Abstract
Although many genes have been shown to play essential roles in learning and memory, the precise molecular and cellular mechanisms underlying these processes remain to be fully elucidated. Here, we present the molecular and behavioral characterization of the Drosophila memory mutant nemy. We provide multiple lines of evidence to show that nemy arises from a mutation in a Drosophila homologue of cytochrome B561. nemy is predominantly expressed in neuroendocrine neurons in the larval brain, and in mushroom bodies and antennal lobes in the adult brain, where it is partially coexpressed with peptidyl α-hydroxylating monooxygenase (PHM), an enzyme required for peptide amidation. Cytochrome b561 was found to be a requisite cofactor for PHM activity and we found that the levels of amidated peptides were reduced in nemy mutants. Moreover, we found that knockdown of PHM gave rise to defects in memory retention. Altogether, the data are consistent with a model whereby cytochrome B561-mediated electron transport plays a role in memory formation by regulating intravesicular PHM activity and the formation of amidated neuropeptides.
Learning and memory represent fundamental examples of individual adaptations in higher organisms. Behavioral genetics approaches make it possible to understand these processes by revealing the specific genetic pathways that regulate the ability of animals to learn and remember. Recent studies in both vertebrate and invertebrate species have demonstrated that the mechanisms of memory formation are highly conserved (1, 2). The relatively small CNS combined with an arsenal of powerful genetic and molecular tools has made Drosophila an excellent model for studying learning and memory and, over the past 30 years, a large number of mutants have been identified that are defective in learning and/or memory [for reviews, see refs. 3 and 4)].
Genetic and pharmacological disruption of Drosophila genes implicated in learning and memory has provided important insights into the signaling pathways involved in memory formation. One of the prominent signaling pathways that has emerged as a central regulator of both learning and memory is the cAMP/protein kinase A signaling pathway. Genes such as dunce, which encodes a cAMP phosphodiesterase, and rutabaga, which encodes a calcium/calmodulin-stimulated adenylyl cyclase, have been shown to affect learning in Drosophila (5, 6). Mutations in dunce and rutabaga lead to increased or decreased cAMP levels, respectively, suggesting that tight temporal and spatial regulation of cAMP concentration is essential for short-term memory. Another Drosophila memory mutant, amnesiac, also appears to affect cAMP signaling. Amnesiac encodes a large protein that is processed to give rise to several predicted peptides. One of these peptides shows weak similarity to pituitary adenylyl cyclase activating peptide (PACAP)-like neuropeptide (7), which, in mammals, has been shown to stimulate cAMP levels through a G-alpha coupled receptor. cAMP levels regulate the activation of PKA, which phosphorylates the transcription factor CREB (the cAMP-response element-binding protein), which in turn plays a role in the formation of long-term olfactory memory (8), as well as cathepsin (9), neurotrypsin (10), and Orb2 (11). Other signaling molecules, such as calcium/calmodulin-dependent protein kinase II (12), protein kinase C (13), and protein kinase G (14), have also been shown to play a role in learning and memory.
Despite considerable progress, it is clear that the relatively small number of genes identified thus far cannot account for the complex behaviors exhibited by a fly, let alone the much more complex behaviors of vertebrates. Therefore, additional studies will be required to identify new molecules and molecular pathways underlying learning and memory.
Here, we report the molecular and behavioral characterization of the mutant no-extended memory (nemy), which was previously isolated in a P-element screen for mutants showing defects in memory after courtship conditioning (15). We provide several lines of evidence to show that the learning and memory deficits observed in nemy flies are due to a mutation of the CG8776 gene encoding for the Drosophila homologue of cytochrome b561 (CytB561). CytB561 is a secretory vesicle membrane protein that shuttles electrons across the vesicle membrane via a process of intravesicular ascorbic acid (AcA) regeneration and is necessary for normal peptidyl α-hydroxylating monooxygenase (PHM) activity. PHM is a key enzyme involved in the biosynthesis of peptide neurotransmitters. We found that nemy is partially coexpressed with PHM and that PHM activity is disrupted in nemy mutants, suggesting a potential role for CytB561 and peptide biosynthesis in learning and memory.
Results
Cloning and Molecular Characterization of nemy.
The first allele, nemyP153, was identified in a P-element screen for mutants showing 3-hour memory defects after courtship conditioning (16). The presence of a single P-element insertion was confirmed by genomic Southern blot analysis. The P-element was shown to be inserted between the genes CG8772 and CG8776. The original annotation in flybase, based on sequences available at the time (before completion of the Drosophila genome project), suggested that the P-element was inserted close to the 3′ end of CG8772 (<400 bp) and >4.5 kb from the 5′ end of CG8776 (SI Text and Fig. S1A). However, sequence analysis of cDNAs isolated from an adult cDNA library revealed the presence of two additional 5′ exons encoding a longer splice form of CG8776 (Fig. S1B), which we refer to as nemyRA. Based on these results, the nemyP153 insertion is located at the 5′ end of the first exon of CG8776 within a noncoding portion of transcribed sequences (Fig. 1).
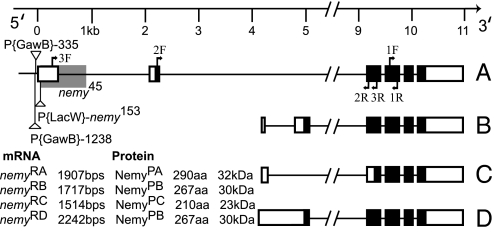
Fig. 1.
Molecular organization of the nemy locus. The exon (box)-intron (line) map is shown below the genomic scale. (A–D) Four alternative transcripts give rise to the 3 proteins (NemyPA, NemyPB, and NemyPC). The gray shading represents the sequence deleted in the nemy45 mutant and black shading indicates the coding region of each transcript. The sites of the different P-element insertions are indicated by triangles, those above the line are oriented 5′ to 3′ left to right, and those below the line have the opposite orientation. Primer pair used for cDNA screening, Northern blot is 1F-1R, for rt-PCR is 2F-2R, and for fluorescent probe synthesis is 3F-3R.
Screening a cDNA library uncovered 11 cDNA clones that corresponded to the gene CG8776. Mapping these cDNAs onto the genomic region revealed four alternatively spliced transcripts that may encode three different proteins (Fig. 1). These transcripts can be distinguished on a northern blot with a probe that is common to all transcripts (Fig. S2). As detailed below, there are several lines of evidence that suggest that the nemy phenotype arises from mutations in the longest splice form, nemyRA. Among these are behavioral and molecular data from independent P-element insertions, precise and imprecise excisions, rescue, and RNAi-mediated knockdown experiments.
nemy Mutants Exhibit Memory Deficits.
Together with the nemyP153 allele (PlacW), we have also used an independent Gal4-containing P-element (PGawB) insertion nemy335, which is located 4 bp upstream of the CG8776 gene (Fig. 1). Using a standard mating scheme to remobilize the P-element from nemyP153, we have also generated both imprecise (nemy45) and precise (nemy+) excisions. The region deleted by the imprecise excision in nemy45 extends 5′ to 3′ removing 743 bp of the longest splice form of nemyRA, including the first exon. The precise breakpoints were confirmed by PCR and sequence analysis. Both P-element insertion lines as well as the precise and imprecise excision lines are fertile and homozygous viable allowing us to test their olfactory learning and memory.
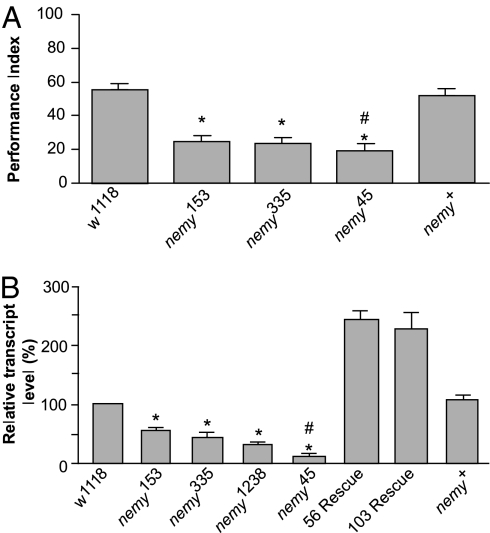
We started by examining 3-h memory performance after olfactory learning in the independent P-element insertion lines (nemyP153 and nemy335) and the precise (nemy+) and imprecise (nemy45) excisions. Consistent with previously reported data (16), the nemyP153 mutant showed a significant defect (P < 0.001) in 3-h memory compared with the w1118 control (Fig. 2A). We also noted a significant reduction (P < 0.001) in 3-h memory performance in the independent P-element insertion line nemy335. When we examined the precise and imprecise excisions, we saw that the nemy45 imprecise excision showed a greater reduction in 3-h memory performance compared with either the precise excision or w1118 (P < 0.001), while these control lines did not significantly differ from each other (Fig. 2A).
Fig. 2.
Mutations in nemy affect memory performance and the expression of CG8776RA. (A) Memory performance in nemy mutants and control lines. Performance indices (PIs) were measured 3 h after a single training session in the odor-shock paradigm. n = 6 PIs per group. (B) The expression level of nemy mRNA is reduced in nemy mutants. Quantitative real time RT-PCR was performed by using total RNA isolated from heads of control (w1118), rescue (56 and 103), P insertional (nemyP153, nemy335 and nemy1238), imprecise excision (nemy45) and precise excision (nemy+) lines. The expression of nemy was normalized relative to rp49 levels. Results represent means ± SEM. Here, and after: *, significant difference from w1118, P < 0.001 (two-sided t test); #, significant difference from nemy+, P < 0.001 (two-sided t test).
To unambiguously verify that the nemy mutant phenotype was due to a disruption of the CG8776 gene, we performed rescue experiments by using the GAL4/UAS system (17). Transgenic flies bearing a UAS construct containing the coding sequence corresponding to the longest splice form of CG8776 (nemyRA) were generated and introduced genetically into the nemyP153 mutant background under the control of a pan-neuronal Gal4 driver (elav-Gal4). The longest splice form of nemy was chosen as the template for the UAS construct because it appears to be predominantly expressed in heads (data not shown) suggesting a link between this isoform and memory. Furthermore, the longest isoform is the only splice form that is affected by the nemyP153 or nemy335 insertions and the nemy45 deletion (see Fig. 1 and Fig. S3).
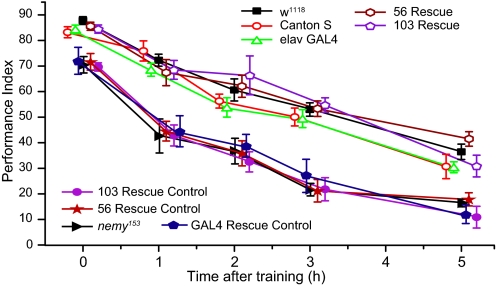
We found that the control lines w1118 and elav-Gal4 both showed similar performance indices to wild-type Canton S at all time points tested. In contrast, the nemyP153 flies showed poor performance indices at all time points tested, with the greatest difference being apparent 3 h after training (Fig. 3). The memory defect observed in nemyP153 was completely rescued by pan-neuronal expression of the nemyRA transgene (two independent UAS-nemy rescue lines: line #56 and line #103) in the nemyP153 background suggesting that disruption of nemyRA is responsible for the nemy mutant phenotype. We saw no rescue in nemyP153 flies containing the UAS-nemyRA transgene when the Gal4-driver was absent (56 and 103 rescue control in Fig. 3) indicating that neuronal expression of nemy is both necessary and sufficient to rescue the memory defects.
Fig. 3.
Pan-neuronal expression of NemyPA rescues memory defects associated with mutations in nemy. Pavlovian olfactory learning and memory were measured at different time points after a single training session. The time course curves for all control (Canton S, w1118 and elav-Gal4) and rescued [elav-Gal4; nemyP153; UAS-nemy+-A (56 or 103)] lines are significantly different from nemyP153 mutant and rescue controls lacking the pan-neuronal driver (nemyP153; UAS-nemy+-A (56 or 103)) or elav-Gal4; nemyP153. All flies were assayed at the same time points: 0, 1, 2, 3, and 5 h after training, however, for convenience of presentation the means and SEM are slightly staggered. n = at least 6 PIs per time point/group.
To ensure that the memory deficit observed in nemy flies was not due to sensorimotor defects, we tested olfactory acuity and shock reactivity. Control and mutant lines showed indistinguishable olfactory avoidance and shock reactivity (SI Text, Table S1). In addition, we found no defects in the gross morphology of the mushroom bodies in nemy mutants (Fig. S4).
nemy Mutations Are Hypomorphic.
To determine whether the behavioral defects in nemy mutants are associated with changes in the levels of nemyRA expression, we performed real-time quantitative RT-PCR analysis of mRNA isolated from adult heads by using primers specific for the long splice form. The expression of nemyRA was significantly reduced compared with the w1118 control line in both the P-element insertion lines and the imprecise excision nemy45 (P < 0.001). However, nemyRA expression was never completely abolished in these lines suggesting that the P-element insertions and the nemy45 mutations represent hypomorphic alleles rather than complete genetic nulls (Fig. 2B). At the same time, the level of expression in two independent rescue lines was elevated more than twofold compared with controls. The expression data are consistent with the results of the behavioral experiments, providing further evidence that the nemy phenotype is due to changes in the expression of nemyRA.
Nemy Is Expressed in Larval and Adult Brains.
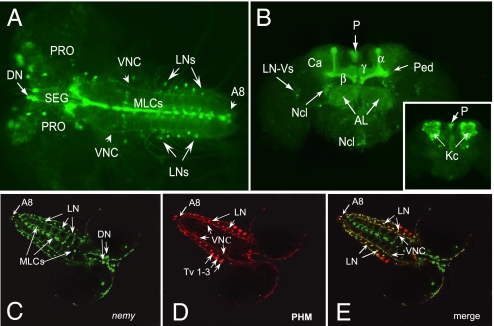
To visualize the expression pattern of nemy, we crossed nemy335 or nemy1238 flies carrying P-Gal4 insertions close to the 5′ end of nemy to flies carrying a UAS-driven enhanced green fluorescent protein (EGFP) transgene. Projections of confocal stacks revealed a complex pattern of expression in a number of different neuron types in the larval CNS (Fig. 4A). GFP expression was detected in several neuronal cell bodies located in the protocerebrum (PRO) and in the subesophageal ganglion (SEG). We also clearly observed a pair of neurons in the anterior part of the brain with descending axons (DN) projecting throughout the brain and ventral ganglion. In the ventral nervous system, GFP expression was observed in midline cells (MLC, Fig. 4A). Prominent GFP fluorescence was also seen in the axons descending down into all of the neuromeres of the ventral nerve cord (VNC), A8 neurons, and several pairs of lateral neurons (LNs), which are assumed to be sensory neurons.
Fig. 4.
nemy is expressed in larval and adult brains and is partially coexpressed with PHM in the larval CNS. (A) GFP expression pattern in larval brain. GFP expression is seen in protocerebrum (PRO), subesophageal ganglion (SEG), a pair of descending neurons (DN), midline cells (MLCs), ventral nerve cord (VNC), lateral neurons (LNs), and a pair of abdominal ganglion neurons (A8). Anterior view (B) and posterior view (inset) of GFP expression pattern in adult brain. Strong expression was observed in antennal lobes (AL) and throughout the mushroom bodies (α-, β-, and γ-lobes; the peduncle (Ped), calyx (Ca) and in the Kenyon cell (Kc) bodies). Additionally, expression was seen in neuron cluster cells (Ncl), ventral lateral neurons (LN-Vs) and in the pars intercerebralis (P). (C) Whole mount fluorescence in situ hybridization (FISH) with a fluorescent-nemy antisense RNA probe. Positive signals for nemy were observed in a pair of descending neurons (DN), midline cells (MLCs), ventral nerve cord (VNC), lateral neurons (LNs), pair of abdominal ganglion neurons (A8). (D) Whole mount (FISH) with DIG-PHM antisense RNA probe. Positive signals for the PHM were detected in the VNC, LN, and A8 neurons. (E) Merge of (C) and (D). Coexpression (yellow) was seen in LNs, VNC, and A8 neurons.
In adult brains (Fig. 4B), GFP expression was found in the glomeruli of the antennal lobes (AL) and in the mushroom bodies (MB), particularly in the α-, β-, and γ-lobes, in the peduncle (Ped), calyx (Ca), and in the Kenyon cell (Kc) bodies. These brain structures are known to be associated with olfactory learning and memory in insects (18, 19). In addition, GFP expression was observed in the pars intercerebralis (P), and in several distinct cell bodies and neuron clusters (Ncl) around the MB, AL, and in the subesophageal neuromeres. Punctate GFP expression was also noted in the minor population of lateral cells that border the optic lobes. The observed GFP pattern matches well with the nemyRA mRNA expression (Fig. S5) suggesting that it is indicative of endogenous nemy expression.
nemy Is Partially Coexpressed with PHM and Regulates Its Activity.
A PSI-BLAST search of current sequence databases with the Nemy amino acid sequence revealed significant similarity with many Cytb561-like sequences from various organisms. In particular, Nemy protein shares 33% identity and 54% similarity with human Cytb561 (Fig. S6). In vertebrates, CytB561 has been shown to act as a transmembrane electron transport protein (20), shuttling electrons across secretory vesicle membranes from cytoplasmic ascorbic acid (AcA) to intravesicular semidehydro ascorbic acid (SDAcA) reducing it again to AcA. In turn, intravesicular AcA serves as a cofactor for DbH (dopamine β hydroxylase) activity in catecholamine storage granules (21) and PHM activity in neuropeptide storage vesicles (22).
If nemy does encode a Drosophila homologue of Cytb561, then it might be expected to be coexpressed with, and modulate the function of DbH and/or PHM. In flies, tyramine β hydroxylase (TbH) is thought to play a similar role to DbH in mammals, converting tyramine to octopamine rather than dopamine to norepinephrine (23). Octopamine synthesis has been linked to reward-based learning in flies (24) but is not thought to play a role in aversive learning (as used in this study). Furthermore, there is no evidence suggesting a role for octopamine in memory. We have, therefore, focused on a potential interaction between nemy and PHM. We first determined whether nemy and PHM are coexpressed within the nervous system. PHM expression in the adult brain is widespread and heterogeneous (25), making it difficult to determine whether nemy and PHM are truly coexpressed in the adult brain (data not shown). We, therefore, performed double labeling experiments on whole mount third instar larval CNS (Fig. 4, C–E). Both nemy and PHM display distinct mRNA expression patterns in the larval CNS. In the case of nemy, obvious staining was found in midline cells (MLC), the ventral nerve cord (VNC), and in a bilaterally paired cell in the abdominal ganglion (A8) (Fig. 4C). In addition, we also reliably observed strong signals in a pair of descending neurons (DN) and in a subset of several distinct protocerebral neurons. Prominent PHM staining (Fig. 4D) was detected in the T1–3 ventral and dorsal neurons as well as in lateral neurons. Coexpression (in yellow) was seen in many lateral and ventral nerve cord cells as well as in A8 pair cells (Fig. 4E). However, no double labeling was detected in the prominent neuroendocrine neurons (e.g., T1–2 neurons). These results demonstrate that PHM mRNA expression partially overlaps with that of nemy in the larval CNS.
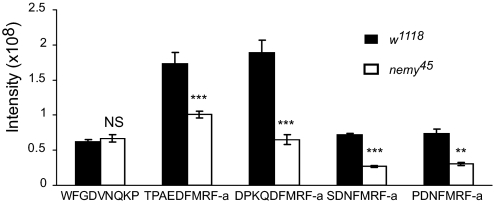
In vertebrates, the amidation process is a specific and necessary modification of peptide hormones, neurotransmitters, and growth factors (26). This process is catalyzed by peptidylglycine α-amidating monooxygenase (PAM). PAM is a bifunctional enzyme with two independent enzymatic domains that catalyze two sequential reactions. First, the rate-limiting enzyme, PHM, hydroxylates the C-terminal Gly residue in peptidylglycine intermediates and then peptidyl-α-hydroxyglycine-α-amidating lyase (PAL) cleaves the intermediates to produce the final amidated peptides (27). In Drosophila, these two enzymatic reactions are encoded by two separate genes (28). To determine whether a functional interaction exists between Nemy and PHM, we quantitated the activity of PHM by measuring the levels of amidated peptides in nemy mutant and control flies using high accuracy mass spectrometric analysis. We focused on four kinds of FMRFamide peptides that are derived from the pro-dFMRF precursor by means of PHM enzymatic activity. We found a significant reduction in the levels of amidated peptides in mutants compared with controls (P < 0.001 for TPAEDFMRFamide, DPKQDFMRFamide, SDNFMRFamide and P < 0.01 for PDNFMRFamide) (Fig. 5). In contrast, we did not find any differences in the level of a nonamidated peptide (WFGDVNQKPI) that served as an internal control. Interestingly, the intensity values for the four FMRFamide peptides reflected, to a certain degree, the number of copies of each peptide in the precursor (29). The highest intensity was observed for DPKQDFMRFamide (five copies per precursor) and lower intensities were observed for both SDNFMRFamide and PDNFMRFamide, which are present in single copies.
Fig. 5.
Nemy regulates the activity of PHM. Comparison of the occurrence of nonamidated and amidated FMRFamide peptides in nemy45 mutant (nonshaded column) and control (shaded column) brains. Peptides extracted from larvae brains were resolved by reverse phase chromatography and identified after tandem mass spectrometry (MS). High resolution Fourier Transform MS (FTMS) was used to quantify the peptides. NS non significant; **, P < 0.01; ***, P < 0.001 (t test). Data combined from 3 independent experiments. Overall n is 150 brains per genotype.
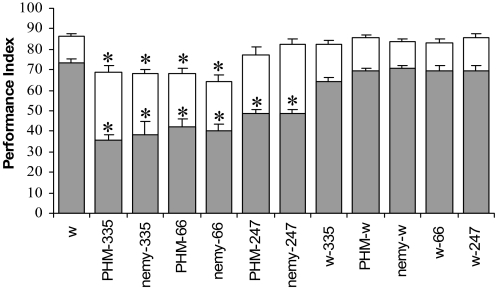
To further demonstrate a link between peptide amidation and learning and memory and to determine the anatomical brain structures that may be responsible for the nemy learning/memory defects, we used RNAi techniques. Specifically, we crossed a UAS-nemy-RNAi or UAS-PHM-RNAi line to specific Gal-4 drivers that would knock down expression of either gene in nemy-positive cells (nemy335), antennal lobes (OK66), or mushroom bodies (MB247) (for the GFP expression pattern of these lines see SI Text and Fig. S7). We found that knockdown of either nemy or PHM expression in nemy-expressing cells or the antennal lobes significantly reduced memory retention when measured immediately or 1 h after training, while knockdown in mushroom body-specific neurons only affected 1-h memory performance (Fig. 6).
Fig. 6.
RNAi-mediated knockdown of nemy and PHM expression in nemy-positive cells (Gal4-nemy335), antennal lobe (Gal4-OK66) or mushroom bodies (Gal4-MB247). Memory retention was tested immediately (nonshaded column) or 1 h (shaded column) after training. *, significant differences between RNAi-Gal4 line and w1118 and its appropriate controls, P < 0.05 at least (t test); n = at least 6 PIs per time point/variant.
Discussion
Using a forward genetic approach, we have identified and characterized nemy, a gene that is required for associative learning and memory in Drosophila. nemy mutants display impaired memory retention in a courtship suppression paradigm (16) and in olfactory conditioning. This suggests that nemy may be involved in the general cellular mechanisms underlying these two forms of associative learning. The evidence presented here demonstrates that the nemy phenotype arises from mutations in the CG8776 gene. First, both the nemyP153 and nemy335 insertions are located in the 5′ region of the CG8776. These insertional lines, as well as the imprecise excision line nemy45 (with a deletion of 743 bp entirely within the CG8776 sequence), show obvious and consistent memory deficits. Second, all nemy mutants display reduced levels of CG8776 expression compared with appropriate controls. Third, both gene expression levels and behavior were completely rescued with a wild-type CG8776 transgene in two independent transgenic lines. Finally, RNAi-mediated knockdown of nemy expression using either nemy-GAL4 or GAL4 lines that drive expression of the nemy RNAi transgene in the antennal lobe or mushroom body cells, also resulted in learning and memory deficits.
nemy encodes a Drosophila homologue of CytB561, which is present in a wide variety of species (30, 31) and encodes for a major transmembrane protein that is found in both small synaptic vesicles and large dense core vesicles (32). In vertebrates, CytB561 is unique among other electron carriers because it does not interact with any other protein. Instead, it uses extravesicular ascorbate as an electron donor for reduction of semidehydroascorbate in the granule matrix with the resulting ascorbate serving as a cofactor for intravesicular DbH and PHM activity (22). PHM performs the rate-limiting step in the C-terminal α-amidation of peptides. α-amidation is a widespread process: In vertebrates, over half of the known neuropeptides require amidation for bioactivity (26), whereas in insects, >90% of neuropeptides show the presence of a C-terminal amide moiety (33). Amidated neuropeptides regulate numerous physiological processes including pain sensation, stress responses, appetite regulation, circadian rhythms, sleep, learning, and memory (for review, see 34).
Our results clearly show that nemy expression is mainly associated with neuropeptidergic neurons within the larval CNS while in the adult brain, nemy is most abundant in AL and MB, which are the main centers for Drosophila learning and memory. Interestingly, PHM-like immunoreactivity was also found among Kenyon cells and in the lobes of MB (25). Similar coexpression of CytB561 and PHM has also been observed in other animals. In the planarian CNS, the distribution of PHM is very similar to that of CytB561 (35). In mammals, immunocytochemical studies have demonstrated that CytB561 is present in many neural and endocrine tissues and that its distribution is correlated with the presence of either catecholamines or amidated peptides in these tissues (36).
Coexpression of nemy and PHM in the Drosophila CNS has important implications for a functional relationship between these two proteins. In fact, we found, using comparable mass spectrometry analysis of amidated neuropeptides, that nemy is required for full activity of PHM. That is, we observe a significant reduction in the levels of amidated peptides in nemy mutants consistent with its proposed role in the regulation of PHM activity. Moreover, we also find that RNAi knockdown of PHM gives rise to similar behavioral defects as those observed in nemy mutants, providing further evidence for a mechanistic link between peptide amidation and learning and memory.
In summary, we propose a model whereby the memory deficits observed in nemy are because of reduced function of CytB561 in specific brain regions that are involved in learning and memory. CytB561 uses ascorbate as an electron donor, and transfers electrons across the secretory vesicle membrane to donate electrons (through intravesicular ascorbate regeneration) for PHM activity, which in turn, is involved in neuropeptide amidation. As such, reduced PHM activity leads to a decrease in the production of bioactive amidated neuropeptides, which may play a role in modulation of the memory process.
Because neuropeptides are also involved in numerous other biological functions, it will be interesting to determine if nemy is also involved in these processes. For example, does nemy contribute to other neuropeptide-related behavioral phenotypes such as circadian rhythm? It will also be interesting to determine whether other nemy transcripts are essential for learning and memory and where they are expressed. Finally, because CytB561 may also affect DbH activity, it will be interesting to determine whether octopamine synthesis is also involved in an alternative nemy-mediated learning/memory pathway.
Materials and Methods
Drosophila Stocks and Genetics.
All stocks were raised on standard fly food, with a 12/12-h light/dark cycle, at 24 ± 1°C and 45–50% relative humidity. The Canton S line was used as the wild-type control. The w1118 control line was outcrossed for 10 generations with Canton S. All other lines including the balancer and GAL4 lines were outcrossed for at least 5 generations with the Cantonized w1118 line. Additional information about used flies strains can be found in SI Text.
Construction of Transgenic Rescue Animals.
The nemy transgenic construct was generated by cloning the coding sequence of CG8776RA (RE 01022 clone) into the pUAST vector. Correct orientation was confirmed by restriction digests and sequencing. Germ-line transformation was carried out by injecting the UAS nemyRA construct into Cantonized w1118 embryos using standard microinjection methods (37). Four independent UAS-transgenic lines (with insertion on the third chromosome) were recovered.
Screening of the cDNA Library and Sequencing.
Approximately 2 × 106 clones from an adult fly cDNA library were screened (gift from Dr. T. Schwarz, Boston, MA). A hybridization probe was amplified by using primers corresponding to common cDNA sequences for all splice variants (for primer sequences, see SI Text). Positive clones contained within pBluescript SK−were excised according to the manufacturer's protocol (Stratagene) by using the ExAssist helper phage system and sequenced. Rapid amplification of cDNA ends (RACE) was performed by using FirstChoice RLM-RACE (Ambion) from full-length, capped mRNA only. Eleven full-length clones were sequenced from both strands and sequence data have been analyzed with the basic local alignment search tool (BLAST) of the National Center for Biotechnology Information. The genome organization was investigated by comparing the cDNA to the Drosophila genomic database (Berkeley Drosophila Genome Project http://www.fruitfly.org).
Real Time RT-PCR.
For Real-Time RT-PCR, total RNA from adult heads was isolated by using TRI REAGENT (Molecular Research Center) according to the manufacturer's instructions, treated with amplification grade DNase1 (Invitrogen), then reverse transcribed by using a SuperScript II reverse transcriptase kit (Roche). Quantitative real-time PCR was performed on an ABI PRISM 7700 (Applied Biosystems) with SYBR Green Reagent (see details in SI Text).
Pavlovian Olfactory Conditioning.
Learning and memory experiments as well as sensorimotor responses were essentially tested as described by Tully and Quinn (38) (for details see SI Text). All training and testing procedures were performed in a climate controlled room with 70% humidity at 25°C under dim red light.
Immunohistochemistry and Fluorescence in situ Hybridization.
Adult and larval brains were dissected in ice-cold PBS, then rinsed and mounted in PBS. Double-labeling fluorescence in situ hybridization (FISH) was performed by using procedures outlined in the protocol on the web site http://www.utoronto.ca/krause/FISH.html. A short description can be found in SI Text.
Imaging and Microscopy.
Fluorescent images were captured with a Leica DM RA2 microscope by using Openlab software version 5 (Improvision) or with an LSM 510 Meta confocal microscope, using LSM 510 software (Zeiss). All images were processed and assembled by using Adobe Photoshop.
Mass Spectrometry.
The brains of 50 third instar larvae were dissected and crushed in an ice-cold methanol/water/formic acid (90:9:1) solution. The supernatants were filtered through Millipore spin-down filters (0.22 μm, 6,000 rpm for 10 min), dried and stored at −20°C until analysis. For all details, see SI Text.
Supplementary Material
Acknowledgments.
We thank Prof. M. Heisenberg, P. Taghert, and E. Skoulakis for their continued interest in these studies and useful discussions; T. Tully for helpful suggestions on the learning and memory assays; W. Trimble for critically reading the manuscript; E. Lecuyer (University of Toronto, Toronto, Canada) for suggestions and supplies for the FISH experiments; and all referees for constructive criticism and suggestions. This work was supported by the Russian Foundation for Basic Research, federal and regional grants from the Russian Academy of Sciences (to N.K.), and Canadian Institutes of Health Research Grant FRN14143 and a grant from the Alzheimer's Association (to G.L.B.). M.F.M is the recipient of a Tier I Canada Research Chair in Molecular Therapeutics, and G.L.B. is the recipient of a Tier I Canada Research Chair in Molecular and Developmental Neurobiology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810698105/DCSupplemental.
References
- 1.Kandel ER. The molecular biology of memory storage: A dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 2.Dubnau J, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 3.Skoulakis EM, Grammenoudi S. Dunces and da Vincis: The genetics of learning and memory in Drosophila. Cell Mol Life Sci. 2006;63:975–988. doi: 10.1007/s00018-006-6023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keene AC, Waddell S. Drosophila olfactory memory: Single genes to complex neural circuits. Nat Rev Neurosci. 2007;8:341–354. doi: 10.1038/nrn2098. [DOI] [PubMed] [Google Scholar]
- 5.Chen CN, Denome S, Davis RL. Molecular analysis of cDNA clones and the corresponding genomic coding sequences of the Drosophila dunce+ gene, the structural gene for cAMP phosphodiesterase. Proc Natl Acad Sci USA. 1986;83:9313–9317. doi: 10.1073/pnas.83.24.9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levin LR, et al. The Drosophila learning and memory gene rutabaga encodes a Ca2+/Calmodulin-responsive adenylyl cyclase. Cell. 1992;68:479–489. doi: 10.1016/0092-8674(92)90185-f. [DOI] [PubMed] [Google Scholar]
- 7.Moore MS, et al. Ethanol intoxication in Drosophila: Genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- 8.Yin JC, et al. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 9.Comas D, Petit F, Preat T. Drosophila long-term memory formation involves regulation of cathepsin activity. Nature. 2004;430:460–463. doi: 10.1038/nature02726. [DOI] [PubMed] [Google Scholar]
- 10.Didelot G, et al. Tequila, a neurotrypsin ortholog, regulates long-term memory formation in Drosophila. Science. 2006;313:851–853. doi: 10.1126/science.1127215. [DOI] [PubMed] [Google Scholar]
- 11.Keleman K, Kruttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- 12.Joiner MlA, Griffith LC. CaM kinase II and visual input modulate memory formation in the neuronal circuit controlling courtship conditioning. J Neurosci. 1997;17:9384–9391. doi: 10.1523/JNEUROSCI.17-23-09384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nogues X. Protein kinase C, learning and memory: A circular determinism between physiology and behaviour. Prog Neuropsychopharmacol Biol Psychiatry. 1997;21:507–529. doi: 10.1016/s0278-5846(97)00015-8. [DOI] [PubMed] [Google Scholar]
- 14.Mery F, Belay AT, So AK, Sokolowski MB, Kawecki TJ. Natural polymorphism affecting learning and memory in Drosophila. Proc Natl Acad Sci USA. 2007;104:13051–13055. doi: 10.1073/pnas.0702923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamyshev NG, et al. Identification of Drosophila mutant with memory defects after acquisition of conditioned reflex suppression of courtship. Neurosci Behav Physiol. 2000;30:307–313. doi: 10.1007/BF02471783. [DOI] [PubMed] [Google Scholar]
- 16.Kamyshev NG, et al. Novel memory mutants in Drosophila: Behavioral characteristics of the mutant nemyP153. BMC Neuroscience. 2002;3:9. doi: 10.1186/1471-2202-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 18.Heisenberg M. Mushroom body memoir: From maps to models. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 19.Davis RL. Olfactory memory formation in Drosophila: From molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- 20.Srivastava M, Duong LT, Fleming PJ. Cytochrome b561 catalyzes transmembrane electron transfer. J Biol Chem. 1984;259:8072–8075. [PubMed] [Google Scholar]
- 21.Menniti FS, Knoth J, Diliberto EJ., Jr. Role of ascorbic acid in dopamine beta-hydroxylation. The endogenous enzyme cofactor and putative electron donor for cofactor regeneration. J Biol Chem. 1986;261:16901–16908. [PubMed] [Google Scholar]
- 22.Kent UM, Fleming PJ. Purified cytochrome b561 catalyzes transmembrane electron transfer for dopamine beta-hydroxylase and peptidyl glycine alpha-amidating monooxygenase activities in reconstituted systems. J Biol Chem. 1987;262:8174–8178. [PubMed] [Google Scholar]
- 23.Wallace BG. The biosynthesis of octopamine - characterization of lobster tyramine beta-hydroxylase. J Neurochem. 1976;26:761–770. doi: 10.1111/j.1471-4159.1976.tb04449.x. [DOI] [PubMed] [Google Scholar]
- 24.Schwaerzel M, et al. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taghert PH, et al. Multiple amidated neuropeptides are required for normal circadian locomotor rhythms in Drosophila. J Neurosci. 2001;21:6673–6686. doi: 10.1523/JNEUROSCI.21-17-06673.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: Peptide alpha-amidation. Annu Rev Neurosci. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- 27.Prigge ST, Mains RE, Eipper BA, Amzel LM. New insights into copper monooxygenases and peptide amidation: Structure, mechanism and function. Cell Mol Life Sci. 2000;57:1236–1259. doi: 10.1007/PL00000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolhekar AS, et al. Neuropeptide amidation in Drosophila: Separate genes encode the two enzymes catalyzing amidation. J Neurosci. 1997;17:1363–1376. doi: 10.1523/JNEUROSCI.17-04-01363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taghert PH. FMRFamide neuropeptides and neuropeptide-associated enzymes in Drosophila. Microsc Res Tech. 1999;45:80–95. doi: 10.1002/(SICI)1097-0029(19990415)45:2<80::AID-JEMT3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 30.Verelst W, Asard H. A phylogenetic study of cytochrome b561 proteins. Genome Biol. 2003;4:R38. doi: 10.1186/gb-2003-4-6-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsubaki M, Takeuchi F, Nakanishi N. Cytochrome b561 protein family: Expanding roles and versatile transmembrane electron transfer abilities as predicted by a new classification system and protein sequence motif analyses. Biochim Biophys Acta. 2005;1753:174–190. doi: 10.1016/j.bbapap.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 32.Fischer von Mollard G, et al. rab3 is a small GTP-binding protein exclusively localized to synaptic vesicles. Proc Natl Acad Sci USA. 1990;87:1988–1992. doi: 10.1073/pnas.87.5.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang N, et al. PHM is required for normal developmental transitions and for biosynthesis of secretory peptides in Drosophila. Dev Biol. 2000;226:118–136. doi: 10.1006/dbio.2000.9832. [DOI] [PubMed] [Google Scholar]
- 34.Strand FL. Neuropeptides: Regulators of Physiological Processes. Cambridge, MA: MIT Press; 1999. [Google Scholar]
- 35.Asada A, Orii H, Watanabe K, Tsubaki M. Planarian peptidylglycine-hydroxylating monooxygenase, a neuropeptide processing enzyme, colocalizes with cytochrome b561 along the central nervous system. FEBS J. 2005;272:942–955. doi: 10.1111/j.1742-4658.2004.04528.x. [DOI] [PubMed] [Google Scholar]
- 36.Pruss RM, Shepard EA. Cytochrome b561 can be detected in many neuroendocrine tissues using a specific monoclonal antibody. Neurosci. 1987;22:149–157. doi: 10.1016/0306-4522(87)90205-3. [DOI] [PubMed] [Google Scholar]
- 37.Spradling AC. P element mediated transformation. In: Roberts DB, editor. Drosophila: A Practical Approach. Oxford, United Kingdom: IRL Press; 1986. pp. 175–197. [Google Scholar]
- 38.Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol [A] 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.