Abstract
Fluorescent proteins (FPs) based on green fluorescent protein (GFP) are widely used throughout cell biology to study protein dynamics, and have extensive use as reporters of virus infection and spread. However, FP-tagging of viruses is limited by the constraints of viral genome size resulting in FP loss through recombination events. To overcome this, we have engineered a smaller (≈10 kDa) flavin-based alternative to GFP (≈25 kDa) derived from the light, oxygen or voltage-sensing (LOV) domain of the plant blue light receptor, phototropin. Molecular evolution and Tobacco mosaic virus (TMV)-based expression screening produced LOV variants with improved fluorescence and photostability in planta. One variant in particular, designated iLOV, possessed photophysical properties that made it ideally suited as a reporter of subcellular protein localization in both plant and mammalian cells. Moreover, iLOV fluorescence was found to recover spontaneously after photobleaching and displayed an intrinsic photochemistry conferring advantages over GFP-based FPs. When expressed either as a cytosolic protein or as a viral protein fusion, iLOV functioned as a superior reporter to GFP for monitoring local and systemic infections of plant RNA viruses. iLOV, therefore, offers greater utility in FP-tagging of viral gene products and represents a viable alternative where functional protein expression is limited by steric constraints or genome size.
Keywords: fluorescence imaging, molecular evolution, photoreceptor, LOV domain
GFP and related FPs have revolutionized the imaging of protein dynamics within living cells (1) and found widespread application in plant virology either as tags for specific viral proteins during infection or as general reporters of cell infection (2, 3). While the study of plant viruses has benefited greatly from the use of GFP, it is known that FP-expressing viruses exhibit reduced infection efficiency that can negatively affect host range relative to the wild-type virus (4–6). The same is true for animal viruses, especially when FPs are attached to structural components of tightly packed virions (7). Moreover, the increased genetic load of plant viruses carrying a FP severely limits both local and systemic spread compared with that of the wild-type virus (6). Attempts have been made to overcome this problem in the extensively studied positive-strand RNA virus, Tobacco mosaic virus (TMV), through molecular evolution of the viral movement protein (MP), a protein critical for cell-to-cell movement of the viral genome (6). An alternative strategy would be to utilize a smaller FP to reduce the genetic load. However, generation of smaller derivatives from GFP-based FPs is unlikely because the β-barrel structure of the protein is intrinsic to its function (8).
Fluorescent peptide ligands represent promising smaller alternatives to GFP-based FPs (9). Yet, the necessity for an exogenous chemical substrate introduces limitations for this approach, particularly in plants where the cell wall poses an additional barrier to permeability (10), prompting us to search for other genetically encoded candidates. The utility of linear tetrapyrrole (bilin)-binding proteins as fluorescent probes in the near infrared region of the spectrum has been recognized (11), as has the potential for flavin-based fluorescent proteins as in vivo reporters (12). The latter are derived from photosensory modules known as light, oxygen or voltage sensing (LOV) domains present in a diverse range of photoreceptors from bacteria, fungi, and plants (13, 14). UV/blue light is detected via the chromophore flavin mononucleotide (FMN) located within the LOV domain, giving the protein a weak intrinsic fluorescence with a maximal emission wavelength at 495 nm (15). Although used successfully to monitor bacterial cell populations (12, 16), the suitability of LOV-based FPs for studying protein localization and trafficking has not been investigated.
In the present study, we examined whether LOV-based FPs could be used as fluorescent reporters of virus infection in plant cells because their relatively small size offers an advantage over GFP. Through the molecular evolution of plant-derived LOV domains, we have isolated a photoreversible FP that can be used effectively to track protein distribution within living cells. This FP, termed iLOV, outperformed GFP as a reporter of plant virus infection and movement, and conferred improved functionality over GFP when fused to proteins required for virus spread. iLOV, therefore, represents a new genetically encoded alternative to GFP-based FPs that exhibits greater utility for monitoring virus infection.
Results and Discussion
Virus-Based Screening Allows the Isolation of LOV Variants with Improved Fluorescence.
To create a LOV-based FP that would be suitable for plants, the LOV2 domain (amino acids 387–496) from Arabidopsis thaliana phototropin 2 (phot2) was chosen because this protein is monomeric (17). Upon UV/blue light excitation, LOV domains undergo a reversible photocycle involving formation of a covalent adduct between the FMN chromophore and a conserved cysteine residue within the protein (18). The photoactive cysteine within LOV2 (Cys426 of Arabidopsis phot2) was replaced with alanine to abolish adduct formation (18, 19) and generate derivative C426A (Fig. 1A).
Fig. 1.
DNA shuffling of phototropin LOV domains. (A) Schematic representation of the shuffling procedure. Arabidopsis phot1 and phot2 consist of a C-terminal serine/threonine kinase domain (KD) and two photosensory LOV domains (LOV1 and LOV2) that bind the chromophore FMN. The conserved cysteine required for LOV-domain photochemistry was replaced with alanine by site-directed mutagenesis before DNA shuffling. Two sequential rounds of DNA shuffling were carried out (R1 and R2, respectively). In R1, shuffled populations were generated by using low fidelity PCR conditions. For R2, high fidelity PCR conditions were used. In each case, the LOV2 domain of Arabidopsis phot2 (C426A) was used as a template scaffold for reassembly. Shuffled populations were subjected to TMV-based expression in tobacco and screened for improved fluorescence under UV light. (B) TMV-based expression of LOV variants in leaves of Nicotiana tabacum. Images were recorded simultaneously under UV illumination to allow direct comparison of green fluorescence. Leaves were either mock inoculated or inoculated with TMV vector expressing the progenitor C426A or the brightest variants from R1 and R2 (914 and 981, respectively) and photographed 3 days post inoculation.
Unfortunately, TMV-based expression of C426A in leaves of Nicotiana tabacum (6) produced fluorescent viral lesions that were barely detectable under UV/blue light (Fig. 1B). Thus, DNA shuffling was performed to enhance the fluorescent properties of C426A. To increase diversity, C426A was shuffled with three other LOV-coding sequences from Arabidopsis containing the photochemically inert cysteine-to-alanine substitution (Fig. 1A). Using C426A as a template scaffold for reassembly, shuffled populations were cloned and screened by virus-based expression in tobacco. Lesions exhibiting enhanced fluorescence relative to that of C426A were excised and pooled, with the exception that the brightest lesion was retained to confirm the veracity of the screening approach [supporting information (SI) Fig. S1]. A second round of DNA shuffling and screening was conducted (Fig. 1A) to establish whether beneficial mutations isolated from the first round could be combined to produce clones with additional improvements and 16 independent clones were obtained. Sequence analysis revealed that a number of amino acid substitutions occurred with high frequency (Table S1), most of which represented substitutions naturally found in LOV domains of phot1 or the LOV1 domain of phot2 (Fig. S2). Recombination of related LOV sequences was not evident in the progeny recovered suggesting that the conditions used favored insertion and recombination of point mutations within the LOV2-coding sequence of phot2. Despite this, sequential shuffling and virus-based screening were successful in producing additive improvements in LOV-mediated fluorescence (Fig. 1B).
Shuffled LOV Variants Show Enhanced Fluorescence and Reduced Photobleaching Characteristics.
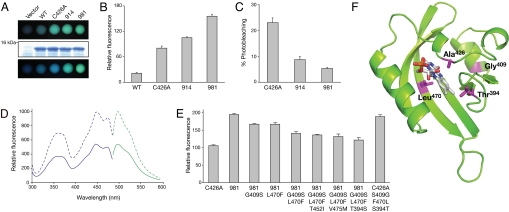
Subsequent analysis was restricted to the brightest variants isolated from the first and second round of shuffling, derivatives 914 and 981, respectively (Fig. 1B). Comparative in vivo fluorescence measurements were performed by using liquid cultures of E. coli (Fig. 2A). Cells expressing derivative 914, and to a greater extent 981, showed improved levels of in vivo fluorescence relative to C426A upon UV irradiation; fluorescence intensities were increased by 1.4-fold and 2-fold, respectively (Fig. 2B), resulting in a 10-fold greater in vivo fluorescence of 981 compared with that of wild-type LOV2 (Fig. 2B). Furthermore, continued exposure to UV light greatly diminished the level of green fluorescence from either C426A or wild-type LOV2 (lower panel in Fig. 2A). C426A showed a 23% reduction in fluorescence after arc lamp irradiation, whereas photobleaching was less apparent for 914 and 981, indicating that they were more photostable (Fig. 2C).
Fig. 2.
Photochemical characterization of shuffled LOV variants expressed in E. coli. (A) In vivo fluorescence in E. coli liquid cultures expressing wild-type Arabidopsis phot2 LOV2 (WT), derivative C426A and shuffled variants 914 and 981 viewed immediately under UV light (Top) or after several minutes of UV irradiation (Bottom). Equal protein levels in E. coli cultures are shown by SDS/PAGE and Coomassie Blue staining using cells transformed with the expression vector only as a control (Middle). (B) Quantification of LOV-mediated in vivo fluorescence in E. coli liquid cultures. Fluorescence intensities of liquid cultures were recorded at 495 nm upon excitation with blue light (450 nm). (C) Fluorescence loss in LOV-expressing E. coli cultures after xenon arc lamp illumination. Fluorescence intensities were recorded as in (B). (D) Fluorescence excitation and emission spectra of purified C426A (solid line) and variant 981 (dashed line). Fluorescence excitation spectra (blue) were recorded by using an emission wavelength of 495 nm, whereas fluorescence emission spectra (green) were recorded by using an excitation wavelength of 450 nm. (E) Reverse mutagenesis and quantification of 981-mediated in vivo fluorescence in E. coli liquid cultures. Point mutations indicated were introduced into 981 and the effect on in vivo fluorescence was assessed as in (B). Selective point mutations were then introduced into the progenitor C426A to confirm their role in enhancing fluorescence emission. (F) Structure of 981 was obtained by homology modeling with the program Swiss Model using the protein structure of Adiantum-capillus-veneris neochrome LOV2 (PDB entry IG28) and visualized by using PyMOL. Amino acid residues contributing to the enhanced fluorescence of 981 are indicated in magenta.
The absorption spectrum for purified 981 was identical to that of C426A (Fig. S3), as was its fluorescence excitation and emission spectra (Fig. 2D). Both proteins showed maximal absorption at 447 nm and maximal fluorescence emission at 497 nm upon excitation with blue light (450 nm). Consistent with our in vivo fluorescence measurements (Fig. 2B), purified 981 showed 2-fold greater fluorescence emission than C426A (Fig. 2D). Determination of fluorescence quantum yields (QF) revealed a QF of 0.32 and 0.44 for C426A and 981, respectively, the latter correlating well with the fluorescence associated with commonly used FPs such as CFP (20) and derivatives of DsRed (21). Because 981 contained multiple amino acid changes (Fig. S2), individual point mutations were introduced to ascertain whether the improved fluorescence properties could be assigned to specific amino acids. This process was simplified by the observation that >60% of the cloned progeny (Table S1), including 914 and 981, contained two substitutions normally found in LOV1 domains: S409G and F470L (22). Reverse mutagenesis of 981 revealed that both amino acid changes contributed to improved fluorescence (Fig. 2E). The generation of triple mutants also indicated that residues Thr394 and, to a lesser degree, Ile452 and Met475 in 981 contribute to fluorescence emission. Introduction of point mutations into C426A further confirmed that Thr394, Gly409 and Leu470 largely account for the improved fluorescence inherent to 981 and provide suitable targets for saturation mutagenesis.
Structural modeling of 981 indicates that Gly409 is distantly located from the FMN cofactor (Fig. 2F) making it difficult to infer the exact impact of this mutation on the chromophore environment, whereas Thr394 and Leu470 are situated in close proximity to the FMN isoalloxazine moiety (Fig. 2F). Although further analysis is required to address how these mutations enhance fluorescence when combined, 981 exhibits many hallmarks of a fluorescence reporter, and is herein referred to as iLOV owing to its improved fluorescent properties.
Improved Fluorescent Properties Make iLOV Suitable for Confocal Imaging in Plant and Mammalian Cells.
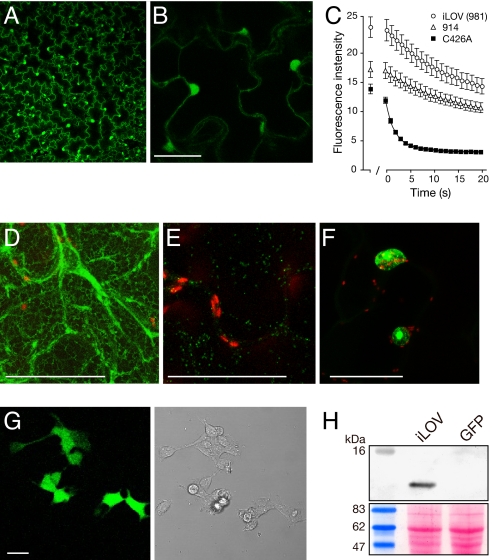
TMV-based expression of iLOV was detectable in tobacco epidermal cells by fluorescence or confocal microscopy (Fig. 3A) in both the cytosol and the nucleus (Fig. 3B). Nuclear localization was used to quantify in vivo fluorescence. As in E. coli (Fig. 2A), iLOV exhibited ≈2-fold increased fluorescence relative to C426A (Fig. 3C). Similar differences in fluorescence intensity were observed upon transient infiltration with Agrobacterium tumefaciens (Fig. S4). Furthermore, C426A fluorescence decreased rapidly after repeated laser scanning to reach a steady-state level indistinguishable from background (Fig. 3C). Fluorescence loss exhibited by iLOV and 914 was reduced by comparison, contributing to an improved level of detection over C426A (Fig. 3C). Prolonged laser scanning under these conditions caused iLOV fluorescence to decrease to only 50%, leaving sufficient signal to still be detectable by confocal imaging (Fig. S5).
Fig. 3.
Expression and subcellular targeting of iLOV in Nicotiana benthamiana and HEK cells. (A) Virus-based expression of free iLOV from a TMV vector. (B) Higher magnification of free iLOV expression showing both cytosolic and nuclear localization. (Scale bar, 50 μm.) (C) Photobleaching kinetics of LOV variants expressed in epidermal cells. LOV-mediated fluorescence from nuclei was used to quantify fluorescence loss in response to repeated scanning at 40% laser power. The first scan was used to focus on nuclei to be imaged and quantified. After 1 min, a series of 20 images was collected every s. Values represent the mean ± SE (n = 21). (D) iLOV targeted to the endoplasmic reticulum with TMV.SP-iLOV-HDEL. (Scale bar, 50 μm.) (E) iLOV targeted to the Golgi from TMV.ST-iLOV (indicated in green). Chloroplast autofluorescence is indicated in red. (Scale bar, 50 μm.) (F) iLOV expressed as a C-terminal fusion to Arabidopsis histone 2B. (Scale bar, 50 μm.) (G) Fluorescence imaging of free iLOV expressed in HEK cells. Bright field image is shown on the right. (Scale bar, 20 μm.) (H) iLOV accumulation in HEK cells detected by Western blotting using anti-iLOV antibody. HEK cells expressing GFP were used as a control. Ponceau S staining of the immunoblot below shows equal protein loading (20 μg).
iLOV fluorescence relies on its FMN chromophore, whereas GFP-based FPs are inherently fluorescent (1). One potential drawback in using iLOV as a reporter could arise from its dependency on a cellular cofactor. However, inclusion of the endoplasmic reticulum (ER) signal peptide and tetrapeptide retention signal, HDEL, showed that fluorescent iLOV protein could be targeted to the lumen of this endomembrane compartment (Fig. 3D). In addition, iLOV was targeted to the trans face of the Golgi apparatus (Fig. 3E) as a fusion protein to the sialyl transferase membrane-spanning domain resulting in fluorescent motile Golgi bodies (Movie S1) indistinguishable from those tagged with GFP (23). Fusion of iLOV to the C terminus of Arabidopsis histone 2B produced distinct labeling of the nucleus and nucleolus (Fig. 3F). Thus, the requirement for FMN does not limit the utility of iLOV as a fluorescent reporter in plant cells, at least for those subcellular compartments examined.
iLOV codon usage was also optimized for expression in human embryonic kidney (HEK) cells. iLOV was readily detectable by both confocal imaging (Fig. 3G) and western analysis using polyclonal antisera raised against iLOV (Fig. 3H), demonstrating that its utility can be extended to mammalian cells.
Photobleaching of iLOV in Vivo Is Reversible.
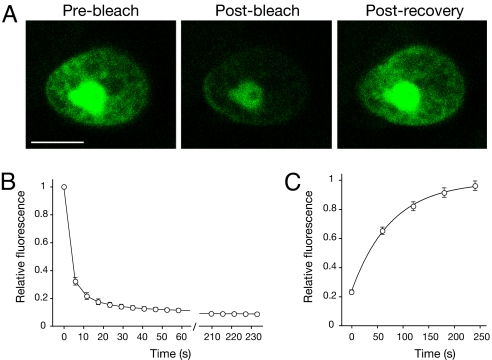
It is well established that GFP bleaches irreversibly under high-intensity imaging conditions (24). Although initially regarded as a problem for imaging, photobleaching of GFP and related FPs has been exploited to follow and quantify protein dynamics within living cells (1). Therefore, it was of interest to establish whether iLOV showed similar photobleaching properties. Nuclear-localized iLOV fused to histone 2B (Fig. 4A) was used to monitor fluorescence recovery after photobleaching (FRAP). Entire nuclei were scanned repeatedly at high laser power to maximize photobleaching (Fig. 4B) and avoid FRAP arising as an influx of molecules from any unbleached regions of the nucleus. iLOV fluorescence recovered fully after photobleaching (Fig. 4C) and time-lapse measurements revealed a half-maximal recovery time of ≈50 s (Fig. 4D). The reversible photobleaching of iLOV likely reflects a photochemical change(s) associated with the FMN chromophore that interchanges between fluorescent and nonfluorescent forms. High light intensities induce the formation of a neutral flavin semiquinone in LOV domains where the photoactive cysteine has been mutated (25) and may account for the observed photoreversible properties. Indeed, the blue fluorescence detected upon prolonged UV irradiation of E. coli cultures expressing C426A (Fig. 2A) would concur with the spectral characteristics of a protein-bound semiquinone (26).
Fig. 4.
Recovery of iLOV fluorescence after photobleaching. (A) iLOV expressed as a C-terminal fusion to Arabidopsis histone 2B was used to quantify fluorescence recovery after photobleaching. Representative images before photobleaching, after bleaching and post recovery are shown. (Scale bar, 5 μm.) (B) Photobleaching kinetics of nuclear-localized iLOV after repeated laser scanning at 88% laser power. A series of 40 scans was performed and one image collected every 6 s. Values represent the mean ± SE (n = 18). (C) Recovery kinetics for iLOV fluorescence after photobleaching. Values represent the mean ± SE (n = 18). Recovery fits to a first exponential and indicates a half-maximal recovery time of 54 s.
iLOV Outperforms GFP as a Reporter of Virus Infection and Movement.
Our main incentive for engineering an LOV-based fluorescent reporter was to overcome the limitations of using GFP for monitoring plant virus infection. Because the coding sequence of iLOV (≈300 bp) is considerably smaller than that of GFP (≈700 bp), we reasoned that the reduced genetic load of iLOV compared with GFP would be less detrimental to virus spread. To test this hypothesis, TMV-based vectors expressing either iLOV (TMV.iLOV) or GFP (TMV.GFP) were compared by inoculation of tobacco leaves. In half-leaf inoculations of reassembled transcripts for both constructs, TMV.iLOV produced pervasive, systemic green fluorescence (Fig. 5A), while TMV.GFP was restricted to local lesions at the sites of inoculation (Fig. 5A). Improved systemic spread of TMV.iLOV over TMV.GFP was also apparent after separate inoculations of plants; after 4 days, all plants inoculated with TMV.iLOV showed extensive systemic movement, whereas only 10% of TMV.GFP-inoculated plants exhibited signs of systemic fluorescence. Similar improvements of systemic spread of TMV.iLOV over TMV.GFP were visualized at later stages of infection (Fig. S6). Reduced genetic load may account for the improved functionality of iLOV over GFP because fusion of iLOV to Arabidopsis histone 2B (≈800 bp) diminished the efficiency of systemic spread (Fig. S7).
Fig. 5.
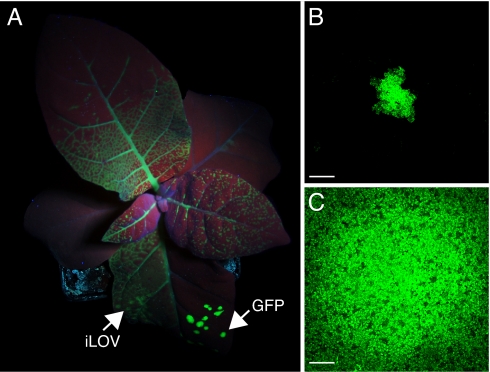
Utility of iLOV as a cytosolic fluorescent reporter for TMV infection. (A) Half-leaf inoculations of Nicotiana tabacum show that systemic spread of TMV.iLOV is extensive 4 days post inoculation (dpi) whereas TMV.GFP is still restricted to primary lesions on the inoculated leaf. Arrows indicate half-leaf inoculation sites. (B) Size of TMV.GFP lesion at 2 dpi. (Scale bar, 500 μm.) Lesions typically measured 687 μm ± 103 (n = 14). (C) Size of TMV.iLOV lesion at 2 dpi. (Scale bar, 500 μm.) Lesions typically measured over 3045 μm ± 113 (n = 8).
Local virus movement during the early stages of infection was also examined. TMV.GFP produced small groups of infected cells (Fig. 5A) whereas multicellular fluorescent lesions were detected for TMV.iLOV (Fig. 5C) indicating that cell-to-cell movement of TMV was less impeded, rendering iLOV a more efficient reporter than GFP for monitoring both local and systemic viral infections. Similarly, increased cell-to-cell movement of iLOV over DsRed was observed at the infection front of lesions created by TMV expressing both fluorescent proteins (Fig. S8), suggesting that iLOV matures faster than DsRed and/or may diffuse into neighboring cells owing to its smaller size.
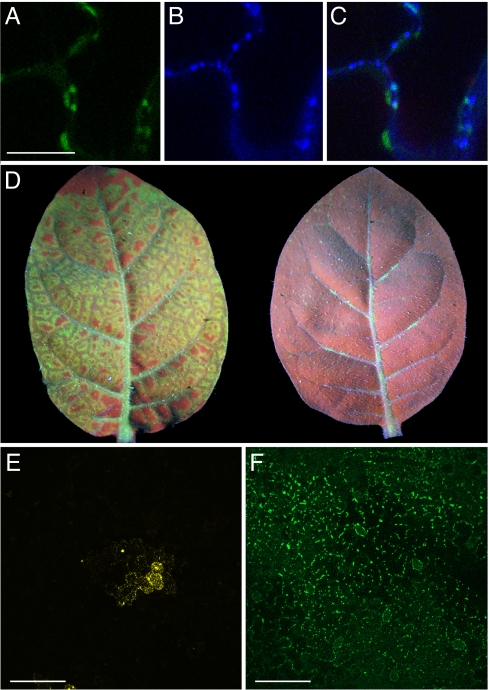
GFP has been used extensively to investigate the function and localization of the TMV movement protein (MP), a 30-kDa-protein essential for cell-to-cell movement of the virus through specialized channels in the cell wall known as plasmodesmata (PD) (27). TMV MP binds single-stranded RNA and accumulates in PD during viral infection (3, 27, 28). Fusion of iLOV to the C terminus of TMV MP produced PD localization in tobacco (Fig. 6 A–C), similar to that reported for MP-GFP (29). Moreover, the systemic spread of TMV expressing MP-iLOV was much greater compared with virus expressing MP-GFP (Fig. 6D). Whereas fusion of GFP is known to compromise the functionality of some viral MPs (30), iLOV may be less disruptive with respect to steric hindrance given its smaller size. Potato mop-top virus (PMTV), unlike TMV, requires the read-through product of the viral coat protein (CPRT) for cell-to-cell movement of the viral genome (31). Fusion of YFP to the C terminus of PMTV CPRT restricted movement of this RNA virus to single or a few cells on the inoculated leaf surface (Fig. 6E), whereas multicellular lesions were observed with virus expressing CPRT-iLOV (Fig. 6F) that localized to punctate bodies at the cell periphery, consistent with its function in cell-to-cell transport of RNA-protein complexes (31).
Fig. 6.
Utility of iLOV as a fluorescent reporter fusion for TMV and PMTV infection. (A) TMV MP-iLOV localization to plasmodesmata. (Scale bar, 20 μm.) (B) Callose staining at plasmodesmata with aniline blue. (C) Colocalization of TMV MP-iLOV fluorescence with aniline blue staining of plasmodesmata. (D) Systemic spread of TMV MP-iLOV and TMV MP-GFP. Upper leaves of Nicotiana tabacum at 4 days post inoculation. TMV MP-iLOV shows extensive systemic spread and unloads from all major vein classes, spreading into neighboring ground tissue (left). TMV MP-GFP by comparison shows no or limited systemic spread unloading only patchily from the midrib and some secondary veins. Leaves were photographed simultaneously to allow direct comparison of green fluorescence intensity. (E) Representative image showing the lesions size produced by PMTV expressing CPRT-YFP 2 days post bombardment of Nicotiana tabacum leaves. (Scale bar, 100 μm.) (F) Lesion size for PMTV expressing CPRT-iLOV visualized as in (E). (Scale bar, 100 μm.)
Further Application of iLOV as a Fluorescent Reporter.
Our findings demonstrate that iLOV represents a new class of genetically encoded FP that outperforms GFP as a reporter for plant virus movement and confers improved viral protein functionality. As many viral replication/movement events occur early in the infection cycle, the slow maturation of the GFP fluorophore limits its use for real-time studies of viral movement processes (29). It will now be important to establish whether iLOV can circumvent this problem. FP genes have predominantly been inserted into the genomes of filamentous plant viruses (28) because packaging constraints hinder their application to spherical viruses (5, 7). The smaller size of the iLOV coding sequence is likely to extend the range of plant and animal viruses that can be fluorescently tagged in vivo and enhance the study of intercellular protein trafficking. It is noteworthy that LOV-mediated fluorescence is extremely stable over a wide pH range (19) and may circumvent the pH sensitivity commonly associated with GFP-related FPs (1). iLOV may also remedy problems associated with dysfunctional GFP fusions given its smaller size, and prove useful for double and triple labeling studies where expression of multiple GFP derivatives can result in gene silencing. Although LOV-based FPs have been used successfully to monitor bacterial cell populations (12, 16), our studies highlight the necessity for protein engineering through DNA shuffling to create variants with improved characteristics that are suitable for fluorescence imaging. In contrast to GFP that bleaches irreversibly under sustained high light intensities (24), iLOV exhibits a latent photochemistry that recovers spontaneously, offering advantages where repeated laser scanning is desired. Although GFP-based FPs are likely to remain the main choice of researchers for the immediate future, we anticipate that iLOV-based fluorescent probes will provide attractive alternatives for specific applications where current genetically encoded FP technologies fall short.
Materials and Methods
Plant Material.
Nicotiana tabacum var Samsun and Nicotiana benthamiana plants were grown from seed and maintained at 22°C with a photoperiod of 16 h. Four- to six-week-old plants were used for viral inoculation.
DNA Shuffling of LOV Coding Sequences.
DNA shuffling was performed as described in ref. 32. Coding sequences for Arabidopsis PHOT1 and PHOT2 were used as PCR templates. The conserved photoactive cysteine residue within the LOV1 and LOV2 domains of Arabidopsis phot1 (Cys234 and Cys512, respectively) and Arabidopsis phot2 (Cys170 and Cys426, respectively) was replaced by alanine using the QuikChange site-directed mutagenesis kit (Stratagene). LOV coding sequences (Fig. 1A) were amplified with specific primers by using TaqDNA polymerase (Invitrogen) as described in ref. 33 and treated with DNase I. DNA fragments (< 100 bp) were reassembled in a 40 cycle PCR and reassembled fragments amplified in a 20 cycle PCR by using primers specific to the LOV2 coding region of Arabidopsis PHOT2. Amplification products were cloned via AscI and XhoI into pTMV.Asc.ΔCP, a derivative of pTMV.Asc (34). Ligated populations were amplified in E. coli resulting in two populations, with complexities of 2,500 and 4,500. Infection foci from each shuffled population were screened to identify lesions with improved fluorescence, and the LOV coding sequences were recovered and subjected to further DNA shuffling and screening as described in the SI Text.
Virus Plasmid Constructs.
The SP-iLOV-HDEL fusion was generated by addition of the signal peptide (SP) and HDEL retention sequences via sequential PCR. For Golgi targeting, the 52 N-terminal amino acids of rat sialyl transferase (ST) were added to the N-terminus of iLOV by sequential PCR. Arabidopsis histone 2B (H2B; At2G37470; RAFL14–57-H12) was obtained from RIKEN BioResource Centre (Tsukuba), amplified by PCR and fused to the N-terminus of iLOV. PCR products were cloned into pTMV.Asc via BssHII and XhoI to create TMV.SP-iLOV-HDEL, TMV.ST-iLOV, and TMV.H2B-iLOV. iLOV was fused to the C-terminus of the TMV MP by PCR cloning and used to replace the MP sequences in pTMV.Asc to create TMV.MP-iLOV. pPMTV-2 (35) was used to create C-terminal protein fusions to PMTV CPRT. Mutagenic PCR was used to convert the leaky stop codon of the CP gene with a tyrosine codon and replace the stop codon of the CPRT protein with NcoI and ApaI sites. Coding sequences for EYFP and iLOV were PCR amplified and cloned into the pPMTV-2 derivative between these sites.
Preparation of Infectious Transcripts and Plant Inoculation.
Runoff transcripts were synthesized from nonlinearized viral plasmids by using the T7 mMessage mMachine kit (Ambion). Transcripts were reassembled with purified TMV coat protein, and manually inoculated to plants as described in ref. 6. Inoculated leaves were examined at 3 days post inoculation unless otherwise stated with a Blak-Ray Model B 100AP lamp (UVP) or under a Leica MZFLIII stereofluorescence microscope fitted with a 1 × objective lens using GFP1 and GFP3 filter sets. To compare the systemic spread of viruses harboring iLOV relative to GFP, two approaches were used. Either TMV.iLOV and TMV.GFP (6) were inoculated onto opposite halves of the same leaf or TMV.iLOV and TMV.GFP were separately inoculated onto 10 plants. For MP fusion comparisons, TMV.MP-iLOV and TMV.MP-GFP (6) were separately inoculated onto 10 plants. PMTV constructs were bombarded as described in ref. 36. Agrobacterium tumefaciens infiltration was performed as described previously (29).
Recombinant Protein Expression and Purification.
For in vivo fluorescence measurements, DNA fragments encoding LOV variants were PCR-amplified and cloned into pCAL-n-EK (Stratagene) via EcoRI and NcoI to create an N-terminal calmodulin-binding peptide fusion. LOV proteins were expressed as described previously (15) using E. coli Rosetta BL21(DE3) pLysS (Novagen). For in vitro spectroscopic measurements, DNA fragments encoding LOV variants were PCR-amplified and cloned into pGEX-4T-1 (GE Healthcare) via EcoRI and SalI to create an N-terminal GST fusion. LOV proteins were expressed as described above and purified by using GST·bind resin (Novagen). Amino acid substitutions were introduced by using the QuikChange Site-directed Mutagenesis kit (Stratagene). All amino acid changes were verified by DNA sequencing.
Mammalian Cell Culture and Expression.
HEK293 cells were grown in Dulbecco's Modified Eagle's Medium (Sigma) containing 10% (vol/vol) FCS, 2 mM l-glutamine, 2 U/ml penicillin, and 2 mg/ml streptomycin at 37°C in a humidity controlled incubator with 5% CO2. iLOV codon usage was optimized for mammalian cell expression by de novo gene synthesis (GenScript) and cloned into pEGFP-N1 (Clontech) via SalI and NotI to replace the GFP encoding sequence. HEK293 cells were transiently transfected by using PolyFect Transfection Reagent (Qiagen). Cells were attached to 1:100 fibronectin-coated cover slips 24 h after transfection. Proteins were extracted in lysis buffer (0.1 M Tris·HCl pH 8.0, 0.1 M NaCl, 0.5% Triton X-100). Recombinant GST-iLOV expressed in E. coli was cleaved by using thrombin to release GST. Denatured iLOV protein was used to generate polyclonal antisera (Scottish National Blood Transfusion Service). Western blotting was performed by using alkaline phosphatase-linked secondary antibodies and 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitro blue tetrazolium (NBT) solution (Sigma) for colorimetric development.
Spectroscopic Analysis.
Fluorescence excitation and emission spectra were recorded by using a PerkinElmer LS-55 luminescence spectrometer. Absorption spectra were measured by using a Shimadzu MultiSpec-1501 diode array spectrometer. Protein concentrations were determined by the Bradford protein assay (Bio-Rad) using BSA as standard. For qualitative in vivo fluorescence measurements, equal densities of E. coli cells (OD600 = 1.5) were harvested by centrifugation and resuspended in Tris·HCl buffer (0.5 M, pH 6.8). Aliquots were illuminated with a Blak-Ray lamp (UVP). For quantitative in vivo fluorescence measurements, E. coli cells (OD600 = 1.5) were harvested by centrifugation, resuspended in Tris·HCl buffer (50 mM, pH 8.0) and diluted 20-fold for analysis. Photobleaching was analyzed by irradiating with a xenon arc flash lamp (Cairn Research Limited) for 1 ms (200 V, 4000 μF). For fluorescence quantum yield determination, FMN (Sigma) was used as a reference standard as reported previously (12, 37).
Fluorescence Imaging.
LOV fluorescence was imaged by using a Leica SP2 confocal laser-scanning microscope with an excitation wavelength of 476 nm. Fluorescence emission was detected between 510 and 550 nm. GFP and YFP were excited at 488 nm and their emission collected between 510–550 nm and 515–535 nm, respectively. For time series measurements, tissue was irradiated briefly to allow image focusing and minimize photobleaching. An initial image was recorded to select a region of interest before a series of images was collected automatically. Fluorescence was quantified by using Leica LCS software. The Leica LCS FRAP application was used for photobleaching and fluorescence recovery measurements (using the TMV.H2B-iLOV construct) with the following settings: 1 pre-bleach scan, 40 bleaching scans every 6 s, 5 post-bleach scans every 60 s.
Supplementary Material
Acknowledgments.
We thank Brian Smith for his help in creating Fig. 2F and Mike Blatt for helpful discussions. J.M.C. is grateful to the Royal Society for the award of a University Research Fellowship. We thank the Gatsby Charitable Foundation and the Scottish Government's Rural and Environmental Department for financial support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.N.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807551105/DCSupplemental.
References
- 1.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe DC, Chapman S, Santa Cruz S. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 1995;7:1045–1053. doi: 10.1046/j.1365-313x.1995.07061045.x. [DOI] [PubMed] [Google Scholar]
- 3.Oparka KJ, Prior DA, Santa Cruz S, Padgett HS, Beachy RN. Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus (TMV) Plant J. 1997;12:781–789. doi: 10.1046/j.1365-313x.1997.12040781.x. [DOI] [PubMed] [Google Scholar]
- 4.Rabindran S, Dawson WO. Assessment of recombinants that arise from the use of a TMV-based transient expression vector. Virology. 2001;284:182–189. doi: 10.1006/viro.2001.0910. [DOI] [PubMed] [Google Scholar]
- 5.Lacomme C, Pogue GP, Wilson TMA, Santa Cruz S. Plant viruses as gene expression vectors. In: Ring CJA, Blair ED, editors. Genetically Engineered Viruses: Development and Applications. Oxford: BIOS Scientific Publishers Ltd; 2001. pp. 59–105. [Google Scholar]
- 6.Toth RL, Pogue GP, Chapman S. Improvement of the movement and host range properties of a plant virus vector through DNA shuffling. Plant J. 2002;30:593–600. doi: 10.1046/j.1365-313x.2002.01308.x. [DOI] [PubMed] [Google Scholar]
- 7.Brandenburg B, Zhuang X. Virus trafficking - learning from single-virus tracking. Nat Rev Microbiol. 2007;5:197–208. doi: 10.1038/nrmicro1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 9.Adams SR, et al. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: Synthesis and biological applications. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 10.Chapman S, Oparka KJ, Roberts AG. New tools for in vivo fluorescence tagging. Curr Opin Plant Biol. 2005;8:565–573. doi: 10.1016/j.pbi.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Fischer AJ, Lagarias JC. Harnessing phytochrome's glowing potential. Proc Natl Acad Sci USA. 2004;101:17334–17339. doi: 10.1073/pnas.0407645101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drepper T, et al. Reporter proteins for in vivo fluorescence without oxygen. Nat Biotechnol. 2007;25:443–445. doi: 10.1038/nbt1293. [DOI] [PubMed] [Google Scholar]
- 13.Briggs WR. The LOV domain: A chromophore module servicing multiple photoreceptors. J Biomed Sci. 2007;14:499–505. doi: 10.1007/s11373-007-9162-6. [DOI] [PubMed] [Google Scholar]
- 14.Christie JM. Phototropin blue-light receptors. Annu Rev Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- 15.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christie JM, et al. Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1. Biochemistry. 2007;46:9310–9319. doi: 10.1021/bi700852w. [DOI] [PubMed] [Google Scholar]
- 17.Nakasako M, Iwata T, Matsuoka D, Tokutomi S. Light-induced structural changes of LOV domain-containing polypeptides from Arabidopsis phototropin 1 and 2 studied by small-angle X-ray scattering. Biochemistry. 2004;43:14881–14890. doi: 10.1021/bi0485530. [DOI] [PubMed] [Google Scholar]
- 18.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 19.Swartz TE, et al. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J Biol Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 20.Patterson G, Day RN, Piston D. Fluorescent protein spectra. J Cell Sci. 2001;114:837–838. doi: 10.1242/jcs.114.5.837. [DOI] [PubMed] [Google Scholar]
- 21.Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 22.Crosson S, Rajagopal S, Moffat K. The LOV domain family: Photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 23.Boevink P, et al. Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 1998;15:441–447. doi: 10.1046/j.1365-313x.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- 24.White J, Stelzer E. Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 1999;9:61–65. doi: 10.1016/s0962-8924(98)01433-0. [DOI] [PubMed] [Google Scholar]
- 25.Kay CW, et al. Blue light perception in plants. Detection and characterization of a light-induced neutral flavin radical in a C450A mutant of phototropin. J Biol Chem. 2003;278:10973–10982. doi: 10.1074/jbc.M205509200. [DOI] [PubMed] [Google Scholar]
- 26.Ghanem M, Fan F, Francis K, Gadda G. Spectroscopic and kinetic properties of recombinant choline oxidase from Arthrobacter globiformis. Biochemistry. 2003;42:15179–15188. doi: 10.1021/bi035435o. [DOI] [PubMed] [Google Scholar]
- 27.Lucas WJ. Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology. 2006;244:169–184. doi: 10.1016/j.virol.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Waigmann E, Ueki S, Trutnyeva K, Citovsky V. The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. Crit Rev Plant Sci. 2004;23:195–250. [Google Scholar]
- 29.Wright KM, et al. Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: Evidence from FRAP. Traffic. 2007;8:21–31. doi: 10.1111/j.1600-0854.2006.00510.x. [DOI] [PubMed] [Google Scholar]
- 30.Thomas CL, Maule AJ. Limitations on the use of fused green fluorescent protein to investigate structure-function relationships for the cauliflower mosaic virus movement protein. J Gen Virol. 2000;81:1851–1855. doi: 10.1099/0022-1317-81-7-1851. [DOI] [PubMed] [Google Scholar]
- 31.Zamyatnin AA, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking. Mol. Plant Microbe Interact. 2004;17:921–930. doi: 10.1094/MPMI.2004.17.8.921. [DOI] [PubMed] [Google Scholar]
- 32.Stemmer WP. DNA shuffling by random fragmentation and reassembly: In vitro recombination for molecular evolution. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung DW, Chen E, Goeddel DV. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 34.Escobar NM, et al. High-throughput viral expression of cDNA-green fluorescent protein fusions reveals novel subcellular addresses and identifies unique proteins that interact with plasmodesmata. Plant Cell. 2003;15:1507–1523. doi: 10.1105/tpc.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savenkov EI, Germundsson A, Zamyatnin AA, Jr, Sandgren M, Valkonen JP. Potato mop-top virus: The coat protein-encoding RNA and the gene for cysteine-rich protein are dispensable for systemic virus movement in Nicotiana benthamiana. J Gen Virol. 2003;84:1001–1005. doi: 10.1099/vir.0.18813-0. [DOI] [PubMed] [Google Scholar]
- 36.Haupt S, et al. Two plant-viral movement proteins traffic in the endocytic recycling pathway. Plant Cell. 2005;17:164–181. doi: 10.1105/tpc.104.027821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Losi A, Ghiraldelli E, Jansen S, Gärtner W. Mutational effects on protein structural changes and interdomain interactions in the blue-light sensing LOV protein YtvA. Photochem Photobiol. 2005;81:1145–1152. doi: 10.1562/2005-05-25-RA-541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.