Abstract
Dynein is a microtubule motor that powers motility of cilia and flagella. There is evidence that the relative sliding of the doublet microtubules is due to a conformational change in the motor domain that moves a microtubule bound to the end of an extension known as the stalk. A predominant model for the movement involves a rotation of the head domain, with its stalk, toward the microtubule plus end. However, stalks bound to microtubules have been difficult to observe. Here, we present the clearest views so far of stalks in action, by observing sea urchin, outer arm dynein molecules bound to microtubules, with a new method, “cryo-positive stain” electron microscopy. The dynein molecules in the complex were shown to be active in in vitro motility assays. Analysis of the electron micrographs shows that the stalk angles relative to microtubules do not change significantly between the ADP·vanadate and no-nucleotide states, but the heads, together with their stalks, shift with respect to their A-tubule attachments. Our results disagree with models in which the stalk acts as a lever arm to amplify structural changes. The observed movement of the head and stalk relative to the tail indicates a new plausible mechanism, in which dynein uses its stalk as a grappling hook, catching a tubulin subunit 8 nm ahead and pulling on it by retracting a part of the tail (linker).
Keywords: electron microscopy, molecular motor
Dynein is a motor protein that hydrolyses ATP and moves toward the minus end of a microtubule (MT). A dynein molecule has 1 to 3 heavy chains, each consisting of 3 domains: a head, a stalk and a tail. ATP is bound and hydrolysed in the head, which has a ring-like structure composed of 6 AAA+ domains. The stalk is an anti-parallel coiled-coil, 10–15 nm long, and has a nucleotide-dependent MT-binding domain at the tip (1). In axonemes, the tail of dynein is fixed on the A-tubule of a doublet MT, together with several intermediate and light chains, whereas the MT-binding domain of the stalk interacts cyclically with the B-tubule of a neighbouring doublet MT. It has been proposed that the nucleotide-dependent binding affinity of the tubulin-binding site at the tip of the stalk is modulated by the 2 alpha helices in the coiled-coil sliding over each other (2). However, it is not known how structural changes that occur in the head during the ATPase cycle are transmitted to the stalk and tail to produce movement of the A-tubule relative to the B-tubule.
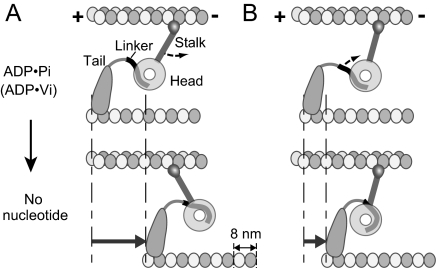
In images obtained by negative stain electron microscopy (EM) of dynein molecules (dynein c) isolated from Chlamydomonas axonemes, the head and stalk appeared to rotate with respect to the tail between different nucleotide states: the stalk and tail project from the head close together in the no-nucleotide state, but ≈90° apart in the ADP·vanadate (Vi) state (3) (see Fig. 1). A subdomain of the tail, called the linker, docks on the surface of the head, and changes in this interaction were proposed to cause the observed positional change of the tail (3, 4). Movement of the tail relative to the head was also detected in fluorescence resonance energy transfer analysis of cytoplasmic dynein (5, 6). Assuming that the observed ADP·Vi and no-nucleotide structures correspond to prepowerstroke and postpowerstroke conformations, respectively, the simplest model based on these results is that the stalk acts as a lever arm (3, 4, 7–9), binding to a MT initially with a tilt toward the minus end, and then rotating toward the plus end to move the MT (Fig. 1A). However, the observed structural differences can also cause movement of a MT without changing the stalk angle relative to the MT, because the tail moves closer to the stalk in the postpowerstroke conformation (Fig. 1B). In the latter model, the stalk is used as a tether, rather than a lever arm (7), and dynein moves by shortening the distance between its head/stalk and the tail-MT attachment.
Fig. 1.
Two possible models based on the structural changes observed with Chlamydomonas dynein c (3). (A) The stalk rotates relative to the MT to pull the MT. (B) The stalk angle is constant, but the movement of the tail with respect to the head and stalk causes relative sliding of the MTs. The resulting MT movement is expected to be smaller in B, but it seems to be enough to produce a shift of ≈8 nm. Note that a longer part of the linker is exposed in the ADP·Pi state than in no-nucleotide.
To test these models, the most direct way would be to observe the angles of the stalks bound to MTs in the two nucleotide states. However, visualizing MT-bound stalks has been difficult. The only reports that clearly showed the MT-bound stalks, so far, are with the quick-freezing, deep-etching (QFDE) techniques. The QFDE images of dynein in axonemes in the absence of nucleotide and with ATP both showed stalks either perpendicular to the MTs or slightly tilted, and the reported angles were not constant (10–14). These results seem to be inconsistent with the stalk-rotating model, but could be due to an artifact of QFDE methods, because QFDE images of muscle fibers showed swelling of the filament lattice, so that myosin cross bridges observed by this method looked almost perpendicular to the filament axis (15). More recently, 3D structures of dynein in axonemes or in the dynein-MT complex were reported by cryo-EM (9, 16–18), but neither of these studies clearly showed the stalks.
Results and Discussion
Visualization of Stalks in a Dynein-MT Complex.
Do stalks really rotate relative to the MTs? To answer this question, we studied the structures of dynein molecules bound to in vitro polymerized MTs. Outer-arm dynein molecules purified from sea urchin sperm flagella were mixed with MTs and observed by EM. Although dynein from some species of sea urchin bound randomly to MTs, others showed patches of regularly bound dynein [supporting information (SI) Fig. S1], such as those observed in previous reports (19–21). The periodical binding patterns were seen mostly between MTs, bundled by dynein cross-bridges. A typical bundle consisted of two to several MTs. We first studied the structure of the complexes by negative stain EM, but found that the dynein stalks were difficult to see (Fig. S2A), probably because a pool of uranyl acetate between two MTs (≈25 nm deep) obscured the stalk structures. By unstained cryo-EM, stalks were visible for some molecules, but the overall contrast of images was low (Fig. S2B).
To see the stalks more clearly, we developed a new method, modifying “cryo-negative staining” (22). The EM specimens were treated with a low concentration of uranyl acetate just before freezing, and examined by conventional cryo-EM methods. The pH of the uranyl acetate solution we used (≈4.8) was low enough that the protein structures should be fixed instantly by uranyl acetate addition. The resulting images (Fig. 2A) show that, although the overall structures and arrangements of dynein and MTs are similar to those observed by negative stain or conventional cryo-EM, the contrast is greatly improved, and stalks of many molecules are now visible. Because this method resulted in positive staining of the proteins, unlike the case of cryo-negative stain, we call it “cryo-positive staining.”
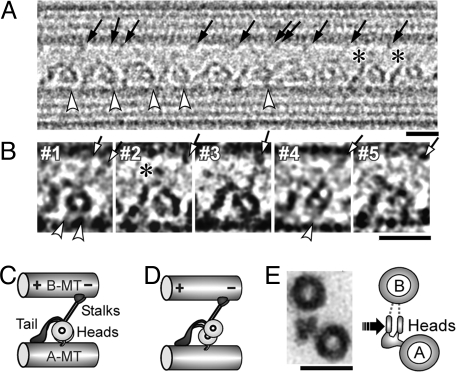
Fig. 2.
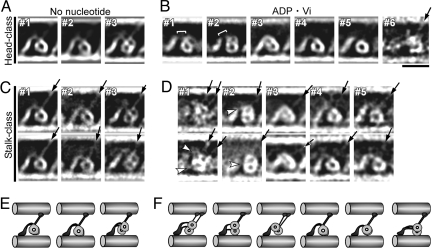
Dynein-MT complex in the no-nucleotide state. (A and B) Cryo-positive stain EM images of dynein purified from S. nudus, bound to MTs in the presence of apyrase. The B-MT minus end is at Right. Dynein molecules are regularly arranged in a single layer between 2 MTs. Individual dynein molecules may show a single ring (B, #1–3), a double ring (B, #4), or be unclear (B, #5). Stalks (arrows), the head-A-MT tethers (white arrowheads), and extra densities on the top of the tails (asterisks) are indicated. Some molecules show 2 stalks. (C and D) Interpretation of the single-ring and double-ring images. (E) Cross sections of the dynein-MT complex embedded in Epon812, with our interpretation of the images. The likely viewing direction of our cryo images is indicated. The micrographs in A, B, and Fig. 4 were Gaussian-filtered to reduce noise. (Scale bars: A and B, 20 nm; C, 50 nm.)
Structure of the Dynein-MT Complex in the No-Nucleotide State.
The cryo-positive stain images of the dynein bound to MTs in the presence of apyrase (we call this the no-nucleotide state) (Fig. 2 A and B) show structures consistent with previously reported dynein c images (3), with a ring-like head, and the stalk and tail emerging from points close to each other. The stalk binds to one of the MTs, which we call the “B-MT,” by analogy to the B tubule of a doublet MT in axonemes, and the tail associates with the other (the “A-MT”) (Fig. 2C). In this article, all figures that show longitudinal views of MTs are presented with the B-MT at the top. Although an outer arm dynein molecule from sea urchin sperm has 2 (α and β) heavy chains, and some molecules do show 2 distinct rings in an “8” shape (Fig. 2B, #4; Fig. 2D), many others show only one, presumably because the two rings are superimposed (Fig. 2B, #1–3; Fig. 2C). The percentages of single- and double-ring appearances were 58% and 16% (n = 932), respectively (see Fig. 4C). The rest showed intermediate structures or did not show clear ring shapes (Fig. 2B, #5). At first sight, the superimposed rings appear different from the previous reports of cryoelectron tomography structures (16, 17), where 2 or 3 rings, like stacked plates (see Fig. 2E), were separately observed. However, we think the difference is simply due to the viewing directions: many of the MTs in our images seem to lie on the EM grids so that the view is parallel to the line through the centers of the rings (arrows in Figs. 2 E and S3D). The stalks, too, seem to be superimposed, so that many of the molecules show a single stalk structure (Fig. 2A). Some others show two separate stalks, but with lower contrast, suggesting that a strong “single stalk” is actually a superposition of two stalks.
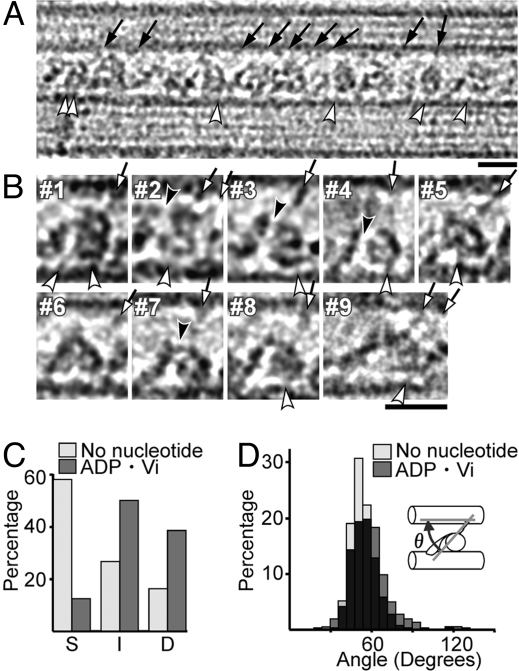
Fig. 4.
Structural changes of dynein with ADP·Vi. (A and B) Cryo-positive stain EM images in the ADP·Vi state, with the B-MT plus end at Left. Individual dynein images (B) show a double ring (#1–6), a single ring (#7), or an intermediate conformation (#8). Most stalks are clearly tilted toward the B-MT minus end (arrows). (C) Populations of dynein molecules in the no-nucleotide and ADP·Vi states showing a superimposed, single ring (S), a double ring (D), or, intermediate or unclear structures (I). (D) Distribution of stalk angles (θ) with respect to MTs. The averaged angles are 54.0 ± 8.7° (n = 451) and 58.6 ± 14.6° (n = 490) for the no-nucleotide and ADP·Vi states, respectively. (Scale bars: 20 nm.)
A notable feature of our images is that a clear view of the stalks always shows them uniformly tilted (Fig. 2A, B, arrows). How does the tilt relate to the polarities of the MTs? We determined many MT polarities from the Moiré patterns of the protofilaments (23), and found that, relative to the tails, the dynein heads and stalks are always closer to and point toward the B-MT minus end (Fig. 3). Our unstained cryo-EM images also show stalks tilted toward the minus end (Fig. S2B), indicating that the observed tilt is not an artifact of uranyl acetate (15) used in our cryo-positive stain procedure. The finding is rather unexpected, because a lever-arm model would predict the stalks in the “postpowerstroke,” no-nucleotide conformation to be tilted toward the plus end (3).
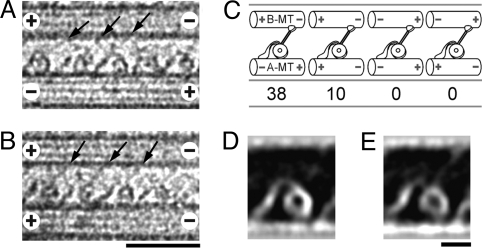
Fig. 3.
Orientation of dynein relative to the MT polarity. (A and B) Cryo-positive stain EM images of dynein cross-bridging anti-parallel (A) or parallel (B) MTs. (C) Proportion of MT pairs with different polarities. Although the polarity of the A-MT is not uniform, all of the dynein molecules are oriented in the same way with respect to the B-MT, with the heads and stalks to the B-MT minus-end side of the tail. The observed head/tail arrangement agrees with that suggested from the QFDE images of dynein in axonemes (11, 12), but inconsistent with the assignment in a recent report on the Chlamydomonas dynein-MT complex (18). (D and E) Averaged images of dynein cross-bridging 2 anti-parallel MTs (D; n = 30) or parallel MTs (E; n = 31), showing no detectable differences. (Scale bars: A and B, 50 nm; D and E, 10 nm.)
Besides the stalks, some images show an extra density extending toward the B-MT from the top of the tail, at a position close to the head (Fig. 2, asterisks). This density does not seem to attach to the MTs and may correspond to one of the light or intermediate chains.
Unlike in axonemes where all doublet MTs have the same polarities, the polarity of A-MTs in our dynein-MT complex was not uniform, and it was more often anti-parallel to the B-MT (Fig. 3C). Cross-bridging of both parallel and anti-parallel MTs was also reported for Tetrahymena outer-arm dynein (24), and seems to happen when dynein is added to preassembled tubulin. To confirm that the dynein molecules in our dynein-MT complex are active in motility, we performed in vitro motility assays. When 1 mM ATP was added to the complex prepared in the same way but using tetramethylrhodamine-labeled MTs, the MTs started sliding along and out of the complex (Movie S1). The averaged velocity (0.64 ± 0.36 μm) was slower than that by the dynein molecules absorbed to the glass surface (3.86 ± 0.75 μm), probably because attachment of the tail to the MT is weak when isolated dynein molecules are added to preassembled MTs. However, MT sliding occurred in all of the complexes we observed (n = 28), showing that the dynein molecules in our dynein-MT complex are functional as motors.
We have also checked to see whether the structures of the dynein molecules between anti-parallel MTs show any differences from those between parallel MTs. First, the averaged images showed no detectable differences (Fig. 3 D and E). They also agreed well with the recently reported 3D structures of a two-headed mutant dynein in Chlamydomonas axonemes (17) (Fig. S3). These results indicate that the polarity of the A-MT has little effect on the structure of the dynein-B-MT complex, and that the structural changes observed here correspond to those of active motors.
Structure of the Dynein-MT Complex in the ADP·Vi State.
How does the dynein-MT complex change its structure with bound nucleotide? We added ATP and vanadate, which are thought to produce a weak binding, ADP·Vi state (25). To capture the complex before the detachment of dynein from MTs, we applied a method described by Frado and Craig (1992) (26), in which ATP·Vi solution and uranyl acetate, both drawn into a pipet tip but separated by an air gap, were squirted on to the sample on an EM grid. In this way, the dynein-MT complex was briefly exposed to ATP·Vi before being fixed by uranyl acetate. The time of exposure to ATP·Vi solution was 120–200 ms (see Methods). Because the MT-activated ATPase rate measured for outer arm dynein from Strongylocentrotus nudus, which we used for most of the EM studies, is 34.5 ± 15.0/s (n = 6) (M. Wada and C.S., unpublished data), and because vanadate was reported to block steady state hydrolysis of ATP immediately after a single turnover of ATP (25), we think that the exposure to the ATP·Vi solution was long enough to put most of the dynein molecules into the ADP·Vi state.
The resulting images (Fig. 4A) show clear differences from those in the no-nucleotide state, although they are more variable than those images. The variability cannot be explained by tilting of some of the MTs out of the plane of the EM grid, because, assuming the arrangement of the rings as in Fig. 2E, a 4-nm relative shift of the rings would require a tilt of the MTs by as much as ≈22°. The most obvious difference between the two nucleotide states is that the majority of the ADP·Vi molecules show double-ring or intermediate conformations (Fig. 4C). The minor, single-ring population in the ADP·Vi images (12%, n = 1,048) and the double-ring population in the no-nucleotide images (16%, n = 932) may both be due to non-uniformity of the nucleotide states; for example, added nucleotides may not have bound to some molecules. Variability in the nucleotide states of the second to the forth AAA subunits may also cause structural non-uniformity (27, 28). A high degree of structural variability in the presence of ATP was also reported for dynein in axonemes (13), and for the Chlamydomonas dynein-MT complex, where 1 of the 3 heads was not visible in the averaged ATP·Vi image (18).
The stalks, too, are more variable in the ADP·Vi images, but they still seem to be tilted toward the B-MT minus end (Fig. 4 A and B, arrows), with a range of angles similar to that in the no-nucleotide states (Fig. 4D). These results suggest that dynein moves MTs without rotating the stalk. Although the stalk angles do not change between the two states, the tails seem to change shape in many of the molecules: In the no-nucleotide state, tails show clear, smoothly curved shapes (Fig. 2 A and B, #1–3); but in the ADP·Vi state they are often like a fishing rod, from which thin, thread-like structures (Fig. 4B, black arrowheads) emerge and connect to the heads. We think that these thread-like structures correspond to the linkers that are proposed to interact with different parts of the head in different nucleotide states (3, 4, 29). Our results indicate that the linkers are exposed more in the ADP·Vi state, possibly because of the changes in the linker-head interaction (Fig. 1).
Another interesting feature observed in both nucleotide states is short filamentous structures connecting the ring to the A-MT (Figs. 2 and 4, white arrowheads). It was proposed that linkage of heads to A-tubules may be important in force production (30). The tethering structures observed here might be one of the components that associate with dynein heavy chains, as proposed for Chlamydomonas dynein (31). Alternatively, they could be the C termini of tubulin subunits interacting with dynein heads. The presence of more than one tether on some molecules supports the latter idea. Such interactions could make it possible for the dynein head to shift its position along the A-MT without completely dissociating from it.
Structural Changes of Dynein Observed in Averaged Images.
To analyze the structural differences statistically, we classified the dynein images by a single particle method (32). When classified according to the head structures, the averaged images of the two major classes in the no-nucleotide state (Fig. 5A, #1 and #2), which include 69% and 24% of the original images, respectively, each show a single ring close to the A-MT. Minor differences between these classes may be due to interaction of the heads with different protofilaments of the A-MT. The shapes of the rings resemble previously reported images of monomeric, inner-arm dynein c in the no-nucleotide state (3) (Fig. 6 A and B), indicating that both rings of an outer-arm dynein have the same orientation. Further classification of the images according to the stalks clearly reveals the stalk positions (Fig. 5C). Confirming the observation of individual images, all of the class-averages in the no-nucleotide state show the stalks tilted toward the B-MT minus end (Fig. 5C, arrows).
Fig. 5.
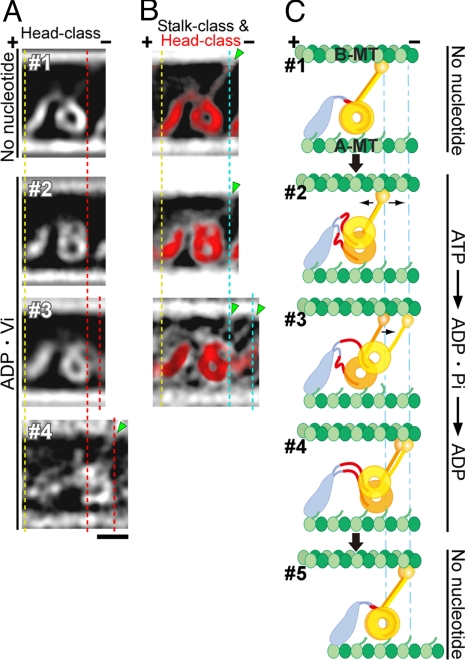
Averaged images after classification. Shown are no-nucleotide (A and C) and ADP·Vi (B and D) averages. A total of 1,062 no-nucleotide images and 988 ADP·Vi images were classified first according to the heads (A and B) and then according to the stalks (C and D). Protein is white. Two selected stalk-classes are shown below each head-class. The numbers of images in averages are: 736, 257, and 69 for #1–3 (A); 188, 144, 74, 73, 72, and 6 for #1–6 (B); between 19 and 269 (C); and 9–40 (D). (E and F) Interpretation of each class-average. Stalks (arrows), and a low-density region between the tail and head (bracket) containing linkers (white arrowheads) are indicated in some images. (Scale bar: 20 nm.)
Fig. 6.
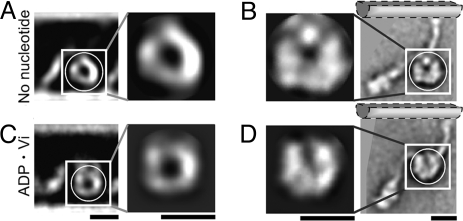
Comparison with Chlamydomonas dynein c images. Our class-averages of sea urchin dynein (A and C) are compared with images from figure 4 of ref. 3 (B and D). (A and B) No-nucleotide state. (C and D) ADP·Vi state. Cross correlation between the masked regions showed that the head regions of the two images agreed best when the dynein c images were rotated as shown in B and D. The hypothetical positions of the B-MT, based on our images with MTs, are indicated. (Scale bars: 10 nm.) [Reproduced with permission from ref. 3 (Copyright 2003, Nature Publishing Group).]
The ADP·Vi images are more variable and it was difficult to sort them into a small number of classes. In major classes where a double ring is observed, one of the two rings stays close to the A-MT, whereas the other ring moves up either to the top-right, top, or top-left of the first head (Fig. 5B, #1–3). In all three classes, the second ring was shifted closer to the B-MT by ≈5 nm, when measured in the 2D images. This does not necessarily mean shortening of the stalks, because the ring may also move in the direction perpendicular to the plane of the figure, which is not detected in the current study. The observed shift is similar to the 3.7-nm movement reported for the Chlamydomonas dynein β heavy chain (18). Our results also agree with the QFDE images of sea urchin axonemes (11), which showed 2, upper and lower, head domains in the presence of ATP and one large globular head domain in the no-nucleotide state (Fig. S4). The most major two classes (Fig. 5B, #1 and #2) of our ATP averages are also the closest images to the QFDE images with ATP, indicating that these classes are representative of the ADP·Pi structures in vivo. The shift of one of the heads suggests that the two heads of a dynein molecule may be in different biochemical states. Although we do not know which of the two heavy chains the observed rings correspond to, the results are consistent with the idea that the two heads of an outer arm dynein have different functions.
Small movements in the stalk add further variability to the ADP·Vi images so that we had to sort the images into many small classes to see clear average images. As in the individual images (Fig. 4 A and B), most of the class averages show the stalks tilted to the minus end (Fig. 5D, arrows), at an angle similar to that in the no-nucleotide state. Some ADP·Vi averages show two stalks nearly parallel to each other. In QFDE studies, the apparent emergence point of the stalk from the heads shifted toward the MT minus end in the presence of ATP (11, 12). The positions of the stalks in our averages are variable, but some of them, especially those in the major two head classes, show at least one of the stalks emerging from the minus end side of the head (Fig. 5D, #1 and #2).
Positional changes are also seen in the tails. Whereas the emergence point of the tail is at the top of the ring in the no-nucleotide state, it appeared to have rotated anti-clockwise in ADP·Vi, so that the stalk and tail are further apart (Fig. 5D, #1 and #2). This is consistent with the changes reported for dynein c (3). As seen in the individual images, some averages show one or two linking structures between the heads and the main part of the tail (Fig. 5D, white arrowheads), which we think correspond to a part of the linker. Exposing more linker would result in a larger distance between the heads and the main part of the tail in the ADP·Vi state. In fact, some classes show a low-density region between the heads and tail, and the heads appear to be shifted toward the B-MT minus end (Fig. 5B, #1; Fig. 7A). Some other classes did not show a minus-end directed shift, but this may be because the weak attachment between the tail and the A-MT in our complex cannot always sustain tension produced by head movement. Structural variability in the tails makes it difficult to determine the precise amount of shift. However, in extreme cases, the heads seem to be shifted by >10 nm, noticeably extending the tail (Fig. 4B, #9 and Fig. 5B, #6). An approximately 12-nm shift of the heads toward the minus end, relative to the A-tubule attachment site, in the presence of ATP was reported for QFDE images of axonemes (11, 13). Our results indicate that the heads move these large distances without a change in the stalk angles.
Fig. 7.
Possible mechanism for dynein movement. (A) Head-class averages characteristic of each nucleotide state are selected from Fig. 5 A and B, and aligned by their A-MT attachment sites (yellow dotted line). In #2 and #3, at least one ring is shifted toward the minus end (red dotted line). (B) Stalk-class averages are superimposed on the head-class averages (red) shown in A to show both stalks and tails clearly. The stalks do not rotate but, together with their heads, shift ≈8 nm (cyan dotted line) toward the minus end in some ADP·Vi averages. (C) A model for dynein stepping (see “Mechanism of Motility” in Results and Discussion). (Scale bar: 10 nm.)
Mechanism of Motility.
The most important finding in our work is that the angle of a bound stalk with respect to the MT changes little between the ADP·Vi and no-nucleotide states. We compared our averaged structures of sea urchin dyneins with the reported dynein c images (3), by shifting and rotating them to find the best correlation. The shapes of the dynein heads appear most similar when the dynein c images are rotated as shown in Fig. 6. With this rotation, the stalks in each nucleotide state have similar angles, both tilting to the minus end. The results disagree with models such as in Fig. 1A, in which the stalk acts as a lever arm, amplifying small structural changes that occur within the head into a large displacement of the MT.
Then, how does dynein pull on a MT? One clue comes from the observed movements of the heads and stalks with respect to the tail. First, compared with heads with no-nucleotide, the heads in some ADP·Vi images were shifted toward the MT minus end by as much as ≈10 nm (Fig. 7A). This movement is probably caused by the change in the way the linker interacts with the head, which separates the emergence points of the stalk and the tail (3, 4, 29), and, at the same time, could increase the net length of the tail (including the linker) (Fig. 1). The separation of the emergence points may cause a clockwise rotation of the ring, and in fact, the point where the stalk emerges from the head appears to shift toward the minus end in our images (Fig. 7B) and in the previous QFDE work (11, 13). The dynein c images oriented as in Fig. 6 B and D also show an apparent shift of the stalk emergence point. However, the rotation does not change the stalk angle relative to the MTs, indicating that movement is produced within the dynein head/tail region, whereas the MT binding domain on the end of the stalk is used like a grappling hook on the end of a tether, to catch and pull on the MT (Fig. 1B).
In the case of the two-headed, outer-arm dynein, each head seems to move using basically the same mechanism. Fig. 7C summarizes a model based on our results plus the previous structural studies. ATP binding to the heads weakens the interaction between the stalk tips and the B-MT (2, 6) so that the stalks are free to slide along the B-MT. ATP-induced structural changes in each head also cause changes in the way in which the linker docks (Fig. 7C, #2) (3–6, 29). As discussed above, this results in (i) separation of the emergence points of the stalk and tail, and (ii) exposure of a longer piece of the linker (red), which both cause increase of the maximum distance between the tail-MT attachment and the stalk, so that the heads can now reach further away. Using the elongated leash, one of the ATP-bound heads (yellow) lifts away from the A-MT and searches for the next binding site on the B-MT (Fig. 7C, #2). Movement of the head away from the A-MT probably accompanies tilt and/or shift out of the plane, so that the length of the stalk can stay constant. After this head binds to the next site (Fig. 7C, #3), the other head (orange) of the dimer also moves toward the B-MT minus end, but this head stays close to the A-MT. This must correspond to the head nearer to the A-MT in Fig. 2E. The observed head-MT tethers, possibly corresponding to the tubulin C terminus, may enable this head to slide along the A-MT while being weakly tethered to it. Once both stalks have bound tightly to the next specific sites on the B-MT (Fig. 7C, #4), structural information transmitted to the heads causes ADP release and redocking of the linkers to the original positions. Because the linkers are retracted and rebind tightly to the heads and the distance between the tail-MT attachment and the head is reduced, the dynein molecule pulls the bound A-MT toward the B-MT minus end (Fig. 7C, #5). Because the heads are positioned on the minus end side of the tail, searching for the next tubulin site is restricted mainly to this direction, but a backward step can also be explained with the same model. Further studies will be necessary to understand which of the two heads corresponds to the α or β heavy chain of the heterodimeric dynein, and how these two heads cooperate to produce force in an axoneme.
The amount of movement expected from this model is consistent with the 8-nm steps that are most commonly observed (33–35). However, larger steps that are occasionally observed can also be explained if a longer part of the linker undocks from the head in the ADP·Pi state. A key point of our proposed model is that motility is caused by changes in the net length of the tail (the distance between the tail-MT attachment and the head), due to changes in the linker-head interaction, and the stalk is used as a tether rather than an active lever arm. It will now be important to study the structural changes in the linker/tail region in more detail, preferably in 3D.
Materials and Methods
Specimen Preparation.
Outer arm dynein was purified from sea urchin (Pseudocentrotus Depressus, Hemicentrotus pulcherrimus, Strongylocentrotus intermedius or S. nudus) sperm flagella as described (27, 36). S. nudus was used for most of the studies. Axonemes were suspended in 0.6 M NaCl buffer (0.6 M NaCl, 2 mM MgSO4, 2 mM glycoletherdiamine-N,N,N′,N′-tetra-acetic acid (EGTA), 1 mM dithiothreitol (DTT), 10 mM Hepes, pH 7.8) and kept on ice for 10 min and centrifuged at 17,000 × g for 10 min. The 0.6 M NaCl extract was centrifuged on a 5% to 20% (wt/vol) sucrose linear density gradient made up in the solution containing 200 mM NaCl, 2 mM MgSO4, 2 mM EGTA, 1 mM DTT, and 10 mM Hepes (pH 7.8), at 190,000 × g for 10 h. After fractionation, the peak fraction obtained from the 0.6 M NaCl extract was used as purified 21 S dynein. MTs were polymerized using pig brain tubulin (37), stabilized with taxol, centrifuged, and resuspended in a solution without GTP (38).
Electron Microscopy.
Dynein was mixed with MTs to give final concentrations of 35 μg/ml and 70 μg/ml, respectively, in a motility buffer [50 mM potassium acetate, 5 mM magnesium acetate, 2 mM EGTA, 0.1 mM EDTA, 20 mM Hepes (pH 7.8)], and incubated for 10 min at room temperature. For the no-nucleotide state, 5 U/ml apyrase (final concentration) was added during incubation. The mixture was then applied onto EM grids coated with a holey carbon film. After removal of the excess solution, a drop of 0.01–0.05% uranyl acetate was added to the mixture, and the grid was rapidly frozen with an ethane slush.
To observe the dynein-MT complex in the ADP·Vi state, ATP and vanadate were added just before fixation with uranyl acetate, using the method described by Frado and Craig (26). First, the dynein-MT complex in motility buffer was applied to a grid. Eighty μl 0.03% uranyl acetate, air, and 20 μl ATP·Vi solution (motility buffer plus 100 μM ATP and 50 μM vanadate) were drawn sequentially into a 200 μl pipet tip, and the contents were squirted onto the grid. In this way, the time interval between addition of ATP·Vi solution and uranyl acetate fixation was 120–200 ms (measured by imaging the pipet tip using a back-illuminated EMCCD camera (Andor Technology), with a time rate of 100 frames per sec).
The grids were examined using a cryo holder (626, Gatan) in a Tecnai F20 electron microscope (FEI) operated at 200 kV. The images were photographed on Kodak SO-163 film at a magnification of 50,000× with a defocus of 4000–4500 nm, and digitized in 7-μm steps using a Zeiss Phodis scanner.
Image Analysis.
First, polarities were determined for 48 pairs of MTs, by checking their Moiré patterns (23). Because we found that the dynein heads are always to the B-MT minus end side of the tail, we used the dynein orientation to assign polarities to the rest of the images. The image sizes were reduced so that a pixel size is 14 μm (corresponding to 2.75Å on the samples). After correction for the contrast transfer function, 1,062 and 988 image segments, each containing a dynein molecule and parts of 2 MTs, were extracted from the images of no-nucleotide and ADP·Vi states, respectively.
The segments were aligned by reference-free algorithms and classified into groups using 2 regions of each dynein molecule, the head and stalk structures, in order. For the first classification based on the arrangement of the two heads, the segments were low-pass filtered and the image area containing dynein heads was emphasized in contrast. The images were then classified by hierarchical cluster analysis. After the initial classification, each segment without low-pass filtering was cross-correlated with the class-averaged image, and only the images with a high correlation coefficient (with >0.65 cross-correlation coefficients) were included to calculate refined class-averages. This refinement procedure was repeated 3 times to produce the final averaged images. For the second step, the images belonging to each head-class were further classified according to the shapes of the stalks using a similar method. To compare our images with the published images, the regions including the heads were extracted from both images, and cross-correlation scores were calculated allowing translation and rotation.
All of the image analysis was performed by using the Extensible and Object-oriented System (Eos) software (32).
Supplementary Material
Acknowledgments.
We thank L. Amos for valuable comments on the manuscript; K. Katoh for EMCCD imaging; Y. Hachikuba for help in motility assays; T. Ishikawa and K. Oiwa for providing their 3D structures of Chlamydomonas dynein; J. Heuser, U. Goodenough, and D. Nicastro for discussion; A. Yoshimura and I. Nakano for contribution in dynein preparation; and K. Yan for technical assistance. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808194105/DCSupplemental.
References
- 1.Gee MA, Heuser JE, Vallee RB. An extended microtubule-binding structure within the dynein motor domain. Nature. 1997;390:636–639. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- 2.Gibbons IR, et al. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J Biol Chem. 2005;280:23960–23965. doi: 10.1074/jbc.M501636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 4.Burgess SA, Knight PJ. Is the dynein motor a winch? Curr Opin Struct Biol. 2004;14:138–146. doi: 10.1016/j.sbi.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. ATP hydrolysis cycle-dependent tail motions in cytoplasmic dynein. Nat Struct Mol Biol. 2005;12:513–519. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- 6.Imamula K, Kon T, Ohkura R, Sutoh K. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc Natl Acad Sci USA. 2007;104:16134–16139. doi: 10.1073/pnas.0702370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gee M, Vallee R. The role of the dynein stalk in cytoplasmic and flagellar motility. Eur Biophys J. 1998;27:466–473. doi: 10.1007/s002490050157. [DOI] [PubMed] [Google Scholar]
- 8.Asai DJ, Koonce MP. The dynein heavy chain: structure, mechanics and evolution. Trends Cell Biol. 2001;11:196–202. doi: 10.1016/s0962-8924(01)01970-5. [DOI] [PubMed] [Google Scholar]
- 9.Mizuno N, Narita A, Kon T, Sutoh K, Kikkawa M. Three-dimensional structure of cytoplasmic dynein bound to microtubules. Proc Natl Acad Sci USA. 2007;104:20832–20837. doi: 10.1073/pnas.0710406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodenough UW, Heuser JE. Substructure of the outer dynein arm. J Cell Biol. 1982;96:798–815. doi: 10.1083/jcb.95.3.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sale WS, Goodenough UW, Heuser JE. The substructure of isolated and in situ outer dynein arms of sea urchin sperm flagella. J Cell Biol. 1985;101:1400–1412. doi: 10.1083/jcb.101.4.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burgess SA, Dover SD, Woolley DM. Architecture of the outer arm dynein ATPase in an avian sperm flagellum, with further evidence for the B-link. J Cell Sci. 1991;98:17–26. doi: 10.1242/jcs.98.1.17. [DOI] [PubMed] [Google Scholar]
- 13.Burgess SA. Rigor and relaxed outer dynein arms in replicas of cryofixed motile flagella. J Mol Biol. 1995;250:52–63. doi: 10.1006/jmbi.1995.0357. [DOI] [PubMed] [Google Scholar]
- 14.Lupetti P, et al. Three-dimensional reconstruction of axonemal outer dynein arms in situ by electron tomography. Cell Motil Cytoskeleton. 2005;62:69–83. doi: 10.1002/cm.20084. [DOI] [PubMed] [Google Scholar]
- 15.Heuser JE, Cooke R. Actin-myosin interactions visualized by the quick-freeze, deep-etch replica technique. J Mol Biol. 1983;169:97–122. doi: 10.1016/s0022-2836(83)80177-6. [DOI] [PubMed] [Google Scholar]
- 16.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa T, Sakakibara H, Oiwa K. The architecture of outer dynein arms in situ. J Mol Biol. 2007;368:1249–1258. doi: 10.1016/j.jmb.2007.02.072. [DOI] [PubMed] [Google Scholar]
- 18.Oda T, Hirokawa N, Kikkawa M. Three-dimensional structures of the flagellar dynein-microtubule complex by cryoelectron microscopy. J Cell Biol. 2007;177:243–252. doi: 10.1083/jcb.200609038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hisanaga S, Hirokawa N. Substructure of sea urchin egg cytoplasmic dynein. J Mol Biol. 1987;195:919–927. doi: 10.1016/0022-2836(87)90495-5. [DOI] [PubMed] [Google Scholar]
- 20.Moss AG, Sale WS, Fox LA, Witman GB. The alpha subunit of sea urchin sperm outer arm dynein mediates structural and rigor binding to microtubules. J Cell Biol. 1992;118:1189–1200. doi: 10.1083/jcb.118.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokota E, Mabuchi I. C/A dynein isolated from sea urchin sperm flagellar axonemes. Enzymatic properties and interaction with microtubules. J Cell Sci. 1994;107:353–361. [PubMed] [Google Scholar]
- 22.Adrian M, Dubochet J, Fuller SD, Harris JR. Cryo-negative staining. Micron. 1998;29:145–160. doi: 10.1016/s0968-4328(97)00068-1. [DOI] [PubMed] [Google Scholar]
- 23.Chrétien D, Kenney JM, Fuller SD, Wade RH. Determination of microtubule polarity by cryo-electron microscopy. Structure. 1996;4:1031–1040. doi: 10.1016/s0969-2126(96)00110-4. [DOI] [PubMed] [Google Scholar]
- 24.Warner FD, Mitchell DR. Polarity of dynein-microtubule interactions in vitro: cross-bridging between parallel and antiparallel microtubules. J Cell Biol. 1981;89:35–44. doi: 10.1083/jcb.89.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu T, Johnson KA. Presteady state kinetic analysis of vanadate-induced inhibition of the dynein ATPase. J Biol Chem. 1983;258:13833–13840. [PubMed] [Google Scholar]
- 26.Frado LL, Craig R. Electron microscopy of the actin-myosin head complex in the presence of ATP. J Mol Biol. 1992;223:391–397. doi: 10.1016/0022-2836(92)90659-8. [DOI] [PubMed] [Google Scholar]
- 27.Inoue Y, Shingyoji C. The roles of noncatalytic ATP binding and ADP binding in the regulation of dynein motile activity in flagella. Cell Motil Cytoskeleton. 2007;64:690–704. doi: 10.1002/cm.20216. [DOI] [PubMed] [Google Scholar]
- 28.Yoshimura A, Nakano I, Shingyoji C. Inhibition by ATP and activation by ADP in the regulation of flagellar movement in sea urchin sperm. Cell Motil Cytoskeleton. 2007;64:777–793. doi: 10.1002/cm.20222. [DOI] [PubMed] [Google Scholar]
- 29.Oiwa K, Sakakibara H. Recent progress in dynein structure and mechanism. Curr Opin Cell Biol. 2005;17:98–103. doi: 10.1016/j.ceb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Lindemann CB, Hunt AJ. Does axonemal dynein push, pull, or oscillate? Cell Motil Cytoskeleton. 2003;56:237–244. doi: 10.1002/cm.10148. [DOI] [PubMed] [Google Scholar]
- 31.Wu H, et al. Solution structure of a dynein motor domain associated light chain. Nat Struct Biol. 2000;7:575–579. doi: 10.1038/76804. [DOI] [PubMed] [Google Scholar]
- 32.Yasunaga T, Wakabayashi T. Extensible and object-oriented system Eos supplies a new environment for image analysis of electron micrographs of macromolecules. J Struct Biol. 1996;116:155–160. doi: 10.1006/jsbi.1996.0025. [DOI] [PubMed] [Google Scholar]
- 33.Hirakawa E, Higuchi H, Toyoshima YY. Processive movement of single 22S dynein molecules occurs only at low ATP concentrations. Proc Natl Acad Sci USA. 2000;97:2533–2537. doi: 10.1073/pnas.050585297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toba S, Watanabe TM, Yamaguchi-Okimoto L, Toyoshima YY, Higuchi H. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc Natl Acad Sci USA. 2006;103:5741–5745. doi: 10.1073/pnas.0508511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang W-JT, Bell CW, Sale WS, Gibbons IR. Structure of the dynein-1 outer arm in sea urchin sperm flagella. J Biol Chem. 1982;257:508–515. [PubMed] [Google Scholar]
- 37.Castoldi M, Popov AV. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- 38.Hirose K, Akimaru E, Akiba T, Endow SA, Amos LA. Large conformational changes in a kinesin motor catalyzed by interaction with microtubules. Mol Cell. 2006;23:913–923. doi: 10.1016/j.molcel.2006.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.