in activated T cells. J Biol Chem. 2001;276:27647–27656. doi: 10.1074/jbc.M009614200. [DOI] [PubMed] [Google Scholar]
- 13.Gerritsen ME, et al. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: Quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 15.Leppa S, Bohmann D. Diverse functions of JNK signaling and c-Jun in stress response and apoptosis. Oncogene. 1999;18:6158–6162. doi: 10.1038/sj.onc.1203173. [DOI] [PubMed] [Google Scholar]
- 16.Ameyar M, Wisniewska M, Weitzman JB. A role for AP-1 in apoptosis: The case for and against. Biochimie. 2003;85:747–752. doi: 10.1016/j.biochi.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Foletta VC, Segal DH, Cohen DR. Transcriptional regulation in the immune system: All roads lead to AP-1. J Leukocyte Biol. 1998;63:139–152. doi: 10.1002/jlb.63.2.139. [DOI] [PubMed] [Google Scholar]
- 18.Colotta F, Polentarutti N, Sironi M, Mantovani A. Expression and involvement of c-fos and c-jun protooncogenes in programmed cell death induced by growth factor deprivation in lymphoid cell lines. J Biol Chem. 1992;267:18278–18283. [PubMed] [Google Scholar]
- 19.Ott RG, et al. JunB is a gatekeeper for B-lymphoid leukemia. Oncogene. 2007;26:4863–4871. doi: 10.1038/sj.onc.1210285. [DOI] [PubMed] [Google Scholar]
- 20.Humbert O, Achour I, Lautier D, Laurent G, Salles B. hMSH2 expression is driven by AP1-dependent regulation through phorbol-ester exposure. Nucleic Acids Res. 2003;31:5627–5634. doi: 10.1093/nar/gkg781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonvin C, et al. Role of the amino-terminal domains of MEKKs in the activation of NF-κB and MAPK pathways and in the regulation of cell proliferation and apoptosis. Cell Signal. 2002;14:123–131. doi: 10.1016/s0898-6568(01)00219-4. [DOI] [PubMed] [Google Scholar]
- 22.Johnson NL, et al. Signal transduction pathways regulated by mitogen-activated/extracellular response kinase kinase kinase induce cell death. J Biol Chem. 1996;271:3229–3237. doi: 10.1074/jbc.271.6.3229. [DOI] [PubMed] [Google Scholar]
- 23.Virgilio L, et al. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc Natl Acad Sci USA. 1998;95:3885–3889. doi: 10.1073/pnas.95.7.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laine J, Kunstle G, Obata T, Sha M, Noguchi M. The protooncogene TCL1 is an Akt kinase coactivator. Mol Cell. 2000;6:395–407. doi: 10.1016/s1097-2765(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 25.Kunstle G, et al. Identification of Akt association and oligomerization domains of the Akt kinase coactivator TCL1. Mol Cell Biol. 2002;22:1513–1525. doi: 10.1128/mcb.22.5.1513-1525.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laine J, Kunstle G, Obata T, Noguchi M. Differential regulation of Akt kinase isoforms by the members of the TCL1 oncogene family. J Biol Chem. 2002;277:3743–3751. doi: 10.1074/jbc.M107069200. [DOI] [PubMed] [Google Scholar]
- 27.Teitell MA. The TCL1 family of oncoproteins: Coactivators of transformation. Nat Rev Cancer. 2005;5:640–648. doi: 10.1038/nrc1672. [DOI] [PubMed] [Google Scholar]
- 28.Andersson S, Davis DL, Dahlback H, Jornvall H, Russell DW. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 29.Palamarchuk A, et al. Akt phosphorylates and regulates Pdcd4 tumor suppressor protein. Cancer Res. 2005;65:11282–11286. doi: 10.1158/0008-5472.CAN-05-3469. [DOI] [PubMed] [Google Scholar]