Abstract
We report the crystal structure of a translation termination complex formed by the Thermus thermophilus 70S ribosome bound with release factor RF2, in response to a UAA stop codon, solved at 3 Å resolution. The backbone of helix α5 and the side chain of serine of the conserved SPF motif of RF2 recognize U1 and A2 of the stop codon, respectively. A3 is unstacked from the first 2 bases, contacting Thr-216 and Val-203 of RF2 and stacking on G530 of 16S rRNA. The structure of the RF2 complex supports our previous proposal that conformational changes in the ribosome in response to recognition of the stop codon stabilize rearrangement of the switch loop of the release factor, resulting in docking of the universally conserved GGQ motif in the PTC of the 50S subunit. As seen for the RF1 complex, the main-chain amide nitrogen of glutamine in the GGQ motif is positioned to contribute directly to catalysis of peptidyl-tRNA hydrolysis, consistent with mutational studies, which show that most side-chain substitutions of the conserved glutamine have little effect. We show that when the H-bonding capability of the main-chain N-H of the conserved glutamine is eliminated by substitution with proline, peptidyl-tRNA esterase activity is abolished, consistent with its proposed role in catalysis.
Keywords: 70S ribosome structure, stop codon recognition, polypeptide release
In bacteria, termination of protein synthesis depends on the type I release factors, RF1 and RF2, which are required for recognition of the stop codons and for hydrolysis of the peptidyl-tRNA ester bond. Our understanding of the mechanism of termination faces three main questions: (i) How are stop codons recognized? Unlike the sense codons, there are no corresponding cognate tRNAs to recognize nonsense codons. Are they recognized directly by the release factors, or indirectly, for example through ribosomal RNA? (ii) What is the mechanism of peptidyl-tRNA hydrolysis? Is the esterase reaction catalyzed directly by the release factors, or by the ribosome? And (iii) How is peptidyl-tRNA hydrolysis coupled to stop codon recognition?
Termination at the UAG stop codon depends on RF1, UGA on RF2, and UAA on either of the two factors (1–3). Thus, although the two release factors have similar overall structures (4–6) and both recognize codons of the general type URR, RF1 is able to discriminate between A and G at the second position whereas RF2 discriminates between A and G at the third position. Determinants for codon specificity were localized to domain 2 of the release factors, in particular to the conserved PxT and SPF motifs of RF1 and RF2, respectively, based on genetic studies in which swapping these motifs was found to switch codon specificity (7, 8). A “tripeptide anticodon” mechanism for stop-codon recognition was proposed, in which the PxT and SPF motifs recognize the corresponding stop codons (7, 8). In a recent 3.2 Å crystal structure of a termination complex containing RF1, Thr-186 of the PxT motif was indeed found to be a critical recognition element of RF1, interacting directly with the UA dinucleotide in the first and second positions of the UAA stop codon (9). The third-position A was seen to be unstacked from the rest of the codon, sandwiched between Ile-192 of RF1 and G530 of 16S rRNA, and recognized separately by interactions with Gln-181 and Thr-194. Stop codon recognition by RF1 also involves a network of interactions with other structural elements of RF1, including critical main-chain atoms and conserved features of 16S rRNA (9).
Many studies have implicated the conserved GGQ motif in domain 3, present in the release factors of all three primary domains of life, in the hydrolysis reaction. Although the side chain of the conserved glutamine has been proposed to play a role in catalysis (10, 11), elimination of its side-chain amide group by mutation of this residue to alanine, for example, confers only a small decrease in catalytic activity (12–14). This was rationalized by the structure of the RF1 termination complex, which showed that the side chain of the glutamine is directed away from the scissile bond, whereas its main-chain amide is positioned to participate in catalysis through product and/or transition-state stabilization (9). This unexpected result also explains why substitutions of the neighboring glycine cause severe defects in peptide release (14–16): introduction of a side chain would block access of the main-chain amide of the glutamine to the reaction center.
The structure of the RF1 complex also suggested a mechanism for how codon recognition is coupled to peptidyl-tRNA hydrolysis. Upon recognition of the UAA stop codon, G530 and A1492 flip out, but A1493, which would clash with domain 2 of RF1 if flipped as in sense codon recognition (17), remains stacked within helix 44; A1913 of 23S rRNA then stacks on A1493 of 16S rRNA. The interface between the rearranged decoding site and the reading head of the factor thus forms a binding site for an altered conformation of the “switch” loop, which links domains 3 and 4 of RF2 (Fig. 1), forming a rigid connector that places domain 3 and its GGQ motif in contact with the peptidyl-tRNA ester linkage in the peptidyl transferase center of the 50S subunit. This scenario is consistent with the observations that deletion of helix 69 of 23S rRNA, whose apical loop contains A1913, results in a specific defect in RF1-dependent peptidyl-tRNA hydrolysis (18), and that paromomycin, which induces flipping out of both A1492 and A1493 (17), and which occupies the site vacated by the flipped A1493, inhibits termination, but not sense codon recognition (19).
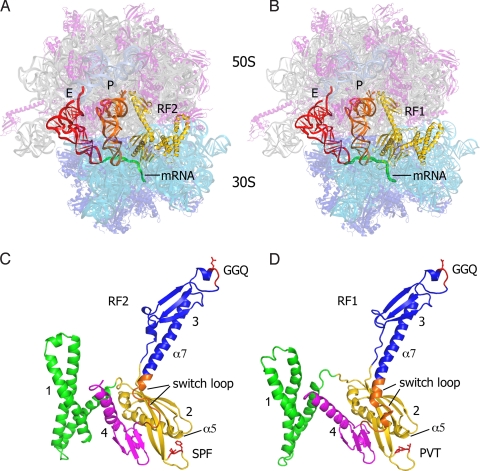
Fig. 1.
Comparison of the structures of the RF1 and RF2 termination complexes. (A) RF2 termination complex (this work), showing RF2 (yellow), P-site tRNA (orange), E-site tRNA (red), mRNA (green), 16S rRNA (cyan), 23S and 5S rRNA (gray), 30S proteins (blue), and 50S proteins (magenta). (B) RF1 termination complex (9); molecular components are colored similarly as in A. (C) RF2 in its ribosome-bound conformation, rotated ≈180° from the view shown in A, with domains numbered. The GGQ and SPF motifs are shown in red, and the switch loop is shown in orange. (D) RF1 in its ribosome-bound conformation. The GGQ and PVT motifs are indicated in red and the switch loop is shown in orange.
Here, we report the crystal structure of a translation termination complex containing release factor RF2 bound in response to a UAA codon, solved at 3Å resolution. The different codon recognition specificities of RF1 and RF2 can be rationalized by structural differences in the decoding center, where Ser-206 of the SPF motif of RF2 interacts directly with the second base and Thr-216 recognizes A3 of the stop codon. Despite considerable sequence divergence in the sequences of the switch loops of RF1 and RF2, the switch loop of RF2 also undergoes a conformational change that is likely involved in coupling codon recognition to the positioning of domain 3 (4). The GGQ motif of RF2 is positioned essentially identically to that seen for the RF1 complex, again implicating the main-chain amide nitrogen of the conserved glutamine in catalysis of peptidyl-tRNA hydrolysis. Finally, when the H-bonding capability of the main-chain N-H of the Gln is eliminated by substitution with proline, activity is abolished, consistent with its proposed role in catalysis.
Results and Discussion
Interaction with the L11 Stalk.
The overall position and conformation of RF2 in the 70S ribosome are similar to those of RF1 (5, 9, 20, 21). The main difference between the two structures is found in the positioning of domain 1 of the factor. In the X-ray structures of the RF1 complex, no contact was observed between domain 1 and the L11 stalk of the 50S subunit (5, 9). By contrast, in the RF2 complex, the distal end of domain 1 is 12 Å closer to the ribosomal A site, placing it in contact with the L11 stalk (Fig. S1), in agreement with low-resolution X-ray studies of the RF2 complex (5). Interactions with 23S rRNA occur at the loops of helices 43 (at A1067) and 44 (at A1095), where the conserved Trp-52 stacks on A1095 (Fig. S1B). The sole protein–protein contacts involve packing of helix α1 of RF2 against the proline-rich helix α1 near the N terminus of L11 (Fig. S1B). The possible biological significance of this difference between the RF1 and RF2 termination complexes is unclear.
Stop Codon Recognition.
In previous studies, the universally conserved nucleotides A530, A1492 and A1493 of 16S ribosomal RNA in the decoding center of the 30S subunit were found to undergo striking rearrangements in response to recognition of a sense codon by cognate tRNA (17, 22), binding of IF1 (23), binding of antibiotics (17, 24, 25) or recognition of a stop codon by RF1 (9). In the RF2 termination complex, G530 and A1492 flip out of their resting states, whereas A1493 remains stacked on the end of helix 44 and A1913 of 23S rRNA stacks on A1493, as observed for the RF1 complex (9) (Fig. 2A and Fig. S2). Also, unlike the conformation seen for sense codons, the third nucleotide of the stop codon (A3) is again found to be unstacked from the first 2 nt and is instead stacked on the flipped G530 of 16S rRNA. Thus, as for the RF1 complex, the first 2 bases of the stop codon are read separately from the third base by RF2.
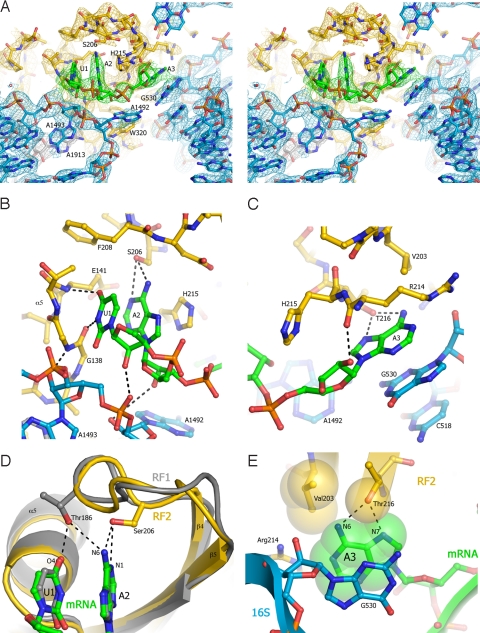
Fig. 2.
Interactions with the UAA stop codon in the decoding center in the RF1 and RF2 termination complexes. (A) Stereoview of the σA-weighted 3Fobs − 2Fcalc electron density map of the stop codon and surrounding elements of RF2 and the ribosome. Electron density is contoured at 1.0 σ for RF2, and at 1.5 σ for rRNA and mRNA, and colored yellow (RF1), green (mRNA), and blue (16S rRNA). (B) Interaction of the hydroxyl group of Ser-206 of the SPF motif of RF2 with A2 of the stop codon. (C) Interaction of Thr-216 of RF2 with A3 of the stop codon. (D) Comparison of the positions of Thr-186 of the PVT motif of RF1 (gray) and Ser-206 of the SPF motif of RF2 (yellow), showing their different modes of recognition of U1 and A2 of the UAA stop codon. The structures of the two termination complexes were globally superimposed. (E) Packing of RF2 around A3 of the UAA stop codon. Val-203 would be positioned to exclude water from H-bonding to O6 of guanine, helping to discriminate against guanine at position 3. The structure model is represented as Van der Waals surfaces for RF2 (yellow) and mRNA (green); 16S rRNA is shown in blue.
Strong specificity for U in the first position (26) is determined by hydrogen bonds between U1 and the backbone carbonyl of Gly-138 and the backbone amide nitrogen of Glu-141 at the tip of helix α5 of RF2, almost identical to the mechanism used by RF1 (Fig. 2B). The main difference is that, whereas Thr-186 of the discriminatory PxT tripeptide motif of RF1 contacts U1 of the stop codon, there is no interaction between U1 and Ser-206 of the SPF motif in RF2. This is a consequence of the different positions of the hydroxyl groups of Thr-186 (of RF1) and Ser-206 (of RF2) in their respective complexes (Fig. 2 B and D). Although the side chain of the serine is not well resolved, its conformation, and those of the clearly resolved features in the surrounding part of domain 2 in our structure (Fig. S3) are virtually identical to those reported for the 1.8 Å structure of free RF2 (4). Whereas the OH of Thr-186 is positioned between U1 and A2, Ser-206 directly faces the Watson–Crick edge of A2, well outside H-bonding distance from U1 (Fig. 2D). Accordingly, Ser-206 is solely occupied with recognition of the second base of the stop codon, and is predicted to be able to H-bond equally well with either A (at N1 and N6) or (by swapping the positions of its hydrogen-bond donor and acceptor moieties) G (at N1 and O6), in keeping with the codon specificity of RF2 for UAA or UGA (Fig. S2 G and H). Conservation of the remaining residues of the SPF (RF2) or PxT (RF1) motifs could be explained by requirements for excluding water from the decoding pocket of the first and second bases, in addition to maintaining the correct fold of the loop.
In the RF1 complex, recognition of the third base (A3) occurs separately from the first 2 bases, and is shared between Gln-181 and Thr-194 (9) (Fig. S2F). Gln-181 is positioned to accept an H-bond from N6 of adenine with its amide carbonyl oxygen and Thr-194 can H-bond to the N6 and N7 positions; for recognition of G in the third position by RF1, the amide group of Gln-181 could donate an H-bond to O6. In the RF2 complex, Thr-216 occupies the same position as Thr-194 of RF1, enabling recognition of adenine; however, no residue equivalent to Gln-181 is found. Moreover, the position of the hydrophobic side chain of Val-203 would prevent H-bonding of the O6 of guanine to water (Fig. 2E). Discrimination against guanine in the third position could be the result of a large free-energy penalty due to desolvation of guanine, whose dipole moment is significantly larger than that of adenine (27, 28). The next 2 nt downstream from the A-site codon are also well ordered, enabling their detailed conformations to be seen here for the first time (Fig. S4). The nucleotide following the stop codon (A4), which has been implicated in translation termination efficiency (29), intercalates between U1196 and C1397 of 16S rRNA. Its Hoogsteen edge faces Ser-211, located on the opposite side of the specificity loop from the SPF motif (Fig. S4B), consistent with the observed cross-linking of RF2 to the base immediately downstream of the stop codon (30).
Communication Between the Decoding and Peptidyl Transferase Centers.
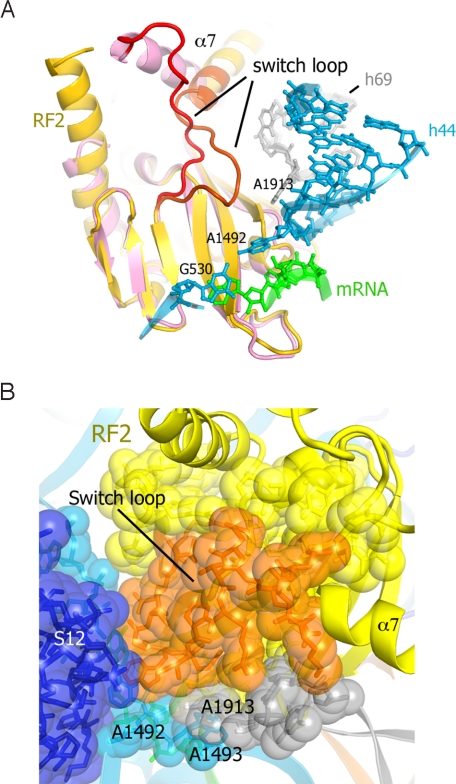
Despite the absence of proof-reading, spontaneous termination is rare (26), implying that recognition of the stop codon and hydrolysis of the peptidyl-tRNA ester bond are strictly coordinated. Comparison of the conformations of the release factors in their free state (4, 6) with that of RF1 in a termination complex (9) led us to propose that recognition of a stop codon stabilizes a rearranged conformation of the switch loop, which directs domain 3 into the PTC. In both the RF1 and RF2 complexes, the binding pocket for the rearranged switch loop is formed in part by flipping A1492 of 16S rRNA and stacking of A1913 of 23S rRNA on A1493. Although the sequence of the switch loop of RF2 diverges sharply from that of RF1 (Fig. S5 A and B), it is similarly packed in the pocket formed by A1492, A1913 and protein S12 (Fig. 3 A and B and Fig. S5), suggesting a conserved mechanism for coordination of codon recognition and peptidyl-tRNA hydrolysis.
Fig. 3.
Rearrangement and packing of the switch loop in the RF2 termination complex. (A) Superposition of domain 2 of free RF2 (pink; ref. 4) on that of ribosome-bound RF2 (yellow; this work). Rearrangement of the switch loop (residues 310–324; red in free RF2 and orange in bound RF2) results in reorientation and extension of α7. (B) Packing of the switch loop in a pocket, which includes A1492 of 16S rRNA, A1913 of 23S rRNA and protein S12 directs helix α7 so that the GGQ docks in the PTC. Components are colored as in Fig. 1A.
Testing the Mechanism of Peptidyl-tRNA Hydrolysis.
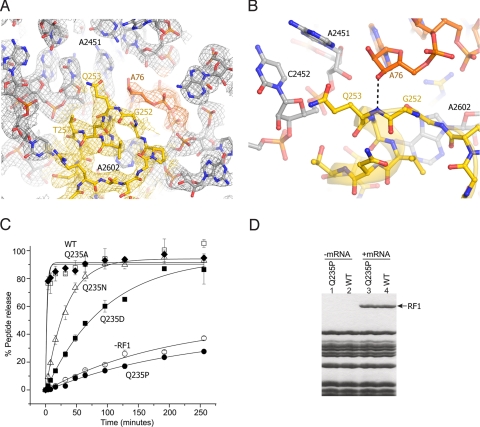
The overall conformation of domain 3 of RF2 in the PTC is very similar to that of RF1 (9). Nucleotide A2602, which was shown to be important for peptide release (31–33), is buried in a pocket formed by RF2 and is blocked from the catalytic center by RF2 (Fig. 4 A and B and Fig. S6). The universally conserved GGQ motif (residues 251–253 in T. thermophilus RF2) contacts the 3′-terminus of the deacylated P-site tRNA (Fig. 4 A and B). As seen for the structure of the RF1 termination complex, the side-chain of Gln-253 is pointed away from the site of catalysis in a pocket formed by A2451, C2452, U2506 of 23S rRNA and ribose 76 of the P-site tRNA, whereas its main-chain amide is within hydrogen-bonding distance of the 3′-OH of A76 (Fig. 4B). In both termination complexes, the main-chain amide of the conserved Gln-253 (Gln-230 in T. thermophilus RF1) is positioned to participate in catalysis of peptidyl-tRNA hydrolysis by forming a hydrogen bond with the leaving 3′OH group and/or with the transition-state oxyanion. Participation of the backbone amide group in catalysis is precedented in the mechanisms of other hydrolases, including proteases, esterases and GTPases, for product stabilization and to stabilize the developing negative charge of the transition state (34, 35).
Fig. 4.
Interactions of the GGQ region of RF2 at the site of catalysis. (A) σA-weighted 3Fobs − 2Fcalc electron density for RF2 (yellow), P-site tRNA (orange) and 23S rRNA (gray), contoured at 1.7 σ. (B) Orientation of Gln-253 of the RF2 GGQ motif. The backbone amide nitrogen of Gln-253 is positioned to H-bond with the 3′-OH of A76 of P-site tRNA, whereas its side chain is oriented away from the reaction site. (C) The Q235P mutation abolishes peptide release activity of E. coli RF1. Model 70S termination complexes assembled with [35S]fMet-tRNAfMet in the P site and mRNA M0–27 (which contains an AUG codon followed a UAA stop codon), were incubated at 37 °C with (open squares) wild-type RF1, or the RF1 mutants (diamonds) Q235A (filled squares) Q235D (triangles) Q235N (filled circles) Q235P, or (open circles) no RF1. Peptide release was monitored by measuring the amount of [35S]fMet extracted into ethyl acetate at the indicated time points. Error bars indicate the range of values from 2 or 3 independent experiments, which were averaged and fit to single-exponential curves to determine the rates of peptide release. (D) Mutant Q235P RF1 binds normally in a stop-codon-dependent manner. Lanes 1 and 2, neither wild-type nor Q235P RF1 binds to ribosomes in the absence of mRNA. Lanes 3 and 4, in the presence of mRNA MO-27, both Q235P and wild-type RF1 bind stoichiometrically to ribosomes.
Previous mutational analysis of the GGQ motif has shown the catalysis to be remarkably robust. For example, mutation of the conserved Gln to Ala or Trp cause only a 4- to 7-fold decrease of activity (14). However, none of the mutations reported so far have tested the importance of the backbone amide group. Accordingly, we have substituted proline for glutamine in the conserved GGQ motif of Escherichia coli RF1 (Gln-235), and tested the ability of the GGP mutant factor to catalyze hydrolysis of fMet-tRNA in response to a UAA stop codon. We directly compared RF1-GGP to other mutants shown by Shaw and Green (14) to have a range of defects: RF1-GGA (down 4-fold), RF1-GGN (7600-fold), and RF1-GGD (9500-fold). The effect of the proline substitution is striking; whereas even the activity of the GGD mutation, shown (14) to have the most drastic defect, is readily measurable, the activity of the Q235P mutant is undetectable, even at long incubation times where spontaneous hydrolysis is observed (Fig. 4C and Table S1). Although catalytically inactive, GGP mutant RF1 retains stoichiometric, codon-dependent binding to ribosomes (Fig. 4D).
Although this result cannot be taken as definitive evidence for the participation of the Gln-235 (Gln-230 or Gln-253 in T. th. RF1 and RF2, respectively) backbone amide in catalysis, because of the possible influence of proline substitution on backbone conformation, it is consistent with our proposal. Deleterious effects of proline substitution have been interpreted as evidence for participation of the backbone amide group in catalysis by serine proteases (36) and GTPases (37). Moreover, concern that introduction of proline into an active site can indirectly affect catalysis is mitigated by high-resolution X-ray structures of a G119P mutant of thrombin (36) and of an A30P mutant of Rab5a (38), in which proline substitutions fail to cause significant conformational changes. In this regard, an in silico test of the effect of the Q253P mutation, in which we modeled the mutant factor, suggests that the proline substitution would be unlikely to induce significant changes in the conformations of RF1 or the surrounding features in the ribosomal PTC (Fig. S7). This can be explained by the similarities between the main-chain (39) torsion angles for Gln-230 in the experimental structure (9) to those typically observed for proline (Fig. S7) in high-resolution crystal structures (39). Beyond these circumstantial arguments, definitive evidence will require determination of the structure of a termination complex containing the mutant factor.
Materials and Methods
Procedures for crystallization and structure determination were similar to those described (9). The structure was determined by molecular replacement followed by refinement yielding R/Rfree of 0.28/0.316 (Table S2). A detailed account of structure determination procedures is given in SI Materials and Methods.
The construction and isolation of mutant RF1 and peptide release assay was as follows: The RF1 gene from E. coli MRE600 was cloned into pET21b (Novagen) to obtain C-terminal 6His-tagged RF1. Mutations at position Q235 were generated by site-directed mutagenesis (40). RF1 proteins were expressed and purified on Ni-NTA agarose resin (Qiagen) using standard procedures, then additionally purified by FPLC chromatography on a 24-mL Superdex 75 gel filtration column (Amersham Pharmacia); proteins were stored in 50 mM Tris·HCl (pH 7.0), 60 mM NH4Cl, 10 mM MgCl2, and 5 mM β-mercaptoethanol (βME) at −80 °C.
30S and 50S subunits were prepared from salt-washed MRE600 ribosomes as described (41, 42). tRNAfMet was aminoacylated as described in ref. 43. 30S subunits were heat-activated at 42 °C for 10 min in Buffer A [50 mM KHepes (pH 7.6), 75 mM NH4Cl, 20 mM MgCl2, 5 mM βME] before use in each assay.
Model 70S ribosome termination complexes were formed by incubating 30S subunits (1 μM), 50S subunits (1.2 μM), M0–27 mRNA (2 μM), and [35S]fMet-tRNAfMet (0.5 μM) for 20 min at 37 °C in Buffer A. The [Mg2+] was reduced to 10 mM by addition of buffer A lacking MgCl2. For peptide release assays, the complex was added to a 6-fold molar excess of RF1 and incubated in Buffer A (10 mM MgCl2) at 37 °C. Aliquots were removed from the reaction at each time point and quenched in 5 vol of 0.1 M HCl; hydrolyzed [35S]fMet was extracted with 1 mL of ethyl acetate, 0.7 mL of which was added to scintillation mixture and counted.
For RF1 binding assays, the 70S complex was formed as above, but with 2 μM [35S]fMet-tRNAfMet, and with or without M0–27 mRNA. As above, each complex was added to a 6-fold excess of RF1, then incubated at 37 °C for 5 min in Buffer A (10 mM MgCl2). Reactions were passed through a Sephacryl S200-HR resin (Sigma) 1-mL spin column, then precipitated with 4 vol of acetone (−20 °C for 1 h), dissolved in loading buffer, and electrophoresed on a 12% acrylamide SDS gel.
Supplementary Material
Acknowledgments.
We thank John Paul Donohue for assistance with data processing and computational support; Eric Maklan for helpful discussions regarding kinetic measurements; and the beamline staffs at SSRL, ALS, and APS for their expert support during screening and data collection. These studies were supported by grants from the National Institutes of Health and the National Science Foundation (to H.F.N.) and a fellowship from the Danish Research Council (to M.L.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited with the Protein Data Bank, www.pdb.org (PDB ID codes 3F1E, 3F1F, 3F1G, and 3F1H).
This article contains supporting information online at www.pnas.org/cgi/content/full/0810953105/DCSupplemental.
References
- 1.Capecchi MR, Klein HA. Characterization of three proteins involved in polypeptide chain termination. Cold Spring Harbor Symp Quant Biol. 1969;34:469–477. doi: 10.1101/sqb.1969.034.01.053. [DOI] [PubMed] [Google Scholar]
- 2.Scolnick E, Tompkins R, Caskey T, Nirenberg M. Release factors differing in specificity for terminator codons. Proc Natl Acad Sci USA. 1968;61:768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scolnick EM, Caskey CT. Peptide chain termination. V. The role of release factors in mRNA terminator codon recognition. Proc Natl Acad Sci USA. 1969;64:1235–1241. doi: 10.1073/pnas.64.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vestergaard B, et al. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol Cell. 2001;8:1375–1382. doi: 10.1016/s1097-2765(01)00415-4. [DOI] [PubMed] [Google Scholar]
- 5.Petry S, et al. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 6.Shin DH, et al. Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J Mol Biol. 2004;341:227–239. doi: 10.1016/j.jmb.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 7.Ito K, Uno M, Nakamura Y. A tripeptide “anticodon” deciphers stop codons in messenger RNA. Nature. 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura Y, Ito K. A tripeptide discriminator for stop codon recognition. FEBS Lett. 2002;514:30–33. doi: 10.1016/s0014-5793(02)02330-x. [DOI] [PubMed] [Google Scholar]
- 9.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 10.Song H, et al. The crystal structure of human eukaryotic release factor eRF1–mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 11.Trobro S, Aqvist J. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol Cell. 2007;27:758–766. doi: 10.1016/j.molcel.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 12.Seit Nebi A, Frolova L, Ivanova N, Poltaraus A, Kiselev L. Mutation of a glutamine residue in the universal tripeptide GGQ in human eRF1 termination factor does not cause complete loss of its activity. Mol Biol (Moscow) 2000;34:899–900. [PubMed] [Google Scholar]
- 13.Seit-Nebi A, Frolova L, Justesen J, Kisselev L. Class-1 translation termination factors: Invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 2001;29:3982–3987. doi: 10.1093/nar/29.19.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw JJ, Green R. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol Cell. 2007;28:458–467. doi: 10.1016/j.molcel.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frolova LY, et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. Rna. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol Cell. 2002;10:789–798. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- 17.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 18.Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell. 2006;23:865–874. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Youngman EM, He SL, Nikstad LJ, Green R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol Cell. 2007;28:533–543. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Rawat UB, et al. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 21.Rawat U, et al. Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. J Mol Biol. 2006;357:1144–1153. doi: 10.1016/j.jmb.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 22.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 23.Carter AP, et al. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 24.Borovinskaya MA, et al. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 25.Francois B, et al. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: Role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 2005;33:5677–5690. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci USA. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu G, Sivanesan D, Kawabata T, Go N. Electrostatic potential of nucleotide-free protein is sufficient for discrimination between adenine and guanine-specific binding sites. J Mol Biol. 2004;342:1053–1066. doi: 10.1016/j.jmb.2004.07.047. [DOI] [PubMed] [Google Scholar]
- 28.Sponer J, Leszczynski J, Hobza P. Structures and energies of hydrogen-bonded DNA base pairs. A nonempirical study with inclusion of electron correlation. J Phys Chem. 1996;100:1965–1974. [Google Scholar]
- 29.Tate WP, et al. Translational termination efficiency in both bacteria and mammals is regulated by the base following the stop codon. Biochem Cell Biol. 1995;73:1095–1103. doi: 10.1139/o95-118. [DOI] [PubMed] [Google Scholar]
- 30.Poole ES, Major LL, Mannering SA, Tate WP. Translational termination in Escherichia coli: Three bases following the stop codon cross-link to release factor 2 and affect the decoding efficiency of UGA-containing signals. Nucleic Acids Res. 1998;26:954–960. doi: 10.1093/nar/26.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amort M, et al. An intact ribose moiety at A2602 of 23S rRNA is key to trigger peptidyl-tRNA hydrolysis during translation termination. Nucleic Acids Res. 2007;35:5130–5140. doi: 10.1093/nar/gkm539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polacek N, et al. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol Cell. 2003;11:103–112. doi: 10.1016/s1097-2765(02)00825-0. [DOI] [PubMed] [Google Scholar]
- 33.Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 34.Wilmouth RC, et al. X-ray snapshots of serine protease catalysis reveal a tetrahedral intermediate. Nat Struct Biol. 2001;8:689–694. doi: 10.1038/90401. [DOI] [PubMed] [Google Scholar]
- 35.Jaeger KE, Dijkstra BW, Reetz MT. Bacterial biocatalysts: Molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol. 1999;53:315–351. doi: 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- 36.Bobofchak KM, Pineda AO, Mathews FS, Di Cera E. Energetic and structural consequences of perturbing Gly-193 in the oxyanion hole of serine proteases. J Biol Chem. 2005;280:25644–25650. doi: 10.1074/jbc.M503499200. [DOI] [PubMed] [Google Scholar]
- 37.Liang Z, Mather T, Li G. GTPase mechanism and function: New insights from systematic mutational analysis of the phosphate-binding loop residue Ala30 of Rab5. Biochem J. 2000;346(Pt 2):501–508. [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu G, et al. High resolution crystal structures of human Rab5a and five mutants with substitutions in the catalytically important phosphate-binding loop. J Biol Chem. 2003;278:2452–2460. doi: 10.1074/jbc.M211042200. [DOI] [PubMed] [Google Scholar]
- 39.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 40.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 42.Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 43.Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.