Abstract
Although the formation of 30-nm chromatin fibers is thought to be the most basic event of chromatin compaction, it remains controversial because high-resolution imaging of chromatin in living eukaryotic cells had not been possible until now. Cryo-electron microscopy of vitreous sections is a relatively new technique, which enables direct high-resolution observation of the cell structures in a close-to-native state. We used cryo-electron microscopy and image processing to further investigate the presence of 30-nm chromatin fibers in human mitotic chromosomes. HeLa S3 cells were vitrified by high-pressure freezing, thin-sectioned, and then imaged under the cryo-electron microscope without any further chemical treatment or staining. For an unambiguous interpretation of the images, the effects of the contrast transfer function were computationally corrected. The mitotic chromosomes of the HeLa S3 cells appeared as compact structures with a homogeneous grainy texture, in which there were no visible 30-nm fibers. Power spectra of the chromosome images also gave no indication of 30-nm chromatin folding. These results, together with our observations of the effects of chromosome swelling, strongly suggest that, within the bulk of compact metaphase chromosomes, the nucleosomal fiber does not undergo 30-nm folding, but exists in a highly disordered and interdigitated state, which is, on the local scale, comparable with a polymer melt.
Keywords: chromatin compaction, polymer melt, chromosome structure, vitreous sections, contrast, transfer function
The degree of mitotic DNA compaction, which is essential for successful segregation of chromatids between daughter cells, is remarkable in that the mitotic chromatid in humans is >10,000 times shorter than the original DNA molecule (1). The most basic DNA compaction in eukaryotes occurs through the wrapping of DNA around octamers of core histone proteins (H2A, H2B, H3, and H4) (2, 3). This step results in a repetitive motif that is composed of nucleosomes alternated with naked “linker DNA,” which is classified as the basic chromatin unit or the first level of DNA compaction (4). The subsequent folding of the basic chromatin fiber into the 0.7-μm thick chromatid seen at metaphase remains unclear, although it is known that condensins and topoisomerase IIα are implicated in this process (for recent reviews, see refs. 5–7). These proteins are variously assumed to mediate the folding of chromatin fibers into radially oriented loops (8–10), a hierarchy of helical structures (11, 12), or an irregular network (13). Despite their disparities, all of these hypotheses assume that before higher-order compaction, the nucleosome chain forms a 30-nm chromatin fiber as the second level of DNA folding.
The concept of the 30-nm fiber in mitotic chromosomes is based on the following 2 observations: (i) fibers of ≈30 nm in diameter have been observed in chromosomes by conventional transmission electron microscopy (TEM) (14, 15); and (ii) X-ray scattering analysis of isolated mitotic chromosomes reveals a diffraction peak that is characteristic of a structure of 30-nm size (16, 17). However, in classical TEM studies, mitotic chromosomes were swollen in hypotonic buffers, and then chemically fixed, dehydrated with alcohol, and embedded into plastic (6, 7). These harsh nonphysiological treatments might have generated artificial de novo folding of chromatin. Also, X-ray scattering experiments have failed to provide unequivocal evidence that the 30-nm signal is derived from chromatin, rather than contaminating cytoplasmic material. Hence, these observations do not provide conclusive evidence for the existence of 30-nm chromatin fibers in chromosomes in vivo.
Cryo-EM of vitreous sections has made it possible to overcome the limitations of the methods described above (18). Cryo-EM is based on vitrification of the water by rapid cooling, which ensures immobilization of all of the macromolecules in the specimen in a close-to-native state (19). Thin vitrified samples, such as layers of macromolecule suspensions, are then directly observed under a cryo-EM without any chemical fixation or staining (19). Bulky samples, like the majority of eukaryotic cells or tissues, must be cut into thin vitreous sections before observation (18). This approach enables a direct high-resolution imaging of cell structures at close-to-native state. However, the interpretation of cryo-EM images is hampered by the effects of the contrast transfer function (CTF), which may cause certain structural features to be artificially amplified or suppressed (20).
Although 30-nm chromatin fibers can be routinely observed by cryo-EM in chromatin suspensions in vitro (21, 22), cryo-EM studies of HeLa S3 cells did not reveal this structural feature in mitotic chromosomes (23). Nevertheless, because the influence of the CTF was not compensated for, it remained unclear whether the absence of 30-nm fibers in cryo-EM images of mitotic chromosomes is characteristic of the native chromatin or an artifact of the imaging properties.
In the present study, we address this uncertainty by analyzing the CTF-corrected cryo-EM images of vitreous sections of human mitotic chromosomes in situ after isolation. We show that within the bulk of the mitotic chromosome, the chromatin fiber exists in a highly disordered and interdigitated state, comparable with a polymer melt. We will also demonstrate that chromosome swelling by reduction of Mg2+ concentration in the buffer results in segregation of the compact structure of the native chromosome into 30-nm fibers.
Results
The Uniform Homogeneous Grainy Texture of Chromosome Images Is Maintained After CTF Correction.
The general appearance of a vitreous section of a mitotic HeLa S3 cell is shown in Fig. 1. The cytoplasm contains grains and various membranous organelles, such as cisterns of the endoplasmic reticulum and mitochondria. The homogeneous domains in the central region of the cell, excluding the globular macromolecular complexes and membranes, are chromosomes.
Fig. 1.
Cryo-EM view of a vitreous section of a mitotic HeLa S3 cell. Chromosomes are recognized by their elongated aspect and uniform texture. Numerous granules, membrane cisterns (mc), vesicles, and mitochondria (m) are evident in the cytoplasm, which is bordered by the cytoplasmic membrane (cm). Oblique striations of the image intensity are the result of knife marks during the sectioning process. A surface contamination with hexagonal ice (h) is also seen. The section thickness is ≈60 nm. (Scale bar, 1 μm.)
The homogeneous grainy texture enables the identification of the HeLa S3 chromosomes at a higher magnification (Fig. 2A, raw image). The figure shows 3 regions of condensed chromosomes separated by a thin branched zone of cytoplasm, which is recognizable by the presence of numerous globular macromolecular complexes. The chromosomes are well delineated, with smooth surfaces that are devoid of complicated convolutions. In addition, the characteristic homogenous grainy texture of the chromosomes becomes better visible at this magnification (25,500×).
Fig. 2.
The characteristic grainy texture of the chromosome is maintained after CTF correction. (A) An area of a mitotic HeLa S3 cell section that contains 3 parts of chromosomes (outlined in white) separated by the cytoplasm, is shown before and after CTF correction. The corresponding 1-DRAPS shown on the right side. The amplitudes are plotted as arbitrary units by using logarithmic scale on the y axis. The y axis units are the same in both plots. (B) A magnified segment of the CTF-corrected chromosomal texture. The inset in B shows simulated top views of 30-nm fibers assembled the interdigitated solenoid (left inset) (22), and 2-start helix (right inset) (27). The simulation was low-pass filtered to make it compatible with the CTF-corrected image of the chromosome. (Scale bars, 200 nm in A and 30 nm in B.)
The image was taken with an objective lens that was defocused at 6.6 μm, to generate sufficient phase contrast. Consequently, this image, as with any phase-contrast TEM image, is affected by the CTF. The CTF results in artificial amplification or suppression of the signal intensity, which, depending on the defocus value, affects different structural features. Although this effect is not easy to discern in the real-space image, it is evident in its computed diffraction pattern. Because the macromolecules are arbitrarily oriented within the sample, for general evaluation the diffraction pattern presented as a power spectrum can be rotationally averaged. The resulting graph [rotationally averaged power spectrum (1-DRAPS)] plots the averaged amplitudes on the y axis and the spacings on the x axis.
The 1-DRAPS of the entire image (Fig. 2A, raw image) had the highest signal peak at ≈11-nm spacing, indicating that structural features of this size predominate. However, because of CTF, the average amplitude dropped dramatically for the 30-nm spacing, which might have resulted in putative 30-nm fibers being hidden.
To compensate for the signal distortions caused by the CTF, several images of the same area were taken at different defocusing values and merged into a single image. This technique is used routinely with single particle cryo-EM to minimize the effects of CTF (24). In the present study, we applied this technique to search more sensitively for any possible higher-order chromatin structure. This type of deconvoluted focal series reconstruction (Fig. 2A, CTF-corrected image) combines the information from 5 images of a mitotic HeLa S3 cell taken at defocus values of between 6.6 and 26.8 μm. The recovery of the 30-nm features was apparent in the 1-DRAPS as a rise of average amplitude for the 30-nm spacing (Fig. 2A, 1-DRAPS of CTF-corrected image). Consistently, the 30-nm features of the cytoplasm, which were faint in the raw image, including globular macromolecular complexes [which most probably correspond to ribosomes (arrows), because of their dimensions], were clearly visible in the reconstructed image (Fig. 2A, CTF-corrected image).
However, no structures of this size were discernible in the chromosomes. The CTF-corrected grainy pattern of the chromosome image consisted of dense dots, which had a next-neighbor distance, directly estimated in the image, of 10–15 nm. The same CTF-correction procedure was performed for other 4 areas of the size of Fig. 2, which were randomly picked in different mitotic HeLa S3 cells. We found no difference in the CTF-corrected texture of chromosomes between the cells. The images did not reveal any obvious higher-order arrangement of the grains. The grainy pattern was essentially homogenous; there were no characteristic grain arrangements extending over distances significantly longer than 15 nm (Fig. 2B).
Because the images in Fig. 2A represent projections of a section of ≈50-nm thickness, which is much thicker than the size of a single nucleosome (≈10-nm diameter and 6-nm thickness), the grains seen in the image are superimpositions of perhaps 4–8 nucleosomes. If these nucleosomes were to be arranged in a regular order, some favorable orientations would reveal characteristic patterns in the images. For example, top views of 30-nm chromatin filaments of a length similar to our section thickness, modeled on the basis of in vitro observations (22), would give rise to characteristic rosette patterns (Fig. 2B Inset). The volume of such a chromatin patch (≈40,000 nm3) corresponds to 1/1000 of the chromatin volume of the section presented in Fig. 2. Given that chromosomes consist of randomly oriented 30-nm filaments, it provides a good chance for 30-nm fibers, if they exist, to be imaged as the characteristic top views. However, these or other ordered patterns were never observed in the condensed chromosomes; we found no authentic 30-nm filaments regularly arranged over a dimension of the order of the section thickness in this or any other section that we viewed (Fig. 2B).
Quantitative Analysis by 1-DRAPS Reveals Characteristic Spacing Peaks that Distinguish Textures of Chromosomes and Cytoplasm.
To quantitatively analyze the chromosome texture, we obtained 1-DRAPS selectively for chromosomal and cytoplasmic areas over a number of images. These spectra were CTF-corrected and averaged into 1 graph to improve the signal-to-noise ratio [for details, see Materials and Methods and supporting information (SI) Text and Fig. S1].
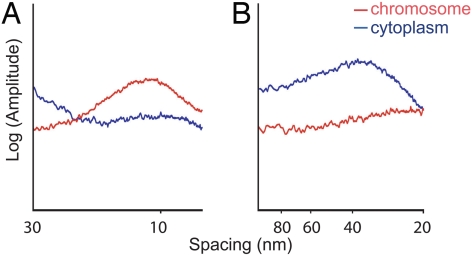
The resulting 1-DRAPS images are shown in Fig. 3. A broad peak of spacing with a maximum of ≈11.3 nm was observed for the chromosomes (Fig. 3A, red), whereas no peak in this range was seen for the cytoplasm (Fig. 3A, blue). The 11.3-nm peak in the chromosomes is consistent with previous studies using optical diffraction (23). There was no evidence of a peak in the 20–80 nm range (Fig. 3B, red) in the chromosomes. However, the 1-DRAPS of the cytoplasm showed a peak of 30–40 nm (Fig. 3B, blue), which may correspond to the dimensions of the globular complexes, which are abundant in the cytoplasm.
Fig. 3.
Averaged 1-DRAPS of chromosomes (red) and cytoplasm (blue) after CTF correction reveals characteristic spacing peaks. Two ranges of spacing are shown: from 7 to 30 nm (A) and from 20 to 100 nm (B). A broad peak of spacing with a maximum of ≈11.3 nm is observed for the chromosomes, whereas no peaks within this range are detected for the cytoplasm. In the 20–80 nm range, the chromosomal texture shows no spacing peaks. In contrast, a peak with maximum at ≈30 nm is detected in the cytoplasm.
The single peak for the ≈11-nm spacing and absence of other peaks in the chromosomes reveal the predominance of an 11-nm structural feature in chromosomes. To understand more clearly the organization underlying the ≈11-nm peak, we carried out gradual swelling of the mitotic chromatin in vitro.
Swelling of Chromosomes Reveals That the ≈11-nm Spacing Peak Results from the Close-Neighbor Distance Between Nucleosomes.
Isolated metaphase HeLa S3 chromosomes frozen in the presence of 5 mM Mg2+ were compact and showed a homogeneous grainy texture (Fig. 4, 5 mM), which was very similar to that observed in mitotic cells (Fig. 2). Consistent with that observed in situ, this texture was characterized by a ≈11-nm spacing peak and by the absence of peaks of large spacing. Decreasing the Mg2+ concentration in the incubation buffer resulted in gradual swelling of the chromosomes. At 1.5 mM Mg2+, there were no significant global changes in chromosome structure, although the peak spacing was slightly displaced (11.8 nm) (Fig. 4, 1.5 mM). At 0.7 mM Mg2+, the mitotic chromosomes were no longer compacted but consisted of irregular chromatin patches extending over regions of ≥100 nm (Fig. 4, 0.7 mM). The peak maximum was further displaced toward the larger dimension (13.7 nm). Lastly, at 0.5 mM Mg2+, the chromosome appeared as irregular filaments formed from discrete grains, which were consistent with nucleosomes in terms of their size and shape (Fig. 4, 0.5 mM). These filaments were reminiscent of the irregular 30-nm fibers observed in vitro (21). The peak in the 7–20 nm range persisted in the 1-DRAPS, with the maximum located at ≈14.3 nm. This value is substantially larger than any dimension of nucleosome, which indicates that the peak results from nucleosome spacing. Because no particular order of spatial positioning of nucleosomes was observed within the filaments, the peak reflects a general preferential distance between the centers of neighboring nucleosomes. The gradual shift of the spacing peak with swelling suggests that the peak has the same origin in open fibers and in partially decompacted and fully compacted native chromosomes. Therefore, the compact native chromosome appears to be a chromatin mass in which nucleosomes are packed in no particular order, albeit with a preferential distance of ≈11 nm.
Fig. 4.
Swelling of isolated mitotic HeLa S3 chromosomes in vitro by decreasing the Mg2+ concentration. The vertical columns of the images show the appearance of chromosomes at the corresponding Mg2+ concentration in the swelling buffer. Each column contains a cryo-EM image of the vitreous section taken at low magnification (LM), high magnification (HM), and the averaged CTF-corrected 1-DRAPS for the spacing range of 8–30 nm. Note that the texture of isolated chromosome in 5 mM Mg2+ is very similar to that of native chromosomes observed in the mitotic cell section (see Fig. 2) and is consistently characterized by a spacing peak with maximum at ≈11 nm. Also noteworthy is the gradual shift of this spacing peak that accompanies the gradual swelling of the chromosomes with decreasing Mg2+ concentration. Complete removal of Mg2+ by EDTA results in fully decompacted chromatin fibers and loss of the spacing peak in the observed range (1 mM EDTA). [Scale bars, 100 nm (LM) and 30 nm (HM).]
In addition to the 14.3-nm peak, the power spectrum of chromosomes swollen at 0.5 mM Mg2+ is characterized by a spacing peak in the range of 40–50 nm (data not shown). It appears that this peak is caused not only by the fiber thickness (≈30 nm), but also by a complicated function of interfibrillar spacing, which shifts this peak to larger distances.
The complete removal of magnesium ions by 1 mM EDTA resulted in further swelling and complete loss of the peak (Fig. 4, 1 mM EDTA). This state resembles the fully open isolated chromatin fibers previously observed in the thin vitrified layer in low-salt buffer (21).
Discussion
We demonstrate that after CTF correction, the images of human mitotic chromosomes in the vitreous sections reflect a compact uniform chromatin mass, in which 30-nm chromatin fibers are not discernible by visual inspection; 1-DRAPS analysis of the chromosome images also gave no indication of 30-nm chromatin fibers.
The previous cryo-EM observations of starfish spermatozoids or isolated chicken erythrocyte nuclei provided conclusive evidence that 30-nm fibers are maintained after the high-pressure freezing and thin sectioning of vitreous material (25). Therefore, the absence of 30-nm fibers in images of chromosomes is not due to a technical aspect of cryo-EM of vitreous sections, but is characteristic of native HeLa S3 metaphase chromosomes.
This phenomenon can be explained by a model that takes into account internucleosome interactions. The formation of a compact 30-nm fiber requires the selective binding of nucleosomes, which are close neighbors on the DNA strand. For example, if the side-to-side binding of a nucleosome to its first neighbor is stabilized, it will give rise to a 30-nm fiber that is organized as a 1-start helix, a solenoid (26). The second neighbor binding will result in a 2-start helix or a zig-zag ribbon fiber (27). Such selective intrafiber nucleosomal associations can be accomplished under diluted conditions, as in in vitro systems, in which interactions between chromatin fibers or between distant segments of the same nucleosome chain are negligible (Fig.5, diluted). However, these interactions become common during chromosome compaction in vivo and, thus, interfere with the formation and/or maintenance of selective intrafiber bonds (Fig. 5, concentrated). In this type of situation, 30-nm folding can exist only if the specific intrafiber nucleosomal binding has higher affinity than other internucleosomal associations. In this case, chromosome swelling first disrupts the weaker bonds, leaving the stronger, specific, intrafiber bonds intact. Consequently, the swelling would give rise to segregation of the 30-nm fibers, which remain compacted. However, this scenario is not compatible with our observations. Initially, the chromosome swells as a uniform mass, and then gradually dissociates into chromatin filaments of rather irregular open zig-zag structures, rather than compact 30-nm fibers. Thus, it appears that within the mitotic bulk of chromatin there is no crucial difference between the forces of interfiber and intrafiber nucleosomal associations. A nucleosome interacts with its neighbors irrespective of the nucleosomal array along the DNA strand. This state is known as a “melt” in polymer physics (28). It means that the nucleosome does not “know” to which fiber it belongs. In this case, no global secondary DNA folding exists, and 30-nm fibers, even if they existed before mitosis, would melt into the uniform mass and lose their structural identity (Fig. 5). The concept of the melt implies dynamic polymer chains (28): polymer chains are constantly moving and rearranging at the local level. Given that it is also valid for the mitotic chromatin, the melt model provides conclusive explanations of the 2 following observations, which are difficult to be interpreted by 30-nm-fiber models.
Fig. 5.
The melt model of mitotic chromosome structure. Under diluted conditions, the flexible nucleosomal fibers may compact through selective close neighbor associations, thus forming the 30-nm chromatin fibers. An increase in chromatin concentration results in interfiber nucleosomal contacts, which interfere with the intrafiber bonds. Nucleosomes of adjacent fibers interdigitate and intermix. The 30-nm folding is disrupted and the nucleosomal fibers melt into a uniform mass. Because there is no difference between the intrafiber and interfiber nucleosome affinities, the nucleosomal filaments return to the open disordered conformation of the diluted state before compaction. Note that the chromatin compaction events are linked to the sequence in the figure to better illustrate the principle of the melt formation. The actual compaction pathway leading to the chromatin melt in vivo is unknown.
The first is the unexpectedly high intra-chromosome diffusion capabilities of many of the soluble factors (29–33). Of these factors, essential structural components of chromosomes, topoisomerase IIα and condensins, are comparable with or larger than nucleosomes. Accordingly, their diffusion within a static chromatin structure would require a sufficient porosity that is not compatible with the highly-dense metaphase chromatin packing (34, 35). The dynamic melt overcomes this problem: constant local movements and rearrangements of the nucleosomes by Brownian motion allow the protein complexes to enter and to move through the bulk of the chromosome. The previously reported high mobility of linker histone H1 in the metaphase chromosomes, suggesting a transient mode of H1 binding, is in agreement with the dynamic state of the chromatin (29).
Second, the melt state overcomes a packing problem. According to available measurements, the local DNA concentration in mammalian mitotic chromosomes is high at ≈170 mg/mL (34, 35). Typical models of chromosome organization based on 30-nm fibers face difficulties in reaching this value, because even a dense packing of 30-nm fibers unavoidably leaves a significant unoccupied space between the fibers (34, 35). The melt, which is a homogeneous dense packing without any empty space, could avoid this problem.
Because, in addition to human cells, similar disordered homogeneous chromatin has been observed by cryo-EM in not only the mitotic, but also the interphase chromatins of rodents and plant cells, the melt may represent the predominant state of compacted chromatin in vivo in general (36, 37). When 30-nm chromatin folding is required locally (e.g., in specific loci) or globally in the entire chromatin of specific cell types (e.g., in starfish spermatozoids; see ref. 25), transition of the homogeneous melt into 30-nm fibers may be caused in vivo through increasing of intrafiber nucleosome affinity (e.g., by histone modifications or binding of specific proteins). A similar effect may be caused by nonphysiological treatments used in conventional TEM, which may account for the 30-nm fibers observed in plastic-embedded chromosomes (9, 10).
Further refinement of the melt model will address the questions of how the chromosome shape is formed and how the structural integrity of the chromosome is maintained in the highly dynamic chromatin state. These advances will be made possible by improved computer modeling and in vivo molecular dynamic measurements, together with the 3D reconstruction of chromatin by using cryo-electron tomography.
Materials and Methods
Preparation of Mitotic Cells and Chromosomes and High-Pressure Freezing.
HeLa S3 cells were grown in RPMI medium 1640 that was supplemented with 10% FBS (Invitrogen). The yield of mitotic cells was increased by adding 0.06 μg/mL colcemide (Sigma-Aldrich) to the culture medium for 4 h. Mitotic cells were collected by shaking and were pelleted by centrifugation at 1000 × g for 5 min. The cell pellet was mixed with 5 volumes of culture medium that contained 20% dextran (40 kDa; Sigma-Aldrich). The mixture was placed on a stack of filter papers and incubated for 5 min in a humid chamber. Concentrated suspensions of cells were rapidly collected from the droplet on the filter paper and placed into specimen carriers before freezing under high pressure (HPM 010; BAL-TEC).
Mitotic chromosome clusters were purified as described previously (9). Vitrification of isolated chromosome clusters, as well as entire cells by high-pressure freezing requires the addition of 20% dextran solution to the swelling buffer. Although dextran with molecular mass of 40 kDa gave satisfactory results for cell vitrification, its presence in the swelling buffer prevented the swelling of chromosomes. We found that 1.5-kDa dextran (Sigma-Aldrich) produced optimal chromosome swelling and sectioning.
To swell the chromosomes, chromosome clusters were diluted in 100 volumes of buffer that contained 10 mM Hepes-KOH (pH 7.5), 20% dextran, and MgCl2 at the indicated concentration. The clusters were then pelleted by centrifugation at 8000 × g for 5 min, and frozen at high pressure by using the Leica EM-PACT machine.
Cryo-Sectioning and Cryo-EM.
Frozen cells were sectioned by using the EM UC-6 FC-6 cryomicrotome (Leica) at −140 °C. Vitrified chromosome clusters were cut at −170 °C, because sectioning at a higher temperature resulted in a strong relief on the section surface. Sections with nominal thickness of 40 nm were produced with a 25° or 35° diamond cutting knife (Diatome). Sections were transferred onto lacy carbon-covered 300-mesh copper grids (Agar Scientific). The grids were transferred to a Gatan cryoholder (Gatan) that was maintained at a temperature below −170 °C and then inserted into a precooled CM100 cryo-electron microscope (FEI), which was equipped with a LaB6 cathode with an accelerating voltage of 100 kV. Electron diffraction was used to check whether the water was vitreous or crystalline. Crystalline sections were discarded. Images were recorded with the TemCam-F224HD charge-coupled-device camera (Tietz Video and Image Processing Systems).
Focal Series Reconstruction and Deconvolution.
Focal series of images of the same area were taken with defocus levels ranging from 6 to 30 μm at 25,000× magnification (0.65 nm per pixel). The images were aligned in Adobe Photoshop 7.0. 1-DRAPS were calculated in SumpS (24) and a Gaussian curve was fitted by using the KaleidaGraph 4.0 software (Synergy Software). The images were denconvoluted and merged in CTFMIX (24). The simulations of topviews of 30-nm fibers were performed in Matlab (The MathWorks). The PDB files of 30-nm fiber models were kindly provided by D. Rhodes.
Quantitative Image Analysis by 1-DRAPS.
Appropriate digital micrographs of chromatin were selected on the basis of optimal defocus levels and minimal cutting distortions and astigmatism. A magnification level of 25,000× (0.65 nm per pixel) with a defocus range from −5 to −10 μm was used for 1-DRAPS with a spacing of 7–20 nm. For the spacing range of 15–40 nm, several groups of images were collected at 10,000× magnification (1.62 nm per pixel), and the defocus was in the range of 37–60 μm. Chromatin areas were excised from the images into irregular regions or into squares of 5122, 10242, or 20482 pixels in Adobe Photoshop 7.0. The total collected ch area was ≈51.7 μm2 for the 7–15 nm range of 1-DRAPS and 280 μm2 for the 15–40 nm range. Excised images were padded into squares of 40962 pixels, and were orientated so that the knife marks were vertical. Images were masked in Fourier space, by using Image J 1.38a (National Institutes of Health, http://rsb.info.nih.gov/ij/), to remove signals that corresponded to knife marks and crevasses (for further details, see SI Text and Fig. S1). We calculated 1-DRAPS for each image by using SumPS (24). CTF correction by deconvolution of individual images was performed by using CTFMIX. Gaussian coefficients were calculated in KaleidaGraph 4.0. All image processing was performed on a Dell Inspiron running Ubuntu 6.06. The averaged deconvoluted 1-DRAPS, which corresponded to individual conditions and cutting conditions, were calculated in CTFMIX and KaleidaGraph 4.0. File format conversion was performed by using EM2EM (Imagic Science).
Supplementary Material
Acknowledgments.
We thank Dr. D. Rhodes (Laboratory of Molecular Biology, Medical Research Council, Cambridge, UK) for providing models of 30-nm fibers, and Drs. A. Stasiak and H. Saibil for their comments. M.E, A.S.F., and J. D. were supported by the 3D-EM Network of Excellence within Research Framework Program 6 of the European Commission. K.M. was supported by a grant-in-aid and the Promotion of X-Ray Free Electron Laser Research of the Ministry of Education, Culture, Sports, Science and Technology, and by a Ministry of Health, Labour, and Welfare grant for Advanced Medical Technology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810057105/DCSupplemental.
References
- 1.Becker WM, Reece JB, Poenie MF. In: The World of the Cell. Becker WM, editor. Menlo Park, CA: Benjamin-Cummings Publishing Company; 1996. pp. 434–435. [Google Scholar]
- 2.Kornberg RD. Chromatin structure: A repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 3.Oudet P, Gross-Bellard M, Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975;4:281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- 4.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 5.Swedlow JR, Hirano T. The making of the mitotic chromosome: Modern insights into classical questions. Mol Cell. 2003;11:557–569. doi: 10.1016/s1097-2765(03)00103-5. [DOI] [PubMed] [Google Scholar]
- 6.Belmont AS. Mitotic chromosome structure and condensation. Curr Opin Cell Biol. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Maeshima K, Eltsov M. Packaging the genome: The structure of mitotic chromosomes. J Biochem. 2007;143:145–153. doi: 10.1093/jb/mvm214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gasser SM, Laroche T, Falquet J, Boy de la Tour E, Laemmli UK. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986;188:613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- 9.Marsden MP, Laemmli UK. Metaphase chromosome structure: Evidence for a radial loop model. Cell. 1979;17:849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- 10.Maeshima K, Eltsov M, Laemmli UK. Chromosome structure: Improved immunolabeling for electron microscopy. Chromosoma V. 2005;114:365–375. doi: 10.1007/s00412-005-0023-7. [DOI] [PubMed] [Google Scholar]
- 11.Belmont A, Sedat J, Agard D. A three-dimensional approach to mitotic chromosome structure: Evidence for a complex hierarchical organization. J Cell Biol. 1987;105:77–92. doi: 10.1083/jcb.105.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kireeva N, Lakonishok M, Kireev I, Hirano T, Belmont AS. Visualization of early chromosome condensation: A hierarchical folding, axial glue model of chromosome structure. J Cell Biol. 2004;166:775–785. doi: 10.1083/jcb.200406049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirier MG, Marko JF. Micromechanical studies of mitotic chromosomes. Curr Top Dev Biol. 2003;55:75–141. doi: 10.1016/s0070-2153(03)01002-0. [DOI] [PubMed] [Google Scholar]
- 14.Gall JG. Chromosome fibers studied by a spreading technique. Chromosoma. 1966;20:221–233. doi: 10.1007/BF00335209. [DOI] [PubMed] [Google Scholar]
- 15.Adolph KW. Organization of chromosomes in mitotic HeLa cells. Exp Cell Res. 1980;125:95–103. doi: 10.1016/0014-4827(80)90193-7. [DOI] [PubMed] [Google Scholar]
- 16.Paulson JR, Langmore JP. Low angle x-ray diffraction studies of HeLa metaphase chromosomes: Effects of histone phosphorylation and chromosome isolation procedure. J Cell Biol. 1983;96:1132–1137. doi: 10.1083/jcb.96.4.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmore JP, Paulson JR. Low angle x-ray diffraction studies of chromatin structure in vivo and in isolated nuclei and metaphase chromosomes. J Cell Biol. 1983;96:1120–1131. doi: 10.1083/jcb.96.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Amoudi A, et al. Cryo-electron microscopy of vitreous sections. EMBO J. 2004;23:3583–3588. doi: 10.1038/sj.emboj.7600366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubochet J, et al. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 20.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. San Diego: Academic; 1996. pp. 34–70. [Google Scholar]
- 21.Bednar J, Horowitz RA, Dubochet J, Woodcock CL. Chromatin conformation and salt-induced compaction: Three-dimensional structural information from cryoelectron microscopy. J Cell Biol. 1995;131:1365–1376. doi: 10.1083/jcb.131.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson PJJ, Fairall L, Huynh VAT, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: Evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDowall AW, Smith JM, Dubochet J. Cryo-electron microscopy of vitrified chromosomes in situ. EMBO J. 1986;5:1395–1402. doi: 10.1002/j.1460-2075.1986.tb04373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conway JF, Steven AC. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J Struct Biol. 1999;128:106–118. doi: 10.1006/jsbi.1999.4168. [DOI] [PubMed] [Google Scholar]
- 25.Woodcock CL. Chromatin fibers observed in situ in frozen hydrated sections. Native fiber diameter is not correlated with nucleosome repeat length. J Cell Biol. 1994;125:11–19. doi: 10.1083/jcb.125.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson PJ, Rhodes D. Structure of the “30 nm” chromatin fibre: A key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Dorigo B, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 28.de Gennes PG. Scaling Concepts in Polymer Physics. Ithaca, NY: Cornell Univ Press; 1979. [Google Scholar]
- 29.Chen D, et al. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christensen MO, et al. Dynamics of human DNA topoisomerases IIalpha and IIbeta in living cells. J Cell Biol. 2002;157:31–44. doi: 10.1083/jcb.200112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliveira RA, Heidmann S, Sunkel CE. Condensin I binds chromatin early in prophase and displays a highly dynamic association with Drosophila mitotic chromosomes. Chromosoma. 2007;116:259–274. doi: 10.1007/s00412-007-0097-5. [DOI] [PubMed] [Google Scholar]
- 32.Tavormina PA, et al. Rapid exchange of mammalian topoisomerase II alpha at kinetochores and chromosome arms in mitosis. J Cell Biol. 2002;158:23–29. doi: 10.1083/jcb.200202053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 34.Daban JR. Physical constraints in the condensation of eukaryotic chromosomes. Local concentration of DNA versus linear packing ratio in higher order chromatin structures. Biochemistry. 2000;39:3861–3866. doi: 10.1021/bi992628w. [DOI] [PubMed] [Google Scholar]
- 35.Daban JR. High concentration of DNA in condensed chromatin. Biochem Cell Biol. 2003;81:91–99. doi: 10.1139/o03-037. [DOI] [PubMed] [Google Scholar]
- 36.Bouchet-Marquis C, Dubochet J, Fakan S. Cryoelectron microscopy of vitrified sections: A new challenge for the analysis of functional nuclear architecture. Histochem Cell Biol. 2006;125:43–51. doi: 10.1007/s00418-005-0093-x. [DOI] [PubMed] [Google Scholar]
- 37.Dubochet J, Sartori Blanc N. The cell in absence of aggregation artifacts. Micron. 2001;32:91–99. doi: 10.1016/s0968-4328(00)00026-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.