Abstract
Although a substantial proportion of plant biomass originates from the activity of vascular cambium, the molecular basis of radial plant growth is still largely unknown. To address whether cytokinins are required for cambial activity, we studied cytokinin signaling across the cambial zones of 2 tree species, poplar (Populus trichocarpa) and birch (Betula pendula). We observed an expression peak for genes encoding cytokinin receptors in the dividing cambial cells. We reduced cytokinin levels endogenously by engineering transgenic poplar trees (P. tremula × tremuloides) to express a cytokinin catabolic gene, Arabidopsis CYTOKININ OXIDASE 2, under the promoter of a birch CYTOKININ RECEPTOR 1 gene. Transgenic trees showed reduced concentration of a biologically active cytokinin, correlating with impaired cytokinin responsiveness. In these trees, both apical and radial growth was compromised. However, radial growth was more affected, as illustrated by a thinner stem diameter than in WT at same height. To dissect radial from apical growth inhibition, we performed a reciprocal grafting experiment. WT scion outgrew the diameter of transgenic stock, implicating cytokinin activity as a direct determinant of radial growth. The reduced radial growth correlated with a reduced number of cambial cell layers. Moreover, expression of a cytokinin primary response gene was dramatically reduced in the thin-stemmed transgenic trees. Thus, a reduced level of cytokinin signaling is the primary basis for the impaired cambial growth observed. Together, our results show that cytokinins are major hormonal regulators required for cambial development.
Keywords: cambial activity, cambium, secondary development, Populus, CYTOKININ OXIDASE
In plants, development of vascular tissues is unique because of its dynamic nature. During embryogenesis, a continuum of provascular tissue is evident between the shoot and root apical meristems. Soon after germination, a subset of these provascular cells differentiates into 2 conductive tissue types, xylem and phloem. Between the xylem and phloem, however, some meristematic cells persist through primary development. On initiation of secondary development, a lateral meristem, vascular cambium, is derived from these procambial cells, together with interfascicular cells in shoots and pericycle cells in root. Secondary vascular xylem and phloem are subsequently produced via cell divisions taking place in the cambium.
Compared with apical meristems, our knowledge about the genetic control of cambium is much less complete. Previously, a radially oriented gradient of basipetal auxin transport has been shown to be present across the cambial zone (1, 2) in accordance with specific expression patterns of auxin signaling-related genes in the region (3). In classic hormone treatment studies, apically applied exogenous auxin was able to reactivate cambium in decapitated shoots (4, 5). Recently, using a transgenic approach based on down-regulating auxin signaling, it was demonstrated that auxin is required for cell proliferation and cell differentiation during cambial development (6). Aside from auxin, several other hormones, including cytokinin (7, 8), gibberellin (9, 5), and ethylene (10), have been implicated in control of cambial activity because of their stimulatory effect on cell division upon hormone treatment. However, there is no indication as to whether these hormones are normally required for cambial activity.
Cytokinin responses in plants are mediated by a signal transduction pathway consisting of components characteristic of bacterial 2-component molecules (11, 12). They have been shown to function in opposite modes, depending on the developmental context, in regulation of apical meristem activity. Based on various genetic and molecular studies (13–17), including systemic overexpression of a catabolic CYTOKININ OXIDASE (CKX) gene, it has become evident that in the shoot, apical meristem cytokinins appear to promote cell proliferation. In contrast, in the root, apical meristem cytokinins appear to inhibit root elongation (14, 16, 18). The negative effect of cytokinins on root development has recently been connected to their function in cell differentiation. Reduction in root cytokinin level delays cell differentiation, leading to a longer proximal meristem zone, and thus enhancement of the root growth (19).
Considering vascular development, cytokinin signaling is required for the pluripotent identity of the procambial cell files during the primary phase of Arabidopsis root development (20–22). Furthermore, cytokinins appear to be required for proliferation of the vascular cell files during primary vascular development in both the root and shoot (14, 20, 21).
To address the mode of cytokinin function in the secondary meristem, vascular cambium, we studied cytokinin signaling during cambial development in the trunks of 2 hardwood tree species, poplar (Populus trichocarpa) and silver birch (Betula pendula). For functional studies, we engineered transgenic poplar trees (P. tremula × tremuloides) to ectopically express a cytokinin-degrading enzyme in the cambial zone for the purpose of repressing cambial cytokinin signaling. Our results indicate that cytokinins are major hormonal regulators required for cambial development.
Results
The CRE Cytokinin Receptor Gene Family Is Conserved Between Herbaceous and Woody Plant Species.
Cytokinins are perceived by histidine kinase receptors, which initiate the cytokinin signal transduction phosphorelay (11, 20, 23). The Arabidopsis genome harbors 3 loci for the cytokinin receptors, CRE1/WOL, AHK2, and AHK3, all of which belong to the superfamily of 2-component regulators (12, 24). The CRE cytokinin receptor gene family has been shown to be conserved between the non-woody plant species Arabidopsis, rice, and maize (25, 26).
To compare cytokinin receptors between herbaceous plants and hardwood trees, we identified genes belonging to this gene family from 2 hardwood tree species, silver birch and poplar. We were able to isolate birch mRNAs for 3 CRE family genes (BpCRE1, BpHK2, and BpHK3). From the sequenced P. trichocarpa genome, we identified 5 genes (PtCRE1a, PtCRE1b, PtHK2, PtHK3a, and PtHK3b) orthologous to the CRE gene family members. One gene (PtHK2) was orthologous to AHK2, 2 (PtHK3a and PtHK3b) to AHK3, and 2 (PtCRE1a and PtCRE1b) to CRE1. From phylogenetic analysis, it is apparent that these genes show high sequence similarity to the 3 Arabidopsis CRE family histidine kinases (Fig. 1A). We were also able to verify that the birch ortholog for Arabidopsis CRE1, BpCRE1, encodes a functional cytokinin receptor. When expressed under the Arabidopsis CRE1 promoter, BpCRE1 was able to complement the phenotype of an Arabidopsis mutant lacking all 3 CRE family genes [supporting information (SI) Fig. S1]. These results further support the view that all flowering plants perceive cytokinin through members of the CRE cytokinin receptor family.
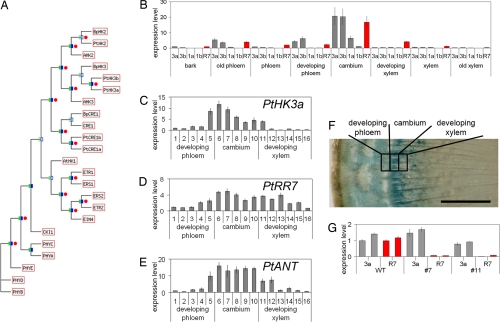
Fig. 1.
Expression of poplar and birch cytokinin receptor genes peaks in the cambial zone of tree trunk. (A) Phylogenetic tree of Arabidopsis 2-component regulators, including CRE family genes WOL/CRE1, AHK2 and AHK3, together with CRE family receptors from B. pendula (BpCRE1, BpHK2, and BpHK3) and P. trichocarpa (PtCRE1a, PtCRE1b, PtHK2, PtHK3a, and PtHK3b). The maximum-likelihood tree is based on an alignment of amino acid sites. Filled green/blue boxes indicate groups with bootstrap support >75%, and open boxes indicate groups with less. Where tree topologies agree, red circles indicate >75% bootstrap support from parsimony analysis of the same alignment. (B) Expression of poplar cytokinin receptor genes PtCRE1a (1a), PtCRE1b (1b), PtHK3a (3a), and PtHK3b (3b), together with an A-type response regulator PtRR7 (R7), across the P. trichocarpa trunk by qRT-PCR. Receptor expression is given relative to the PtHK3a level in bark and PtRR7 relative to itself in bark (error bars = SE). Expression of PtHK3a (C), PtRR7 (D), and PtANT (E) across the P. trichocarpa cambial zone in 16 24-μm sections. Expression is given relative to the developing phloem section 1. (F) Expression of pBpCRE1::GUS peaks in the cambial zone of P. tremula × tremuloides stem. (Scale bar: 1 mm.) (G) Expression of PttHK3a and PttRR7 in whole-stem samples of WT and pBpCRE1::AtCKX2 transgenic P. tremula × tremuloides lines 7 and 11. Expression is given relative to the WT. Two trees were analyzed per line.
Cytokinin Signaling Genes Are Expressed in the Cambial Zone.
To explore the potential role of cytokinins during cambial development, we studied the expression of cytokinin signal transduction components across the radius of the tree trunk. In both silver birch and poplar, members of the cytokinin receptor gene family are expressed across the cambial zone (Fig. 1 B and C and Fig. S2H). In poplar, PtHK3a and PtHK3b show the highest cambial expression, peaking in the same zone as the marker gene for cambial cell identity, PtANT (27) (Fig. 1 B, C, and E and Fig. S3). A similar pattern was seen through the expression of GUS under the BpCRE1 promoter in both transgenic birch (Fig. S2G) and poplar (Fig. 1F). Also, the expression of a cytokinin primary response gene from poplar, encoding an A-type response regulator PtRR7 (28), peaked in the cambial zone along with the receptor genes (Fig. 1 B and D and Fig. S3). The expression of cytokinin receptors in vascular cambium suggests that cytokinin signaling participates in a regulation function in this meristem.
In addition to the cambial meristem, CRE family genes are expressed in apical meristems. By in situ hybridization, BpCRE1 was shown to be expressed in both birch shoot and root apical meristems (Fig. S2 A, B, E, and F). This pattern was reproduced by expressing GUS under the BpCRE1 promoter in transgenic birch (Fig. S2 C and D) and poplar (data not shown). The observed BpCRE1 expression pattern resembles CRE1 expression in Arabidopsis apical meristems (20, 21), further indicating that the function of cytokinin receptors is highly conserved among different plant species.
pBpCRE1::AtCKX2 Poplars Display Several Phenotypes Diagnostic for Reduced Cytokinin Responsiveness.
We engineered transgenic poplar (P. tremula × tremuloides) trees that were compromised in cytokinin signaling during cambial development. These trees expressed a cytokinin catabolic gene from Arabidopsis cytokinin oxidase 2, under the promoter of a birch cytokinin receptor, BpCRE1. CKX family enzymes irreversibly degrade active cytokinin species (29), and they have been successfully expressed under the systemic 35S promoter to reduce the general cytokinin levels in several plant species (13, 14, 30, 31). The BpCRE1 promoter was chosen for its high cambial expression (Fig. 1F); as a promoter driving the expression of a cytokinin receptor gene, it would presumably direct the expression of the cytokinin-degrading enzyme to the location of cytokinin perception.
The regeneration capacity of pBpCRE1::AtCKX2 transgenic plants was partially compromised, apparently indicating the importance of cytokinin action for plant development. Fewer transgenic lines were obtained from the same number of stem segments transformed with the pBpCRE1::AtCKX2 construct than with the pBpCRE1::GUS construct (data not shown). We were able to obtain 11 pBpCRE1::AtCKX2 lines. Four were WT-like, and 7 showed a distinct phenotype during in vitro growth; they had stunted shoots with short internodes and small dark-green leaves. Both the apical growth of the shoots and internode elongation were severely retarded, whereas root growth was extensive (data not shown). These observed phenotypic alterations in vitro resemble those reported for the p35S::CKX plants in tobacco and Arabidopsis (13, 14).
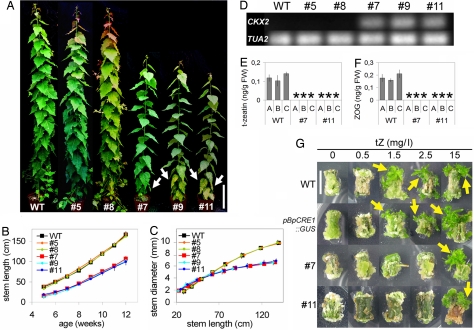
We were able to further propagate 3 (lines 7, 9, and 11) of the 7 lines with the stunted in vitro phenotype. When these 3 lines were grown in soil, their growth improved, although not to the WT level (Fig. 2A). Both the apical and radial growth of lines 7, 9, and 11 was reduced compared with the WT or with the WT-like lines 5 and 8 (Fig. 2 B and C). However, the radial growth of lines 7, 9, and 11 was more compromised than apical growth; these trees had thinner trunks than WT trees of similar height (Fig. 2C). We next analyzed the status of AtCKX2 expression in the greenhouse-grown pBpCRE1::AtCKX2 lines. Strong AtCKX2 expression correlated with the phenotype of reduced growth in the thin-stemmed lines 7, 9, and 11, whereas no AtCKX2 expression was detected in either of the analyzed WT-like lines 5 and 8 (Fig. 2A, C, and D).
Fig. 2.
pBpCRE1::AtCKX2 poplar lines with a high expression level of the transgene display reduced cytokinin content and responsiveness, together with a thin-stemmed phenotype. (A) WT and pBpCRE1::AtCKX2 poplar lines 5, 8, 7, 9, and 11 (4-month-old trees). Lines 7, 9, and 11 with a strong transgene expression have elongated internodes and show premature leaf senescence (indicated by white arrows). (Scale bar: 20 cm.) Height vs. age ratio (B) and width vs. height ratio (C) in WT and pBpCRE1::AtCKX2 poplar lines 5, 8, 7, 9, and 11 (n = 3). pBpCRE1::AtCKX2 poplar lines 5 and 8 have a WT-like phenotype, and lines 7, 9, and 11 display a thin-stemmed phenotype (error bars = SD). (D) AtCKX2 and PtTUA2 expression in WT and pBpCRE1::AtCKX2 poplar lines 5, 8, 7, 9, and 11 by qRT-PCR shown in a gel. Both t-zeatin (E) and ZOG (F) are below the detection limit (*) in the shoot of lines 7 and 11. Three biological replicates (A–C) are shown per line (error bars = SD). (G) Cytokinin responsiveness assay. Medium with indole acetic acid (IAA) at 0.5 mg/L IAA and 0, 0.5, 1.5, 2.5, or 15 mg/L t-zeatin. A low cytokinin-to-auxin ratio induces root regeneration, and a high cytokinin-to-auxin ratio enhances shoot regeneration. The following lines were analyzed: WT, pBpCRE1::GUS, and pBpCRE1::AtCKX2 lines 7 and 11. WT and pBpCRE1::GUS regenerate shoots from medium to high cytokinin concentrations (1.5–15 mg/L), whereas lines 7 and 11 regenerate shoots only in a high (15 mg/L) cytokinin concentration. Emerging shoots are indicated by yellow arrows. (Scale bar: 1 cm.)
The greenhouse-grown thin-stemmed lines 7, 9, and 11 also had longer internodes than WT and displayed premature leaf senescence, an indication of impaired cytokinin action (Fig. 2A). To study if ectopic AtCKX2 expression affects cytokinin levels, we measured the cytokinin content of the thin-stemmed lines 7 and 11. Compared with WT, the levels of trans-zeatin (t-zeatin), one of the biologically active cytokinin species, and its storage form, zeatin-O-glucoside (ZOG), were reduced below detection limit in the stem of these lines (Fig. 2 E and F). The level of another biologically active cytokinin, isopentenyladenine, was reduced only in line 11 (Fig. S4). Levels of other metabolic forms of cytokinin were not reduced from WT levels in either line (Fig. S4). Free bases are preferable substrates of AtCKX2 (32) and the reduction of either isopentenyladenine and/or t-zeatin is consistent with earlier p35S::CKX transgenic tobacco and Arabidopsis studies (13, 14). However, high levels of other metabolic forms have not been observed, which may be attributable to the cell type-specific promoter used in our study. The reduced t-zeatin and ZOG content indicates that ectopic AtCKX2 expression leads to a reduced content of active cytokinin species, which results in the observed thin-stemmed phenotype.
To further evaluate the effect of ectopic AtCKX2 expression on cytokinin signaling in the thin-stemmed lines, we tested their cytokinin responsiveness. In the classic cytokinin responsiveness assay (33), a low cytokinin-to-auxin ratio induces root regeneration from plant segments and a high cytokinin-to-auxin ratio promotes shoot regeneration instead. In this assay, lines 7 and 11 showed reduced cytokinin responsiveness compared with WT and a pBpCRE1::GUS line (Fig. 2G). A low cytokinin-to-auxin ratio is also known to support apical dominance by inhibiting axillary bud outgrowth, whereas a high ratio reduces it by facilitating bud outgrowth (34). Decapitated pBpCRE1::AtCKX2 trees produced fewer new shoots from the axillary buds than the control trees, indicating enhanced apical dominance (data not shown). Taken together, our data indicate that the pBpCRE1::AtCKX2 trees are compromised in cytokinin-regulated developmental processes.
Dissecting the Effects of Cytokinins on Radial Versus Apical Growth.
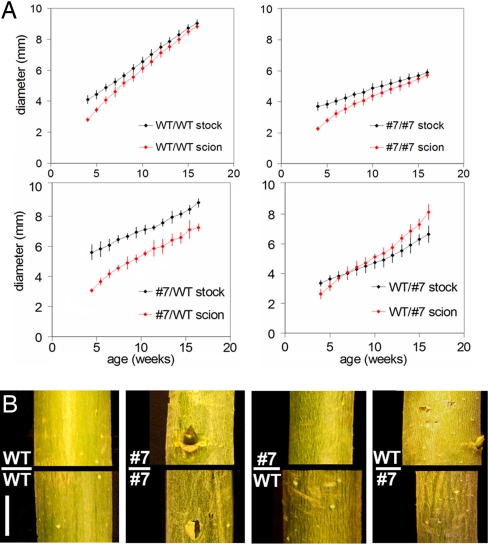
We further examined the apical and radial growth of several pBpCRE1::AtCKX2 lines in greenhouse conditions. In the shoot apex of the transgenic lines 7 and 11, 0.36 ± 0.02 leaf primordia per day were produced compared with 0.59 ± 0.05 in WT (average ± SD, 4- to 12-week-old plants, n = 3), indicating somewhat reduced activity of the shoot apical meristem. To address the impact of reduced apical growth on the radial growth in the thin-stemmed pBpCRE1::AtCKX2 trees, we studied the relation between apical and radial growth by means of a grafting experiment. One transgenic thin-stemmed line (line 7) was reciprocally grafted to WT (n = 3) (Fig. 3 A and B and Fig. S5). In the WT/WT and line 7/7 grafts, the stock and scion reached the same diameter (9.0 ± 0.1 SD vs. 8.8 ± 0.1 mm and 5.9 ± 0.2 SD vs. 5.7 ± 0.2 mm, respectively) during 4 months of linear growth. In contrast, the thin-stemmed scion did not reach the diameter of the WT stock (7.2 ± 0.3 SD vs. 8.9 ± 0.4 mm), whereas the WT scion outgrew the diameter of the thin-stemmed stock (8.1 ± 0.5 SD vs. 6.7 ± 0.4 mm). The data show that the WT stem was not transformed to the thin-stemmed phenotype by a pBpCRE1::AtCKX2 apex, nor was the thin-stemmed phenotype rescued to WT by a WT apex. Thus, compromised cytokinin activity during radial growth rather than the reduced activity of apical meristem appears to be the major determinant of the thin-stemmed phenotype.
Fig. 3.
Relation between the apical and radial growth in pBpCRE1::AtCKX2 poplars. (A) Reciprocal grafts with WT and pBpCRE1::AtCKX2 line 7. Graphs show scion and stock diameter vs. age. Diameter was measured 5 cm above and below the scion/stock junction. WT/WT (self-grafting): both the WT scion and stock part reach the same diameter. Line 7/7 (self-grafting): both the line 7 scion and stock part reach same diameter. Line 7/WT (line 7 scion, WT stock): line 7 scion part does not reach diameter of the WT stock. WT/line 7 (WT scion, line 7 stock): WT scion outgrows the line 7 stock. Average from 3 individual grafted trees is shown in each graph. (B) Grafted trees shown 5 cm above and below the scion/stock junction 6 months after the grafting. (Scale bar: 5 mm.)
Reduced Cambial Activity in pBpCRE1::AtCKX2 Poplars.
To study the effect of reduced cytokinin signaling on cambial growth-related cell division and differentiation, we analyzed the vascular anatomy of the thin-stemmed lines. The cambial cells were seen as undifferentiated, thin-walled, flat cells localized between differentiating xylem and phloem cells. In the pBpCRE1::AtCKX2 trees, the vascular cambium consists of fewer cell layers than in WT. The reduction was evident when comparing internodes of similar position or similar diameter. The 20th WT internode with a diameter of 7.3 mm contained 16.4 ± 0.11 (average ± SE) apparently undifferentiated cells per 1 cambial cell file (n = 100 cambial cell files, data from 50 files from 2 separate trees combined). Line 7 contained, on average, 10.5 ± 0.1 apparently undifferentiated cells in the 20th internode, and line 11 contained 10.7 ± 0.1 cells. When comparing internodes of similar diameter (7.3 mm) with WT, line 7 contained 11.0 ± 0.1 cells and line 11 contained 11.4 ± 0.1 cells (Fig. 4). The significantly (P < 0.001) reduced number of undifferentiated cells in the cambial cell files indicates that fewer cell divisions occur in the cambial layer of the transgenic trees than in WT trees.
Fig. 4.
pBpCRE1::AtCKX2 poplars have a reduced number of cambial cells. (A) Cross sections of WT, line 7, and line 11 stem. The stem thickness and internode number at the section site are indicated. The cambial cell file is indicated with dots. The number of undifferentiated cambial cells (marked between asterisks) per cell file is reduced in lines 7 and 11 compared with WT (see text for details). (Scale bar: 0.2 mm [0.1 mm in Insets].) (B) Number of undifferentiated cambial cells was reduced in the thin-stemmed pBpCRE1::AtCKX2 lines. Frequency distributions of cell numbers in cambial cell files in the 20th WT internode (Ø 7.3 mm) and in internodes of pBpCRE1::AtCKX2 lines 7 and 11 of the same position (20th from the apex) or the same diameter (Ø 7.3 mm) are shown (n = 100 cambial files, data from 50 files from 2 separate trees combined). The difference in distributions was statistically significant (P < 0.001) between WT and pBpCRE1::AtCKX2 lines.
We also analyzed wood anatomy of the pBpCRE1::AtCKX2 trees. Thin-stemmed lines had slightly shorter xylem fibers than WT (P < 0.05), whereas the fiber width was not significantly different from WT (Fig. S6A). The opposite was observed with vessel dimensions: vessels were slightly wider in WT than in the thin-stemmed lines (P < 0.05; Fig. S6B), whereas no significant difference was observed in vessel length.
Reduced Cambial Cytokinin Signaling in pBpCRE1::AtCKX2 Poplars.
To associate reduced cambial activity in the pBpCRE1::AtCKX2 poplars with cytokinin function, we analyzed the status of cambial cytokinin signaling. In the trunks of thin-stemmed lines 7 and 11, the expression level of PttRR7 was dramatically reduced compared with WT, whereas the expression of cytokinin receptor PttHK3a was essentially the same as in WT (Fig. 1G). This indicates that a reduced level rather than a restricted spatial domain of cytokinin signaling is the primary basis for the impaired cambial growth of pBpCRE1::AtCKX2 trees.
Discussion
Understanding the regulation of the radial growth that underlies wood development is of great importance for the future use of tree products as a renewable resource. To understand the role of various phytohormones in regulation of wood development, it is important to investigate the consequences of their reduced action. Here, we have taken a transgenic approach to reduce cytokinin levels in P. tremula × tremuloides by driving the expression of a cytokinin catabolic gene (AtCKX2) using the promoter of a birch gene (BpCRE1) that encodes a cytokinin receptor. As a consequence, we were able to show that reduced levels of cytokinins and their storage forms (t-zeatin and ZOG) are produced in selected transgenic lines that strongly express the transgene. Furthermore, these lines display various symptoms indicative of reduced cytokinin action: impaired shoot regeneration in tissue culture, enhanced apical dominance, enhanced leaf senescence, and impaired apical growth of shoot. Concerning the longer internodes and enhanced leaf senescence, the affected lines differ from the earlier reported tobacco and Arabidopsis p35S::CKX lines, which display short internodes and nonaccelerated leaf senescence (13, 14).
Our experimental focus here has been whether cytokinins are required for the secondary phase of vascular development characterized by the activity of vascular cambium, a stem cell population that orchestrates plant radial growth. The transgenic trees with high AtCKX2 expression have significantly impaired radial growth. Radial growth in these lines appears to be more affected than apical growth. Furthermore, by grafting, we have shown that the defects in the apical growth cannot explain the reduced radial growth. A detailed anatomical characterization of the transgenic lines reveals that the number of undifferentiated cell files in the cambial zone is reduced in the transgenic plants with high AtCKX2 expression. Taken together, our data show that cytokinins are required for vascular cambium function in controlling radial growth. In this respect, the cambial meristem function resembles the shoot apical meristem, which is also positively regulated by cytokinin signaling (14–17), and differs from the root apical meristem, which is characterized by negative cytokinin regulation (14, 16, 18, 19).
Using transgenic approaches to impair auxin signaling during cambial development, it has recently been shown that auxin is required for both normal cambial cell proliferation and differentiation of xylem cells during secondary growth (6). In our transgenic trees with high AtCKX2 expression levels, we also observed slight differences in wood anatomy: fiber lengths and vessel widths were slightly reduced. It remains to be studied, however, whether or not these differences can be attributed to the altered rate of cell proliferation in the cambial zone.
During primary vascular development, cytokinins appear to be required for both cell proliferation and cell specification (19–22). We have shown here that during secondary development, their major function is the regulation of cell proliferation. Thus, cytokinins appear to have diverse roles during vascular and meristem development, perhaps dependent on how they interact with other growth regulators. Our finding of the proliferative role of cytokinins in regulating cambial development in radial stem growth may contribute to the development of more efficient plant biomass production systems in the future.
Materials and Methods
Cloning of Birch Genes.
Short cDNA fragments corresponding to BpCRE1 and BpHK2 genes were amplified by PCR using degenerate primers. A partial BpHK3 1.4-kb fragment was identified from a birch leaf cDNA library (J. Kangasjärvi, personal communication). A birch genomic library (Y.H., unpublished data) was screened with BpCRE1 cDNA fragment as a probe. The genomic clone for BpCRE1 was isolated and subcloned for sequencing. A genomic fragment containing the BpCRE1 gene was concatenated from 2 different clones with 5 kb of 5′ upstream sequence and a coding region with 11 exons (GenBank EU583454). cDNAs representing BpCRE1 (EU583455), BpHK2 (EU583456), and BpHK3 (EU583457) were cloned using RT-PCR and 5′ RACE techniques. The sizes of predicted proteins encoded by the cDNAs were 1,004, 1,260, and 1,066 amino acids, respectively.
Phylogenetic Analyses.
Phylogenetic analyses were performed on an amino acid alignment of selected 2-component receptor genes. Two phylogeny reconstruction methods were used: parsimony and maximum likelihood. Details are provided in SI Materials and Methods.
Constructs.
To generate the pBpCRE1::GUS construct, a 4,723-kb fragment containing the BpCRE1 promoter was isolated from a genomic DNA clone and cloned into pBI101 GUS-reporter vector. For the pBpCRE1::AtCKX2 construct, Arabidopsis CKX2 gene (At2g19500) was amplified by PCR from genomic DNA and cloned into pGEM-T Easy vector downstream of the BpCRE1 promoter. Details are provided in SI Materials and Methods.
Transgenic Plants.
B. pendula clone BPM5 was transformed with pBpCRE1::GUS and pBpCRE1::AtCKX2 using an Agrobacterium-mediated method (35). P. tremula × tremuloides clone T89 was transformed with pBpCRE1::GUS and pBpCRE1::AtCKX2 by an Agrobacterium-based method (36). Details of the tissue culture are provided in SI Materials and Methods.
Quantitative RT-PCR.
For poplar quantitative PCR analysis, samples were collected from two 8-month-old P. trichocarpa “Nisqually-1” trees (data shown from 1 tree). Birch quantitative RT-PCR (qRT-PCR) analysis is described in SI Materials and Methods. Expression of putative poplar cytokinin receptors PtHK2, PtHK3a, PtHK3b, PtCRE1a, and PtCRE1b and of a putative A-type response regulator, PtRR7 (28), was analyzed across the trunk. A tangential cryosectioning protocol (27) was used to section the stem into fractions representing “bark,” “old phloem,” “phloem,” “developing phloem,” “cambium,” “developing xylem,” “xylem,” and “old xylem.” Anatomical cross sections representing each fraction were cut with a razor blade. No detectable expression of PtHK2 was observed in any of the fractions by the 2 primer pairs used (data not shown). Expression of the other receptors was very weak outside the cambial zone. The cambial zone of 2 trees was further divided into 16 24-μm cryosections. Details are provided in SI Materials and Methods. The P. trichocarpa TUBULIN ALPHA 2 (PtTUA2) gene was used as a reference for relative quantification. The identity of cambial cells was verified by a marker diagnostic for cambium (AINTEGUMENTA, PtANT) (27). Details about markers for cambial zone tissues are provided in SI Materials and Methods. Expression of the transgene in 6-month-old P. tremula × tremuloides lines (WT and pBpCRE1::AtCKX2 lines 5, 7, 8, 9, and 11) was verified by qPCR analysis for AtCKX2, and reaction products for AtCKX2 and PtTUA2 were run into 2% (w/v) agarose gel for visualization. In qPCR analysis of PttRR7/PttHK3a expression from the stem of WT and lines 7 and 11, the primers for PtTUA2, PtRR7, and PtHK3a were used. Expression studies of PttRR7/PttHK3a and cytokinin analyses were conducted from the same sample material.
Grafting Experiment.
A poplar (P. tremula × tremuloides) reciprocal grafting experiment with WT and pBpCRE1::AtCKX2 line 7 was done essentially as described elsewhere (37). Details are provided in SI Materials and Methods.
Histological Techniques and a Physiological Assay.
Plastic sectioning for anatomical samples (Arabidopsis and poplar) was performed as described elsewhere (20). Histochemical staining for GUS activity was carried out according to ref. 38, except that 20 mM ascorbic acid was added to the assay solution to prevent browning of the tissues. For the cytokinin responsiveness assay, 20 stem segments (Ø 0.5 cm, 1 cm in length) were cut from greenhouse poplars per line (WT, pBpCRE1::AtCKX2 lines 7 and 11, pBpCRE1::GUS), surface sterilized, and grown for 2 months on a 0.5 Murashige and Skoog Basal Medium with 0.5 mg/L IAA and 0, 0.5, 1.5, 2.5, or 15 mg/L t-zeatin.
Cytokinin Analysis.
Cytokinins were analyzed from stem tissue representing the apical part of greenhouse-grown poplar shoot. The 10 topmost internodes were used for analysis, and the shoot apical meristem, leaf nodes, and leaves were removed. Three individual trees were analyzed per line. Cytokinins were extracted, purified, and analyzed by liquid chromatography electrospray ionisation tandem mass spectrometry (LC-ESI-MS/MS) as described elsewhere (39).
Statistical Analysis of Cambial Cell Numbers and Xylem Cell Dimensions.
The number of undifferentiated cambial cells was calculated from 50 cambial cell files from 2 trees per line (Ø 5.5 mm in lines 7 and 11 and Ø 7.3 mm in WT and lines 7 and 11). Xylem cells were macerated according to the method described in ref. 6 from stem samples with Ø 7.3 mm. Two hundred fiber cells and 100 vessel cells were measured from 2 trees per line (WT, line 7, and line 11). The data from 2 trees were combined for statistical analysis. Details of statistical analysis are provided in SI Materials and Methods.
Acknowledgments.
We thank Katja Kainulainen and Kjell Olofsson for excellent technical assistance, Jaakko Kangasjärvi and Raili Ruonala (University of Helsinki) for BpHK3 EST and for advice, Anna Karlberg (Umeå Plant Science Center) for PtANT primers, and Ron Sederoff for comments. This research was supported by the Academy of Finland and Tekes. P.T. and K.D. were supported by the Czech Ministry of Education (MSM 6198959216).
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [Betula pendula CYTOKININ RECEPTOR 1 (BpCRE1) genomic EU583454, cDNA EU583455; HISTIDINE KINASE 2 (BpHK2) cDNA EU583456; HISTIDINE KINASE 3 (BpHK3) cDNA EU583457; and TUBULIN ALPHA (BpTUA) cDNA FJ228477].
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805617106/DCSupplemental.
References
- 1.Uggla C, Mellerowicz EJ, Sundberg B. Indole-3-acetic acid controls cambial growth in Scots pine by positional signaling. Plant Physiol. 1998;117:113–121. doi: 10.1104/pp.117.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuominen H, et al. A radial concentration gradient of indole-3-acetic acid is related to secondary xylem development in hybrid aspen. Plant Physiol. 1997;115:577–585. doi: 10.1104/pp.115.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moyle R, et al. Environmental and auxin regulation of wood formation involves members of the Aux/IAA gene family in hybrid aspen. Plant J. 2002;31:675–685. doi: 10.1046/j.1365-313x.2002.01386.x. [DOI] [PubMed] [Google Scholar]
- 4.Savidge RA. Auxin and ethylene regulation of diameter growth in trees. Tree Physiol. 1988;4:401–414. doi: 10.1093/treephys/4.4.401. [DOI] [PubMed] [Google Scholar]
- 5.Björklund S, Antti H, Uddestrand I, Moritz T, Sundberg B. Cross-talk between gibberellin and auxin in development of Populus wood: Gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J. 2007;52:499–511. doi: 10.1111/j.1365-313X.2007.03250.x. [DOI] [PubMed] [Google Scholar]
- 6.Nilsson J, et al. Dissecting the molecular basis of the regulation of wood formation by auxin in hybrid aspen. Plant Cell. 2008;20:843–855. doi: 10.1105/tpc.107.055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loomis RS, Torrey JG. Chemical control of vascular cambium initiation in isolated radish roots. Proc Natl Acad Sci USA. 1964;52:3–11. doi: 10.1073/pnas.52.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saks Y, Feigenbaum P, Aloni R. Regulatory effect of cytokinin on secondary xylem fiber formation in an in vivo system. Plant Physiol. 1984;76:638–642. doi: 10.1104/pp.76.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Little CH, Odén PC. Control of longitudinal and cambial growth by gibberellins and indole-3-acetic acid in current-year shoots of Pinus sylvestris. Tree Physiol. 1997;17:715–721. doi: 10.1093/treephys/17.11.715. [DOI] [PubMed] [Google Scholar]
- 10.Junghans U, Langenfeld-Heyser R, Polle A, Teichmann T. Effect of auxin transport inhibitors and ethylene on the wood anatomy of poplar. Plant Biol (Stuttg) 2004;6:22–29. doi: 10.1055/s-2003-44712. [DOI] [PubMed] [Google Scholar]
- 11.Inoue T, et al. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- 12.Hwang I, Chen HC, Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002;129:500–515. doi: 10.1104/pp.005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werner T, Motyka V, Strnad M, Schmülling T. Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA. 2001;98:10487–10492. doi: 10.1073/pnas.171304098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner T, et al. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15:2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higuchi M, et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA. 2004;101:8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyawaki K, et al. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA. 2006;103:16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurakawa T, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 18.Riefler M, Novak O, Strnad M, Schmülling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dello Ioio R, et al. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007;17:678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- 20.Mähönen A-P, et al. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mähönen A-P, et al., editors. Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr Biol. 2006;16:1116–1122. doi: 10.1016/j.cub.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 22.Mähönen A-P, et al., editors. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–96. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 23.Nishimura C, et al. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell. 2004;16:1365–1377. doi: 10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakimoto T. Perception and signal transduction of cytokinins. Annu Rev Plant Biol. 2003;54:605–627. doi: 10.1146/annurev.arplant.54.031902.134802. [DOI] [PubMed] [Google Scholar]
- 25.Ito Y, Kurata N. Identification and characterization of cytokinin-signalling gene families in rice. Gene. 2006;382:57–65. doi: 10.1016/j.gene.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 26.Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H. Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiol. 2004;134:1654–1661. doi: 10.1104/pp.103.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrader J, et al. A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell. 2004;16:2278–2292. doi: 10.1105/tpc.104.024190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramírez-Carvajal GA, Morse AM, Davis JM. Transcript profiles of the cytokinin response regulator gene family in Populus imply diverse roles in plant development. New Phytol. 2008;177:77–89. doi: 10.1111/j.1469-8137.2007.02240.x. [DOI] [PubMed] [Google Scholar]
- 29.Werner T, Köllmer I, Bartrina I, Holst K, Schmülling T. New insights into the biology of cytokinin degradation. Plant Biol (Stuttg) 2006;8:371–381. doi: 10.1055/s-2006-923928. [DOI] [PubMed] [Google Scholar]
- 30.Yang S, Yu H, Xu Y, Goh CJ. Investigation of cytokinin-deficient phenotypes in Arabidopsis by ectopic expression of orchid DSCKX1. FEBS Lett. 2003;555:291–296. doi: 10.1016/s0014-5793(03)01259-6. [DOI] [PubMed] [Google Scholar]
- 31.Galuszka P, et al. Cytokinin oxidase/dehydrogenase genes in barley and wheat: Cloning and heterologous expression. Eur J Biochem. 2004;271:3990–4002. doi: 10.1111/j.1432-1033.2004.04334.x. [DOI] [PubMed] [Google Scholar]
- 32.Galuszka P, et al. Biochemical characterization of cytokinin oxidases/dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabacum L. J Plant Growth Regul. 2007;26:255–267. [Google Scholar]
- 33.Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol. 1957;11:118–130. [PubMed] [Google Scholar]
- 34.Ward SP, Leyser O. Shoot branching. Curr Opin Plant Biol. 2004;7:73–78. doi: 10.1016/j.pbi.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Keinonen-Mettälä K, Pappinen A, von Weissenberg K. Comparisons of the efficiency of some promoters in silver birch (Betula pendula) Plant Cell Rep. 1998;17:356–361. doi: 10.1007/s002990050406. [DOI] [PubMed] [Google Scholar]
- 36.Nilsson O, et al. Spatial pattern of cauliflower mosaic virus 35S promoter-luciferase expression in transgenic hybrid aspen trees monitored by enzymatic assay and non-destructive imaging. Transgenic Res. 1992;1:209–220. [Google Scholar]
- 37.Ruonala R, Rinne PL, Kangasjärvi J, van der Schoot C. CENL1 expression in the rib meristem affects stem elongation and the transition to dormancy in Populus. Plant Cell. 2008;20:59–74. doi: 10.1105/tpc.107.056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jefferson RA, Kavanagh TA, Bewan MW. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nordström A, et al., editors. Derivatization for LC-electrospray ionization-MS: A tool for improving reversed-phase separation and ESI responses of bases, ribosides, and intact nucleotides. Anal Chem 7. 2004;6:2869–2877. doi: 10.1021/ac0499017. [DOI] [PubMed] [Google Scholar]