Abstract
The positions of protons are not available in most high-resolution structural data of biomolecules, thus the identity of proton storage sites in biomolecules that transport proton is generally difficult to determine unambiguously. Using combined quantum mechanical/molecular mechanical computations, we demonstrate that a pair of conserved glutamate residues (Glu 194/204) bonded by a delocalized proton is the proton release group that has been long sought in the proton pump, bacteriorhodopsin. This model is consistent with all available experimental structural and infrared data for both the wild-type bacteriorhodopsin and several mutants. In particular, the continuum infrared band in the 1,800- to 2,000-cm−1 region is shown to arise due to the partially delocalized nature of the proton between the glutamates in the wild-type bacteriorhodopsin; alternations in the flexibility of the glutamates and electrostatic nature of nearby residues in various mutants modulate the degree of proton delocalization and therefore intensity of the continuum band. The strong hydrogen bond between Glu 194/204 also significantly shifts the carboxylate stretches of these residues well <1,700 cm−1, which explains why carboxylate spectral shift was not observed experimentally in the typical >1,700-cm−1 region upon proton release. By contrast, simulations with the proton restrained on the nearby water cluster, as proposed by several recent studies [see, for example, Garezarek K, Gerwert K (2006) Functional waters in intraprotein proton transfer monitored by FTIR difference spectroscopy. Nature 439:109], led to significant structural deviations from available X-ray structures. This study establishes a biological function for strong, low-barrier hydrogen bonds.
Keywords: infrared spectroscopy, proton pumping, water cluster, quantum mechanical/molecular mechanical simulations
The transport and storage of protons in a controlled fashion is crucial to many fundamental biological processes such as energy transduction and signaling (1). Although many proteins that play an important role in such contexts have been identified, understanding in the regulatory mechanism of proton transport and storage remains incomplete. For instance, although the celebrated Grotthuss mechanism (2) is often the assumed proton transport pathway, the complementary “proton hole” scheme that involves hydroxide may also play a role in some systems (3). Another important mechanistic issue concerns the identity of proton storage site(s), which is difficult to determine unambiguously because protons are invisible in the crystal structure of most biomolecules. Indeed, although titratable amino acids are the common suspect for proton storage sites, the unconventional possibility of storing proton in water clusters has also been raised in several recent studies (4–7).
An excellent example that has been discussed extensively in the literature is the light-driven proton pump, bacteriorhodopsin (bR). In bR, the light-activated isomerization of its all-trans chromophore to the 13-cis configuration initiates a photocycle that eventually transports protons from the cytoplasmic to the extracellular side of the protein (8). Under physiological conditions, after the first proton transfer from the Schiff base to Asp-85, a proton is released to the extracellular bulk during the L-to-M transition. This released proton does not originate from Asp-85 (9) but from the residue initially termed as XH, which more recently is referred to as the proton release group (PRG).
The precise chemical nature of the PRG has been a long-standing puzzle in the bR community. Earlier IR studies have assigned the highly conserved Glu-204 and/or Glu-194 as the PRG (10, 11). Gerwert and coworkers carried out careful analysis of the IR spectra using the WT bR and a collection of mutants. Based on the results of these studies, they refuted the classical proposal because the expected spectra shifts in carboxylate groups upon Glu deprotonation were not observed for the L-to-M transition in the WT bR. Instead, they noted the decay of a continuum band near 2,000 cm−1 during the rise of the M state, which is reminiscent of the IR signature of protonated water clusters (12). Accordingly, it was proposed that the proton is stored on a water cluster trapped in a region surrounded by Glu-194, Glu-204, and Arg-82 (4, 5).
Although it is well-accepted that water in different protonation states plays a crucial role during proton transfers in both liquid and biomolecules (13), assigning the excess proton to a water cluster rather than the more basic Glu residues seems counterintuitive, especially considering the close proximity of the positively charged Arg-82 (Fig. 1A). Nevertheless, continuum electrostatics calculations that explicitly considered a titratable water dimer in the relevant region (14) indicated that the proton may preferentially exist in the Zundel form (H5O2+) with the 2 Glu residues (194 and 204) ionized to stabilize the positive charge. The continuum electrostatics calculations, however, were based on simple point-charge models and therefore are not conclusive; the close distances between Glu-194, Glu-204 and the relevant water molecules indicate that a quantum mechanical treatment of these residues is warranted for a quantitative consideration of the protonation pattern in this region. Car–Parrinello molecular dynamics (CPMD)-based quantum mechanical/molecular mechanical (QM/MM) simulations (6) found that a protonated water cluster in the PRG region generated a continuum IR band qualitatively consistent with the experimental observation, providing further support to the idea of involving water clusters rather than amino acid side chains as the proton storage site. However, the QM region included only the protonated water molecules and therefore did not allow proton transfer to the Glu residues. In other words, these calculations (6) did not show that the protonated water cluster is stable in the bR environment and does not deprotonate to the nearby Glu residues. Further, the observation of a short distance (<3 Å, see below) between Glu-204/Glu-194 side chains in high-resolution X-ray structures for both the bR ground state and the L state is consistent with the idea of trapping a proton between these 2 residues (15, 16); considering the uncertainty in hydrogen-bonding distances even in high-resolution X-ray structures (17), however, the structural data alone do not provide a conclusive statement.
Fig. 1.
Structural features of the proposed proton release group (PRG) region (4, 5, 10, 11) in X-ray structures of the WT bacteriorhodopsin. (A) Spatial location of key residues and water molecules in the PRG region based on a ground-state structure (PDB ID code 1C3W). The retinal chromophore is shown in the transparent line form; water molecules resolved in the X-ray structure are shown as spheres. (B) Comparison of various X-ray structures for bacteriorhodopsin in the L state (see SI for a similar figure for the ground state) for the PRG region: 1E0P in orange, 1O0A in green, 1UCQ in blue, 1VJM in cyan, and 2NTW in magenta (see Table 1 for references). The figure is generated by using VMD (47).
Here, we performed combined QM/MM molecular dynamics simulations with both the protonated water cluster and the Glu-194/204 side chains treated at the QM level; the QM level is either the popular density functional theory (DFT) (18), B3LYP (19), with the 6–31+G(d,p) basis (20), or an approximate DFT method, SCC-DFTB (21). At both QM/MM levels, we find that an excess proton on a water cluster is highly unstable and quickly (≈1 ps) moves to become partially delocalized between the Glu-194/Glu-204 residue pair. IR spectra calculations (see Materials and Methods) show that it is this strong hydrogen-bonding interaction that gives rise to the continuum band as observed experimentally for the WT bR. In addition, the vibrational frequencies for the carboxylate stretch are significantly red-shifted for Glu-194/204 due to the strong interaction mediated by the delocalized proton, which explains why the characteristic spectra shifts in carboxylate groups upon Glu deprotonation were not observed experimentally in the expected region of 1,700 cm−1 for the L-to-M transition in the WT bR.
Results and Discussion
Structural Features of WT bR X-Ray Crystal Structures.
It is instructive to first examine the structural feature of the PRG region in available X-ray crystal structures. The position of key residues (Arg-82, Glu-194/204), the hydrogen-bonding pattern between them and the number of resolved water molecules are largely consistent among the structures (Fig. 1B). In particular, as shown in Table 1, the pair of conserved Glu residues (Glu 194/204) in the PRG region are spatially very close in most X-ray structures, with the shortest O O distances substantially <3 Å. Among the ground-state structures, only that proposed by Luecke et al. (PDB ID code 1C3W) (22) has the distance slightly longer 3 Å. Among the L-state structures, the major outlier is the recently proposed structure of Lanyi and coworkers (2NTW) (23), where the significantly longer Glu-194–Glu-204 distance is due to the extracellular movement of the Arg-82 side chain and rotation of the Glu-194 side chain, most likely representing the structure after proton release (i.e., an early M state). The high-resolution structure PDB ID code 1O0A (24) has the retinal in the 13-cis, 15-syn conformation; previous study of proton transfer between the Schiff base and Asp 85 (25) found that such a configuration is not suited for active proton pumping. These considerations lead us to the choice of using 1UCQ as the starting structure of the L-state simulations in this work [see supporting information (SI) for additional discussions]. Although the resolution of this structure is not very high (2.4 Å), the fact that all key residues and water molecules in the PRG region are in the proper locations suggests that this is a sensible choice for the current purpose.
O distances substantially <3 Å. Among the ground-state structures, only that proposed by Luecke et al. (PDB ID code 1C3W) (22) has the distance slightly longer 3 Å. Among the L-state structures, the major outlier is the recently proposed structure of Lanyi and coworkers (2NTW) (23), where the significantly longer Glu-194–Glu-204 distance is due to the extracellular movement of the Arg-82 side chain and rotation of the Glu-194 side chain, most likely representing the structure after proton release (i.e., an early M state). The high-resolution structure PDB ID code 1O0A (24) has the retinal in the 13-cis, 15-syn conformation; previous study of proton transfer between the Schiff base and Asp 85 (25) found that such a configuration is not suited for active proton pumping. These considerations lead us to the choice of using 1UCQ as the starting structure of the L-state simulations in this work [see supporting information (SI) for additional discussions]. Although the resolution of this structure is not very high (2.4 Å), the fact that all key residues and water molecules in the PRG region are in the proper locations suggests that this is a sensible choice for the current purpose.
Table 1.
Comparison of various ground-state and L-state crystal structures of the region around Glu194-Glu204 residues in bacteriorhodopsin
| State | PDB ID code (Res*) | Glu194(Oε)- Glu204(Oε) distance†, Å | No. of waters around Glu194-Glu204 residues | Ser193(Oγ) Glu204(Oε) distance, Å | Ser193(N) Glu204(Oε) distance, Å |
|---|---|---|---|---|---|
| Ground | 1C3W (1.55 Å) (22) | 3.0 | 3 | 2.6 | 2.8 |
| Ground | 1QHJ (1.90 Å) (40) | 2.4 | 4 | 3.4 | 2.8 |
| Ground | 1KGB (1.65 Å) (41) | 2.5 | 3 | 2.6 | 2.7 |
| Ground | 1IW6 (2.30 Å) (42) | 2.5 | 3 | 2.9 | 2.7 |
| Ground | 1C8R (1.80 Å) (43) | 2.6 | 3 | 2.6 | 2.7 |
| L | 1E0P (2.10 Å) (44) | 3.1 | 3 | 4.2 | 3.2 |
| L | 1O0A (1.62 Å) (24) | 3.1 | 3 | 2.6 | 2.9 |
| L | 1UCQ (2.40 Å) (45) | 2.6 | 3 | 3.0 | 2.7 |
| L | 1VJM (2.30 Å) (46) | 2.4 | 3 | 3.4 | 2.8 |
| L | 2NTW (1.53 Å) (23) | 3.8 | 3 | 2.3 | 2.9 |
*Resolution of the X-ray structure in Å.
†The shortest distances involving the Oε atoms in Glu194/204 are reported.
Structural Features of the WT bR in QM/MM Simulations.
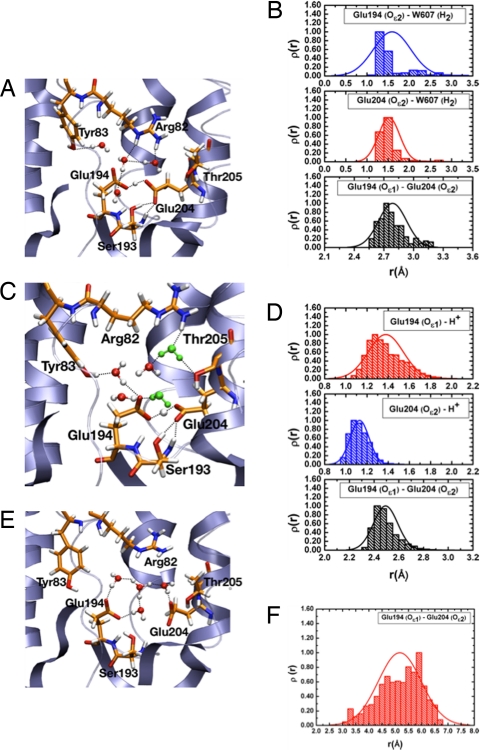
During B3LYP/MM simulations, the excess proton on the water cluster in the PRG region is highly unstable and quickly (≈1 ps) moves to become delocalized between the Glu-194/Glu-204 residue pair and remains there for the rest of the 5-ps trajectory (Fig. 2A). The same behavior is confirmed at the SCC-DFTB/MM level with 5 independent simulations, each at the much longer time scale of 2 ns (Fig. 2C). The histograms for the minimal distance between the excess proton and the 2 Glu residues (Fig. 2 B and D) confirm that the excess proton is shared by the pair of Glu residues with a slight preference over Glu-204. Further, the average distance of the 2 oxygens of Glu-204 and Glu-194 is ≈2.5 Å at both QM/MM levels, in good agreement with distances found in the crystal structures (Table 1). As shown by snapshots in Fig. 2 A and C, the carbonyl groups of the glutamates form stable hydrogen bonds with the surrounding residues like Tyr-83, Ser-193 and water molecules. During multiple simulations 2 bulk waters consistently become involved in this hydrogen-bond network (Fig. 2C), indicating the protein has flexibility to accommodate more waters than shown by the crystal structure.
Fig. 2.
Representative results from QM/MM simulations for the WT bacteriorhodopsin in the L state. (A) A snapshot of the PRG region from a B3LYP/6–31+G**/CHARMM simulation (≈5 ps) with a large QM region (see Materials and Methods); note that the excess proton is shared between Glu-194 and Glu-204 side chains. (B) Histograms for key distances involving the excess proton and Glu-194/Glu-204 side chains, which demonstrate that the excess proton is shared between the 2 Glu residues. (C and D) The same as A and B, but at the SCC-DFTB/MM level (4 independent 2-ns trajectories). Although all simulations started with 3 water molecules in the active site according to the X-ray structure (1UCQ), 2 water molecules (in green) are consistently observed to move to the region during the nanosecond-scale simulation. (E and F) A snapshot of the PRG region from a B3LYP/6–31+G**/CHARMM simulation (≈10 ps) with only water molecules in the PRG region (plus an excess proton) treated as QM (E), which leads to O O distances between Glu-194/204 significantly longer than the X-ray result (Table 1) (F). For additional analysis at both B3LYP/MM and SCC-DFTB/MM levels, see SI.
O distances between Glu-194/204 significantly longer than the X-ray result (Table 1) (F). For additional analysis at both B3LYP/MM and SCC-DFTB/MM levels, see SI.
As an independent evaluation of the original proposal of protonating the water cluster (4), we performed both B3LYP/MM and SCC-DFTB/MM simulations with only the protonated water cluster (H+·(H2O)4) treated as QM whereas the rest of the system as MM (i.e., same as ref. 6). Without any constraint, the protonated water cluster samples predominately the Eigen-like form (Fig. 2E), which is different from the observation of dominant Zundel form in the CPMD simulations (6); the Zundel form is favored in our simulations only if additional spatial constraints are applied to the water molecules (see SI). The key observation is that, in all cases, the distance between the anionic Glu-194/204 oxygens increases to values significantly larger than that in the various X-ray structures (e.g., compare O O histogram in Fig. 2F to Table 1). These changes are much larger than the expected uncertainties in the X-ray structures at the respective resolution (17), which strongly indicates that the protonated water cluster model is unlikely to reflect reality.
O histogram in Fig. 2F to Table 1). These changes are much larger than the expected uncertainties in the X-ray structures at the respective resolution (17), which strongly indicates that the protonated water cluster model is unlikely to reflect reality.
Spectral Features of the WT bR.
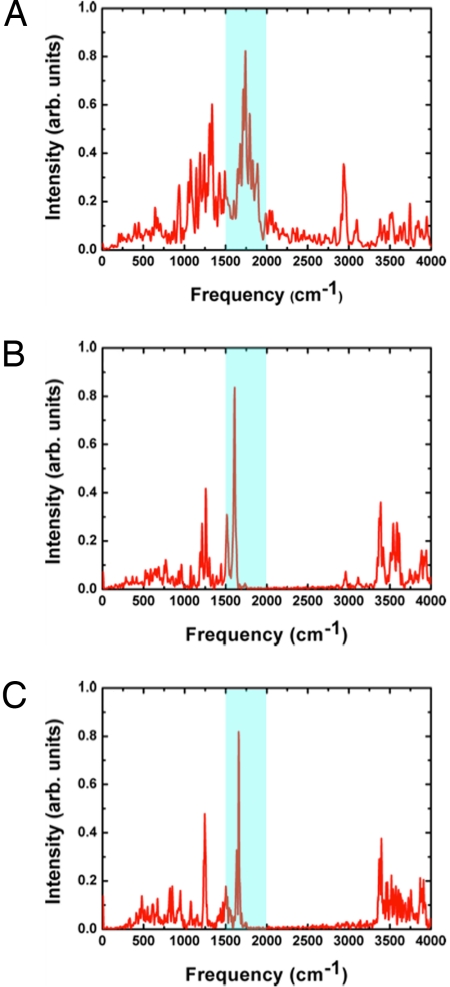
The computed IR spectra of the QM atoms at the SCC-DFTB/MM level (see SI) clearly show the continuum band in the 1,800- to 2,000-cm−1 region (Fig. 3A), in agreement with the experimental observation (4). This continuum band is absent (Fig. 3 B–C) when 1 of the Glu-194/204 residues is treated by using molecular mechanics and is therefore unable to share the excess proton. These findings confirm that the continuum band arises due to the partially delocalized nature of the proton between the 2 Glu residues, consistent with the recent analysis of small molecules involving an “intermolecular proton bond” (26); in other words, the diffuse vibrational band of shared proton is not unique to protonated water clusters. The partially delocalized nature of the proton is also reflected in peaks ≈2,900 cm−1, which likely correspond to OH vibrations and become substantially higher in frequency (≈3,300–3,400 cm−1) if 1 of the Glus is treated with molecular mechanics. Another important consequence of the strong hydrogen bonding interaction is that the νCOOH band of Glu-194/204 is significantly red-shifted to <1,700 cm−1, falling in the range of amide bands; such strong red-shifts of IR bands are also observed in gas-phase models with a shared proton at both the SCC-DFTB and DFT levels (see SI). This explains why the νCOOH bands have not been identified in the previous FTIR studies (16, 27), which led those authors to pursue alternative models for the PRG.
Fig. 3.
Computed IR spectra for the WT bacteriorhodopsin in the L state from SCC-DFTB/MM simulations (5 independent 2-ns trajectories). (A) Computed IR spectra for the QM atoms (Glu-194/204 side chains, the excess proton and 3 water molecules treated with SCC-DFTB); the spectra region of most importance here is highlighted in blue. (B and C) the continuum band in 1,800–2,000 cm−1 disappears if only 1 of the Glu side chains (Glu 194 in B and Glu-204 in C) is treated with QM, which confirms that the continuum band arises because of the delocalized nature of the excess proton.
Structural and Spectral Features of bR Mutants.
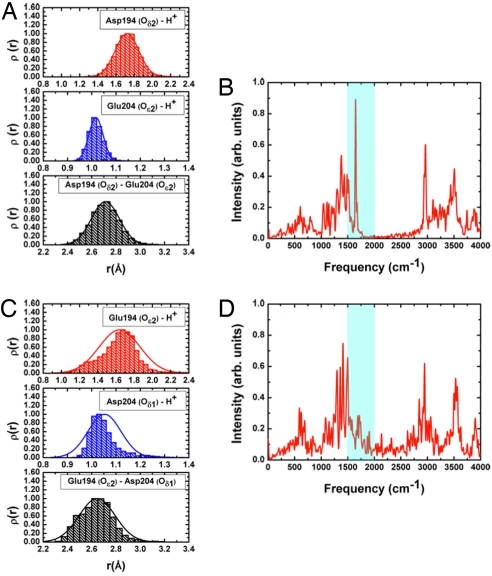
Several mutants have been studied by using the FTIR technique (4, 27) to shed light on the detailed structure of the PRG. A critical test for our shared proton model is, therefore, whether it is able to reproduce these findings. The first set of mutants involve mutation of 1 of the key Glu residues to aspartate: E194D and E204D. In our simulations for the E194D mutant, the excess proton is largely localized on Glu-204 as indicated by the distance histograms (Fig. 4A). The negatively charged Asp-194 is stabilized by Tyr-83, Ser-193, and surrounding water molecules. The lack of proton delocalization between Glu-204 and Asp-194 leads to the absence of the continuum band in the computed IR spectra (Fig. 4B), in agreement with the experimental observation (4).
Fig. 4.
Representative results from SCC-DFTB/MM simulations for 2 mutants of bacteriorhodopsin (5 independent 2-ns trajectories each) where 1 of the key Glu residues is mutated to an aspartic. (A and B) The E194D mutant. (C and D) The E204D mutant. The distance histograms indicate that the excess proton is largely localized on Glu-204 in E194D but weakly delocalized between Glu-194/Asp-204 in E204D; this is because of the higher flexibility of Glu-194 (see main text). As a result, the continuum band in the 1,800- to 2,000-cm−1 region is absent in the computed IR spectra for E194D (B) but present in E204D (D), in agreement with the experiment (4).
Interestingly, the situation is somewhat different in the E204D mutant simulations. In these simulations, although the proton remains localized mostly on Asp-204, the higher flexibility of Glu-194 results in a weak sharing of proton with Asp-204, as indicated by the histograms for the minimal distance between the excess proton and these 2 residues (compare Fig. 4 A and C). Therefore, a weak continuum band around the 1,800- to 2,000-cm−1 region is observed in the calculated spectra (Fig. 4D), also in agreement with experimental observation (4). The negative charge on Glu-194 is stabilized by Tyr-83 and active-site waters, whereas the hydrogen bonds between Ser-193 and Asp-204 are negligibly affected by the mutation of residue 204.
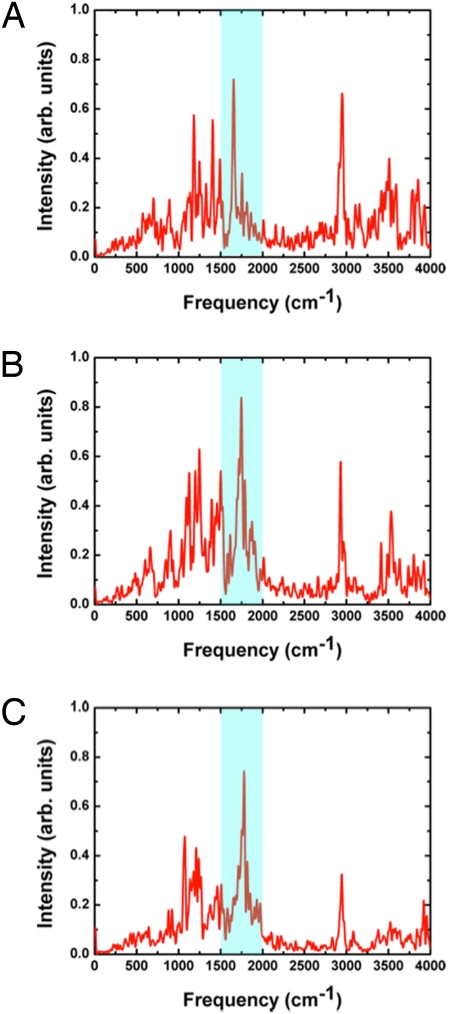
The second set of mutants involve mutations near the 2 Glu residues: S193A, Y83F, and R82Q (Fig. 5). In all these simulations, the excess proton is delocalized between Glu-194 and Glu-204, similar to the WT bR. Correspondingly, the diffuse band near 1,800–2,000 cm−1 is observed for all 3 mutants (Fig. 5), although with different intensities, in agreement with experimental observations (4). Specifically, replacing the hydroxyl group of Ser-193 with a hydrophobic methyl group increases the flexibility of the Glu-204 side chain and hence elongates the average distance between Glu-194 and Glu-204 compared with the WT bR; this is consistent with the weaker continuum band found in the experimental spectra for the S193A mutant. By contrast, replacing tyrosine with phenylalanine at position 83 negligibly affects the continuum band as also has been observed experimentally (4). This is because the lack of hydrogen bonding from Phe-83 to Glu-194 is compensated by active-site water molecules. In the R82Q mutant, the excess proton remains shared between the Glu-194/Glu-204 residues, which gives rise to the continuum band in the 1,800- to 2,000-cm−1 region. The stability of the 2 negatively charged Glu residues is due both to the delocalized proton and hydrogen-bonding interactions involving nearby residues, which include active-site water molecules that are hydrogen-bonded with the Gln-82 side chain.
Fig. 5.
Computed IR spectra for 3 additional key mutants (5 independent 2-ns trajectories each) studied experimentally (4). (A) S193A. (B) Y83F. (C) R82Q. For all mutants, the excess proton remains partially delocalized between Glu-194 and Glu-204 during SCC-DFTB/MM simulations, thus the continuum band in the 1,800- to 2,000-cm−1 region is observed in all cases, in agreement with experimental observations.
Concluding Remarks.
In summary, our QM/MM simulations indicate that a model in which the excess proton is stored in a partially delocalized fashion between the 2 conserved Glu residues produces IR spectral features consistent with experiments for both the WT and several key mutants of bR. This model is chemically more intuitive compared with the proposal (4) that involves a protonated water cluster near a positively charged Arg and 2 more basic Glu residues; the water cluster model also leads to structural features less consistent with available X-ray data. Conceptually, our model does not disqualify the functional importance of water molecules in proton transfer. Rather, our finding emphasizes that proton delocalization is not limited to cases involving water molecules as in the commonly discussed Zundel/Eigen cations. In fact, by using an “intermolecular proton bond” (26) or a “low-barrier hydrogen-bond” (28, 29) to stabilize 2 otherwise repulsive groups (e.g., deprotonated Glu residues in bR) might be advantageous from the perspective of balancing significant binding affinity and rapid proton release, which clearly is of functional importance to biomolecular ion pumps. Therefore, we expect that proton-mediated amino acid pairs exist as an important functional motif, especially in those involved in proton transports. It is worthwhile to emphasize explicitly this type of motif because protons are rarely seen in X-ray structures, although very short distances (<2.8 Å) between heavy atoms are useful indications. Unique IR and NMR features induced by the delocalized proton (28, 29) are notable signatures, although, as this study highlights, they need to be carefully analyzed for a proper structural interpretation.
Materials and Methods
Combined QM/MM calculations have been carried out by using 2 QM methods: B3LYP/6–31+G(d,p) as implemented in QChem (30) and SCC-DFTB (21). The calculations are done with the CHARMM program (31) and the relevant QM/MM modules (32, 33). SCC-DFTB is an approximate DFT method, which is ≈3 orders of magnitude more efficient compared with the standard implementations of DFT. Thus, the SCC-DFTB-based QM/MM simulations allow for nanosecond-scale sampling rather than the picosecond scale at the B3LYP/MM level. Various applications have shown that SCC-DFTB is able to reproduce DFT results with sufficient accuracy in many systems (34, 35). For the present study, the ability of SCC-DFTB to reproduce strongly hydrogen bonded structures and proton affinities as well as the qualitative spectral features of various water clusters have been shown in recent studies (36, 37) (also see SI for additional benchmarks).
To be consistent with the experimental IR studies (4), MD simulations are carried out starting from the L-state X-ray structure (PDB ID code: 1UCQ, see Results and Discussion and SI for the choice of the starting structure). The treatment of long-range electrostatics is done by using the generalized solvent boundary potential (GSBP) (38) implemented in CHARMM (39). The IR spectrum for the QM region is computed by the Fourier transform of the dipole autocorrelation function collected from SCC-DFTB/MM molecular dynamics trajectories; multiple independent simulations are carried out for statistical significance. The nuclear quantum effect on the computed IR spectra is approximated with a harmonic quantum correction factor as done in the recent study of water clusters (37). See SI for additional technical details and discussions. Additional analysis of the various X-ray structures for bacteriorhodopsin in the ground (bR) and L states, gas-phase benchmark calculations, details for the QM/MM simulations and additional results at SCC-DFTB/MM and B3LYP/MM levels with different QM regions are included in the SI.
Supplementary Material
Acknowledgments.
P.P. thanks Michael Gaus and Maike Clemens for stimulating discussions. N.G. thanks Yang Yang and Shuo Yang for assistance in QCHEM/CHARMM calculations and M. Hoffmann for discussions. Computational resources from the National Center for Supercomputing Applications at the University of Illinois are greatly appreciated. This work was partially supported by National Science Foundation Grant CHEM-CAREER-0348649 (to Q.C.). Q.C. also acknowledges a Research Fellowship from the Alfred P. Sloan Foundation. M.E. acknowledges funding from the German Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810712105/DCSupplemental.
References
- 1.Alberts B, et al. Molecular Biology of the Cell. New York: Garland; 1994. [Google Scholar]
- 2.de Grotthuss CJT. Sur la décomposition de l'eau et des corps qu'elle tient en dissolution à l'aide de l'électricité galvanique. Ann Chim. 1806;58:54–73. [Google Scholar]
- 3.Riccardi D, et al. “Proton holes” in long-range proton transfer reactions in solution and enzymes: A theoretical analysis. J Am Chem Soc. 2006;128:16302–16311. doi: 10.1021/ja065451j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garczarek F, Brown LS, Lanyi JK, Gerwert K. Proton binding within a membrane protein by a protonated water cluster. Proc Natl Acad Sci USA. 2005;102:3633–3638. doi: 10.1073/pnas.0500421102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garczarek F, Gerwert K. Functional waters in intraprotein proton transfer monitored by FTIR difference spectroscopy. Nature. 2006;439:109–112. doi: 10.1038/nature04231. [DOI] [PubMed] [Google Scholar]
- 6.Mathias G, Marx D. Structures and spectral signatures of protonated water networks in bacteriorhodopsin. Proc Natl Acad Sci USA. 2007;104:6980–6985. doi: 10.1073/pnas.0609229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Sharpe MA, Qin L, Ferguson-Miller S, Voth GA. Storage of an excess proton in the hydrogen-bonded network of the d pathway of cytochrome c oxidase: Identification of a protonated water cluster. J Am Chem Soc. 2007;129:2910–2913. doi: 10.1021/ja067360s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oesterhelt D, Stoeckenius W. Functions of a new photoreceptor membrane. Proc Natl Acad Sci USA. 1973;70:2853–2857. doi: 10.1073/pnas.70.10.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerwert K, Souvignier G, Hess B. Simultaneous monitoring of light-induced changes in protein side-group protonation, chromophore isomerization, and backbone motion of bacteriorhodopsin by time-resolved Fourier-transform infrared spectroscopy. Proc Natl Acad Sci USA. 1990;87:9774–9778. doi: 10.1073/pnas.87.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown L, et al. Glutamic acid 204 is the terminal proton release group at the extracellular surface of bacteriorhodopsin. J Biol Chem. 1995;270:27122–27126. doi: 10.1074/jbc.270.45.27122. [DOI] [PubMed] [Google Scholar]
- 11.Balashov SP, et al. Glutamate-194 to cysteine mutation inhibits fast light-induced proton release in bacteriorhodopsin. Biochemistry. 1997;36:8671–8676. doi: 10.1021/bi970744y. [DOI] [PubMed] [Google Scholar]
- 12.Headrick JM, et al. Spectral signatures of hydrated proton vibrations in water clusters. Science. 2005;308:1765–1769. doi: 10.1126/science.1113094. [DOI] [PubMed] [Google Scholar]
- 13.Ball P. Water as an active constituent in cell biology. Chem Rev. 2008;108:74–108. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- 14.Spassov VZ, Luecke H, Gerwert K, Bashford D. pKa calculations suggest storage of an excess proton in a hydrogen-bonded water network in bacteriorhodopsin. J Mol Biol. 2001;312:203–219. doi: 10.1006/jmbi.2001.4902. [DOI] [PubMed] [Google Scholar]
- 15.Essen LO, Siegert R, Lehmann WD, Oesterhelt D. Lipid patches in membrane protein oligomers: Crystal structure of the bacteriorhodopsin–lipid complex. Proc Natl Acad Sci USA. 1998;95:11673–11678. doi: 10.1073/pnas.95.20.11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zscherp C, Schlesinger R, Tittor J, Oesterhelt D, Heberle J. In situ determination of transient pKa changes of internal amino acids of bacteriorhodopsin by using time-resolved attenuated total reflection Fourier-transform infrared spectroscopy. Proc Natl Acad Sci USA. 1999;96:5498–5503. doi: 10.1073/pnas.96.10.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris TK, Mildvan AS. High-precision measurement of hydrogen bond lengths in proteins by nuclear magnetic resonance methods. Proteins Struct Funct Genet. 1999;35:275–282. doi: 10.1002/(sici)1097-0134(19990515)35:3<275::aid-prot1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 18.Parr RG, Yang WT. Density-Functional Theory of Atoms and Molecules. New York: Oxford Univ Press; 1989. [Google Scholar]
- 19.Becke AD. Density-functional thermochemistry. iii. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 20.Krishnan R, Binkley JS, Seeger R, Pople JA. Self-consistent molecular orbital methods. xx. A basis set for correlated wavefunctions. J Chem Phys. 1980;72:650–654. [Google Scholar]
- 21.Elstner M, et al. Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties. Phys Rev B. 1998;58:7260–7268. [Google Scholar]
- 22.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 Å resolutions. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 23.Lanyi JK, Schobert B. Structural changes in the L photointermediate of bacteriorhodopsin. J Mol Biol. 2007;365:1379–1392. doi: 10.1016/j.jmb.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanyi JK, Schobert B. Mechanism of proton transport in bacteriorhodopsin from crystallographic structures of the K, L, M-1, M-2, and M-2′ intermediates of the photocycle. J Mol Biol. 2003;328:439–450. doi: 10.1016/s0022-2836(03)00263-8. [DOI] [PubMed] [Google Scholar]
- 25.Bondar AN, Fischer S, Smith JC, Elstner M, Suhái S. Key role of electrostatic interactions in bacteriorhodopsin proton transfer. J Am Chem Soc. 2004;126:14668–14677. doi: 10.1021/ja047982i. [DOI] [PubMed] [Google Scholar]
- 26.Roscioli JR, McCunn LR, Johnson MA. Quantum structure of the intermolecular proton bond. Science. 2007;316:249–254. doi: 10.1126/science.1138962. [DOI] [PubMed] [Google Scholar]
- 27.Rammelsberg R, Huhn G, Lübben M, Gerwert K. Bacteriorhodopsin's intramolecular proton-release pathway consists of a hydrogen-bonded network. Biochemistry. 1998;37:5001–5009. doi: 10.1021/bi971701k. [DOI] [PubMed] [Google Scholar]
- 28.Perrin CL, Nielson JB. “Strong” hydrogen bonds in chemistry and biology. Annu Rev Phys Chem. 1997;48:511–544. doi: 10.1146/annurev.physchem.48.1.511. [DOI] [PubMed] [Google Scholar]
- 29.Cleland WW, Ferry PA, Gerlt JA. The low barrier hydrogen bond in enzyme catalysis. J Biol Chem. 1998;273:25529–25532. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 30.Shao Y, et al. Advances in methods and algorithms in a modern quantum chemistry program package. Phys Chem Chem Phys. 2006;8:3172–3191. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- 31.Brooks BR, et al. CHARMM: A program for macromolecular energy, minimization and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 32.Woodcock L, et al. Interfacing Q-CHEM and CHARMM to perform QM/MM reaction path calculations. J Comput Chem. 2007;28:1485–1502. doi: 10.1002/jcc.20587. [DOI] [PubMed] [Google Scholar]
- 33.Cui Q, Elstner M, Kaxiras E, Frauenheim Th, Karplus M. A QM/MM implementation of the self-consistent charge density functional tight binding (SCC-DFTB) method. J Phys Chem B. 2001;105:569–585. [Google Scholar]
- 34.Elstner M. The SCC-DFTB method and its application to biological systems. Theor Chem Accounts. 2006;116:316–325. [Google Scholar]
- 35.Riccardi D, et al. Development of effective quantum mechanical/molecular mechanical (QM/MM) methods for complex biological processes (Feature Article) J Phys Chem B. 2006;110:6458–6469. doi: 10.1021/jp056361o. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Yu H, York D, Cui Q, Elstner M. Extension of the self-consistent-charge density-functional tight-binding third-order expansion of the density functional theory total energy an introduction of a modified effective coulomb interaction. J Phys Chem A. 2007;111:10861–10873. doi: 10.1021/jp074167r. [DOI] [PubMed] [Google Scholar]
- 37.Yu H, Cui Q. The vibrational spectra of protonated water clusters: A benchmark for SCC-DFTB. J Chem Phys. 2007;127:234504. doi: 10.1063/1.2806992. [DOI] [PubMed] [Google Scholar]
- 38.Im W, Berneche S, Roux B. Generalized solvent boundary potential for computer simulations. J Chem Phys. 2001;114:2924–2937. [Google Scholar]
- 39.Schaefer P, Riccardi D, Cui Q. Reliable treatment of electrostatics in combined QM/MM simulation of macromolecules. J Chem Phys. 2005;123 doi: 10.1063/1.1940047. 014905–014914. [DOI] [PubMed] [Google Scholar]
- 40.Belrhali H, et al. Protein, lipid and water organization in bacteriorhodopsin crystals: A molecular view of the purple membrane at 1.9 Å resolution. Structure (London) 1999;7:909–917. doi: 10.1016/s0969-2126(99)80118-x. [DOI] [PubMed] [Google Scholar]
- 41.Facciotti MT, et al. Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle. Biophhys J. 2001;81:3442–3455. doi: 10.1016/S0006-3495(01)75976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsui Y, et al. Specific damage induced by X-ray radiation and structural changes in the primary photoreaction of bacteriorhodopsin. J Mol Biol. 2002;324:469–481. doi: 10.1016/s0022-2836(02)01110-5. [DOI] [PubMed] [Google Scholar]
- 43.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution. Science. 1999;286:255–260. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- 44.Royant A, et al. Helix deformation is coupled to vectorial proton transport in the photocycle of bacteriorhodopsin. Nature. 2000;406:645–648. doi: 10.1038/35020599. [DOI] [PubMed] [Google Scholar]
- 45.Kouyama T, Nishikawa T, Tokuhisa T, Okumura H. Crystal structure of the L intermediate of bacteriorhodopsin: Evidence for vertical translocation of a water molecule during the proton pumping cycle. J Mol Biol. 2004;335:531–546. doi: 10.1016/j.jmb.2003.10.068. [DOI] [PubMed] [Google Scholar]
- 46.Edman K, et al. Deformation of helix C in the low temperature L-intermediate of bacteriorhodopsin. J Biol Chem. 2004;279:2147–2158. doi: 10.1074/jbc.M300709200. [DOI] [PubMed] [Google Scholar]
- 47.Humphrey W, Dalke A, Schulten K. VMD—Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.