Abstract
The Cse4p-containing centromere regions of Candida albicans have unique and different DNA sequences on each of the eight chromosomes. In a closely related yeast, C. dubliniensis, we have identified the centromeric histone, CdCse4p, and shown that it is localized at the kinetochore. We have identified putative centromeric regions, orthologous to the C. albicans centromeres, in each of the eight C. dubliniensis chromosomes by bioinformatic analysis. Chromatin immunoprecipitation followed by PCR using a specific set of primers confirmed that these regions bind CdCse4p in vivo. As in C. albicans, the CdCse4p-associated core centromeric regions are 3–5 kb in length and show no sequence similarity to one another. Comparative sequence analysis suggests that the Cse4p-rich centromere DNA sequences in these two species have diverged faster than other orthologous intergenic regions and even faster than our best estimated “neutral” mutation rate. However, the location of the centromere and the relative position of Cse4p-rich centromeric chromatin in the orthologous regions with respect to adjacent ORFs are conserved in both species, suggesting that centromere identity is not solely determined by DNA sequence. Unlike known point and regional centromeres of other organisms, centromeres in C. albicans and C. dubliniensis have no common centromere-specific sequence motifs or repeats except some of the chromosome-specific pericentric repeats that are found to be similar in these two species. We propose that centromeres of these two Candida species are of an intermediate type between point and regional centromeres.
Keywords: chromatin, chromosome segregation, kinetochore, nucleosome, pericentric
Faithful chromosome segregation during mitosis and meiosis in eukaryotes is performed by a dynamic interaction between spindle microtubules and kinetochores. The kinetochore is a proteinaceous structure that forms on a specific DNA locus on each chromosome, termed the centromere (CEN). Centromeres have been cloned and characterized in several organisms from yeasts to humans. Interestingly, there is no centromere-specific cis-acting DNA sequence that is conserved across species (1). However, centromeres in all eukaryotes studied to date assemble into specialized chromatin containing a histone H3 variant protein in the CENP-A/Cse4p family. Members of this family are called centromeric histones (CenH3s) and are regarded as possible epigenetic markers of CEN identity (1, 2). The Saccharomyces cerevisiae centromere, the most intensively studied budding yeast centromere, is a well-defined, short (125-bp) region (hence called a “point” centromere) and consists of two conserved consensus sequences (centromere DNA elements, CDEs), CDEI (8 bp) and CDEIII (25 bp) separated by CDEII, a 78- to 86-bp nonconserved AT-rich (> 90%) “spacer” sequence (3). CDEI is not absolutely necessary for mitotic centromere function (4). Retention of a portion of CDEII is essential for CEN activity, but changes in length or base composition of CDEII cause only partial inactivation (4, 5). The S. cerevisiae CenH3, ScCse4p, has been shown to bind to a single nucleosome containing the nonconserved CDEII and to flanking CDEI and CDEIII regions (6). CDEIII is absolutely essential: centromere function is completely inactivated by deletion of CDEIII or even by single base substitutions in the central CCG sequence. Centromeres of most other eukaryotes, including the fission yeast Schizosaccharomyces pombe, are much longer and more complex than those of S. cerevisiae and are called “regional” centromeres (3). The centromeres of S. pombe are 40–110 kb in length and organized into distinct classes of repeats that are further arranged into a large inverted repeat. The nonrepetitive central region, also known as the central core (cc), contains a 4- to 7-kb nonhomologous region that is not conserved in all three chromosomes (3). The CenH3 homolog in S. pombe, Cnp1p, binds to the central core and the inner repeats (7). However, the central domain alone cannot assemble centromere chromatin de novo, but requires the cis-acting dg/K repeat present at the outer repeat array to promote de novo centromere assembly (8, 9). Several experiments suggest that unlike in S. cerevisiae, no unique conserved sequence within S. pombe centromeres is sufficient for establishment and maintenance of centromere function, although flanking repeats play a crucial role in establishing heterochromatin that is important for centromere activity (10).
Several lines of evidence suggest that primary DNA sequence may not be the only determinant of CEN identity in regional centromeres. Studies in a pathogenic budding yeast, Candida albicans, containing regional centromeres suggest that each of its eight chromosomes contains a different, 3- to 5-kb nonconserved DNA sequence that assembles into Cse4p-rich centromeric chromatin (11, 12). C. albicans centromeres partly resemble those of S. pombe but lack any pericentric repeat that is common to all of its eight centromeres (12, 13). Therefore, the mechanisms by which CenH3s confer centromere identity, are deposited at the right location, and are epigenetically propagated for several generations in C. albicans without any centromere-specific DNA sequence remain largely unknown.
A recent study of several independent clinical isolates of C. albicans reveals that, despite having no centromere-specific DNA sequence motifs or repeats common to all of its eight centromeres, centromere sequences remain conserved and their relative chromosomal positions are maintained (12). As a first step toward understanding the importance of cis-acting CEN DNA sequences in centromere function in C. albicans, we have identified and characterized centromeres of a closely related pathogenic yeast, C. dubliniensis, which was identified as a less pathogenic independent species in 1995 (14). We reasoned that CEN DNA comparisons between related Candida species might uncover properties that were not evident from interchromosomal comparisons of C. albicans CEN sequences alone. Moreover, functional characterization of centromeres of these two related Candida species may be helpful in understanding the evolution of centromeres. Several studies indicate that both CEN DNA and its associated proteins in animals and plants are rapidly evolving, although the relative position of the centromere is maintained for a long time (15).
Here, we report the identification and characterization of Cse4p-rich centromere sequences of each of the eight chromosomes of C. dubliniensis. Comparative genomic analysis of CEN DNA sequences of C. albicans and C. dubliniensis reveals no detectable conservation among Cse4p-associated CEN sequences. Nonetheless, the lengths of Cse4p-enriched DNAs assembled as specialized centromeric chromatin and their relative locations in orthologous regions have been maintained for millions of years. A genomewide analysis also reveals that centromeres are probably the most rapidly evolving genomic loci in C. albicans and C. dubliniensis.
Results
Synteny of Centromere-Adjacent Genes Is Maintained in C. albicans and C. dubliniensis.
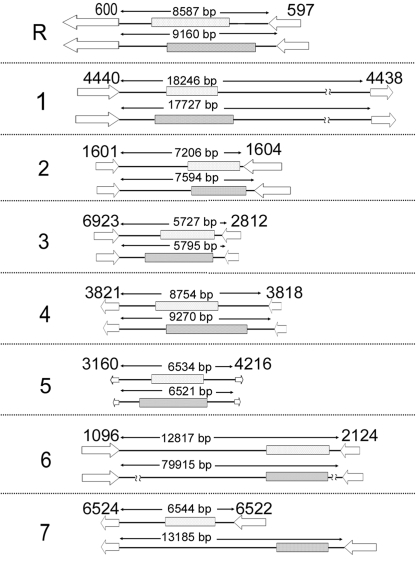
C. albicans and C. dubliniensis diverged ∼20 million years ago from a common ancestor (12). Gene synteny (collinearity) is maintained almost throughout the genome in these two organisms. Therefore, we examined potential orthologous CEN regions in C. dubliniensis by identifying ORFs of C. dubliniensis with homology to CEN-proximal ORFs of C. albicans. C. dubliniensis homologs of C. albicans ORFs that are adjacent to centromere regions were identified by BLAST analysis of the C. dubliniensis genome database available at the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/c_dubliniensis). The homology of amino acid sequences coded by CEN-adjacent genes in C. albicans and C. dubliniensis ranges from 81 to 99% [supporting information (SI) Table S1]. The synteny of these genes is maintained in all chromosomes except chromosome 6 (Fig. 1 and Fig. S1). C. albicans CEN6 is flanked by Orf19.1097 and Orf19.2124. Since there is no Orf19.1097 homolog in C. dubliniensis, we identified the C. dubliniensis homolog of Orf19.1096, the gene adjacent to Orf19.1097 in C. albicans. The distance between Orf19.1096 and Orf19.2124 is 12.8 kb in C. albicans as opposed to 80 kb in C. dubliniensis. A systematic analysis of this 80-kb region of C. dubliniensis reveals that two paracentric inversions followed by an insertion between the Orf19.1096 homolog and its downstream region occurred in C. dubliniensis at the left arm of the orthologous pericentric region as compared to C. albicans (Fig. S1).
Fig. 1.
Orthologous Cse4p-rich centromere regions in C. albicans and C. dubliniensis. On the basis of BLAST analysis, the putative homologs of C. albicans CEN-adjacent ORFs in C. dubliniensis were identified. Chromosome numbers are shown on the left (R through 7). The top line for each chromosome denotes C. albicans centromere regions and the bottom line corresponds to the orthologous regions in C. dubliniensis. The dotted and cross-hatched boxes correspond to Cse4p-binding regions in C. albicans (12) and C. dubliniensis, respectively (see text and Table S2). Only one homolog is shown for each chromosome of C. albicans and C. dubliniensis. ORFs and the direction of transcription of corresponding ORFs are shown by open arrows. Only those ORFs that have homologs in both C. albicans and C. dubliniensis are shown. The number on the top of each arrow corresponds to the C. albicans assembly 19 ORF numbers (for example, orf19.600 is shown as 600). The lengths of CEN-containing intergenic regions of C. albicans and orthologous regions in C. dubliniensis are shown. This analysis was done on the basis of Assembly 20 of the Candida albicans Genome Database and the present version (May 16, 2007) of the Candida dubliniensis Genome Database.
The Centromeric Histone Protein of C. dubliniensis (CdCse4p) is localized at the Kinetochore.
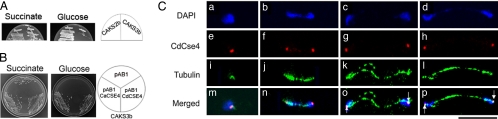
CenH3 proteins in the Cse4p/CENP-A family have been shown to be uniquely associated with centromeres in all organisms studied to date (1). Using CaCse4p as the query in a BLAST analysis against the C. dubliniensis genome, we identified the centromeric histone of C. dubliniensis, CdCse4p (see Materials and Methods). This histone is found to be highly similar (97% identity over 211 aa) to CaCse4p (Fig. S2). CdCse4p codes for a 212-aa-long predicted protein with a C-terminal (amino acid residues 110–212) histone-fold domain (HFD). The HFD of Cse4p in C. albicans and C. dubliniensis is identical (Fig. S2B). To examine whether CdCse4p can functionally complement CaCse4p, we have expressed CdCSE4 from its native promoter (pAB1CdCSE4) cloned in an ARS2/HIS1 plasmid (pAB1) in a C. albicans strain (CAKS3b) carrying the only full-length copy of CaCSE4 under control of the PCK1 promoter (see SI Text). The ability of the strain CAKS3b carrying pAB1CdCSE4 to grow as well as the same strain carrying a control plasmid pAB1CaCSE4 on glucose medium (where endogenous CaCSE4 expression is suppressed) suggests that CdCse4p can complement CaCse4p function and hence codes for the centromeric histone in C. dubliniensis (Fig. 2B). We further examined the subcellular localization of CdCse4p in C. dubliniensis strain Cd36 by indirect immunofluorescence (see Materials and Methods). Indirect immunofluorescence microscopy using affinity-purified polyclonal anti-Ca/CdCse4p antibodies (against aa 1–18 of CaCse4p/CdCse4p) (16) revealed bright dot-like signals in all cells. The dots always colocalized with nuclei stained with DAPI (Fig. 2C). Each bright dot-like signal represents a cluster of 16 centromeres. Unbudded G1 cells exhibited one dot per cell, while large-budded cells at later stages of the cell cycle exhibited two dots that cosegregated with the DAPI-stained nuclei in daughter cells (Fig. 2C). The localization patterns of CdCse4p appear to be identical to those of CaCse4p in C. albicans at corresponding stages of the cell cycle (16). Coimmunostaining of fixed Cd36 cells with anti-tubulin and anti-Ca/CdCse4p antibodies showed that CdCse4p signals are localized close to the spindle pole bodies, analogous to typical localization patterns of kinetochore proteins in S. cerevisiae and C. albicans (Fig. 2C). Together, these results strongly suggest that CdCse4p is the authentic centromeric histone of C. dubliniensis.
Fig. 2.
Localization of CdCse4p at the kinetochore of C. dubliniensis. (A) The C. albicans strain CAKS3b was streaked on media containing succinate and glucose and incubated at 30 °C for 3 days. (B) CAKS3b is transformed with pAB1, pAB1CaCSE4, and pAB1CdCSE4. These transformants were streaked on plates containing complete media lacking histidine with succinate or glucose as the carbon source. (C) C. dubliniensis strain Cd36 was grown in YPD and fixed. Fixed cells were stained with DAPI (a–d), anti-Ca/CdCse4p (e–h), and anti-tubulin (i–l) antibodies. The intense red dot-like CdCse4p signals were observed in unbudded (e) and at different stages of budded cells (f–h). Corresponding spindle structures are shown by coimmunostaining with anti-tubulin antibodies (i–l). Arrows indicate the position of spindle pole bodies in large-budded cells at anaphase. (Scale bar, 10 μm.)
Centromeric Chromatin on Various C. dubliniensis Chromosomes Is Restricted to a 3- to 5-kb Region.
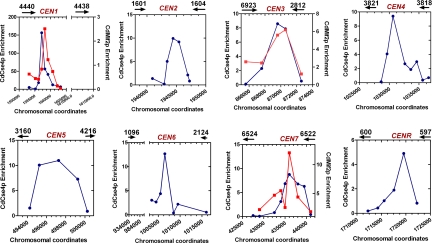
We performed standard chromatin immunoprecipitation (ChIP) assays with anti-Ca/CdCse4p antibodies to assay for enrichment of CdCse4p on putative CEN regions (orthologous to C. albicans CENs) in C. dubliniensis strain Cd36 (see Materials and Methods). The immunoprecipitated DNA sample was analyzed by PCR using a specific set of primers designed from the putative CEN sequences (Table S2). These regions are, indeed, found to be associated with CdCse4p (Fig. 3 and Fig. S3). This ChIP–PCR analysis precisely localized the boundaries of CdCse4p binding to a 3- to 5-kb region on each chromosome (Fig. 3). However, as mentioned earlier, the homologs of two genes adjacent to the CEN6 region in C. albicans are 80 kb apart in chromosome 6 of C. dubliniensis because of chromosome rearrangement (Fig. S1). Since other CEN regions of C. dubliniensis are present in ORF-free regions that are >3 kb, we first identified all of the intergenic regions ≥3 kb, to find CEN6 in this 80-kb region. The ChIP–PCR analysis using specific primers from such regions delimited Cse4p binding to a 3.6-kb region that is adjacent to the C. albicans Orf19.2124 homolog in C. dubliniensis (Fig. 3 and Fig. S3; not all ChIP data are shown). Thus, we have successfully identified CdCse4p-rich CEN regions and determined the boundaries of centromeric chromatin in all eight chromosomes in C. dubliniensis. We also find that the relative distance of Cse4p-rich centromeric chromatin from orthologous neighboring ORFs is similar in both species in most cases (Fig. 1).
Fig. 3.
Two evolutionarily conserved key kinetochore proteins, CdCse4p (CENP-A homolog) and CdMif2p (CENP-C homolog) bind to the same regions of different C. dubliniensis chromosomes. Standard ChIP assays were performed on strains Cd36 and CDM1 (CdMif2-TAP-tagged strain) using anti-Ca/CdCse4p or anti-Protein A antibodies and analyzed with specific primers corresponding to putative centromere regions of C. dubliniensis to PCR amplify DNA fragments (150–300 bp) located at specific intervals as indicated (Table S2). Graphs show relative enrichment of CdCse4p (blue lines) and CdMif2p (red lines) that mark the boundaries of centromeric chromatin in various C. dubliniensis chromosomes. PCR was performed on total, immunoprecipitated (+Ab), and beads-only control (−Ab) ChIP DNA fractions (see Fig. S3 and Fig. S4). The coordinates of primer locations are based on the present version (May 16, 2007) of the Candida dubliniensis genome database. Enrichment values are calculated by determining the intensities of (+Ab) minus (−Ab) signals divided by the total DNA signals and are normalized to a value of 1 for the values obtained for a noncentromeric locus (CdLEU2) and plotted. The chromosomal coordinates are marked along the x-axis while the enrichment values are marked along the y-axis. Black arrows show the location of ORFs and arrowheads indicate the direction of transcription.
The Evolutionarily Conserved Kinetochore Protein CENP-C Homolog in C. dubliniensis, CdMif2p Binds Preferentially to CdCse4p-Associated DNA.
Proteins in the CENP-C family are shown to be associated with kinetochores in a large number of species (17). Using CaMif2p as the query sequence, we identified the CENP-C homolog (CdMif2p) in C. dubliniensis (see Materials and Methods). CdMif2p shows 77% identity and 5% similarity in a 516-aa overlap. CdMif2p codes for a 520-aa-long predicted protein in which the CENP-C box (amino acid residues 275–297) is 100% identical in C. albicans and C. dubliniensis (see Fig. S4). We constructed a strain (CDM1) to express CdMif2p with a C-terminal tandem affinity purification (TAP) tag (18) from its native promoter in the background of one wild-type copy of CdMIF2 (see SI Text). The subcellular localization patterns using polyclonal anti-Protein A antibodies in the C. dubliniensis strain (CDM1) at various stages of the cell cycle are very similar to those observed for CdCse4p (Fig. S4). We analyzed binding of TAP-tagged CdMif2p in the strain CDM1 by standard ChIP assays using anti-Protein A antibodies (Fig. 3 and Fig. S4). This experiment suggests that CdMif2p binds to the same 3-kb CdCse4p-rich region of three different chromosomes (chromosomes 1, 3, and 7) in C. dubliniensis (Fig. 3 and Fig. S4). Binding of two different evolutionarily conserved kinetochore proteins CdCse4p and CdMif2p at the same regions strongly implies that these regions are centromeric.
Comparative Sequence Analysis Between C. albicans and C. dubliniensis Reveals That Cse4p-Rich Centromere Regions Are the Most Rapidly Evolving Loci of the Chromosome.
Pairwise alignment of CdCse4p-rich sequences on different chromosomes (Table S3) with one another reveals no homology. To compare orthologous CEN regions of C. albicans and C. dubliniensis, we performed pairwise alignments using Sigma (19) and DIALIGN2 (20). These programs assemble global alignments from significant gapless local alignments. Sigma detects no homology in Cse4p-binding regions. DIALIGN2, with default parameters, reports a little homology; but when we compare known nonorthologous sequence (namely, CEN sequences from nonmatching chromosomes), it reports almost identical results (Table 1). In other words, it finds no homology beyond what it would with the “null hypothesis” of unrelated sequence. Similar results were obtained with other sequence alignment programs. We conclude there is no significant homology in the orthologous Cse4p-containing CEN regions in C. albicans and C. dubliniensis, even though the CEN regions are flanked by orthologous, syntenous ORFs. However, neighboring (pericentric) ORF-free regions, located between the Cse4p-binding regions and CEN-adjacent ORFs, do exhibit a higher degree of homology compared to Cse4p-rich regions. We count mutation rates only in aligned blocks (ignoring insertions and deletions); DIALIGN2 aligns 68% of these regions, with a mutation rate of 36%, while Sigma aligns 37% of the regions, with a mutation rate of 22% in aligned regions. Much of the conservation occurs toward the outer ends of these regions, that is, near the bounding ORFs.
Table 1.
Comparison of mutation rates in Cse4p-binding and other genomic noncoding regions in C. albicans and C. dubliniensis
| Cse4p-binding (%) | Cse4p-binding (shuffled) (%) | Pericentric (%) | Intergenic (%) | |
|---|---|---|---|---|
| Total bases | 26,836 | 26,836 | 40,280 | 593,782 |
| Aligned (DIALIGN2) | 12,440 (46) | 11,650 (43) | 27,684 (68) | 530,847 (89) |
| Mutated (DIALIGN2) | 7,624 (61) | 7,201 (62) | 10,229 (36) | 154,473 (29) |
| Aligned (Sigma) | 0 | 0 | 15,015 (37) | 334,363 (56) |
| Mutated (Sigma) | 0 | 0 | 3,323 (22) | 57,548 (17) |
The fraction of bases aligned by Sigma and DIALIGN2 and the mutation rates within the aligned regions, for Cse4p-binding regions, pericentric regions, and intergenic regions are shown. Also shown, as a null hypothesis, are numbers for ″shuffled″ Cse4p-binding regions, where regions from different nonorthologous chromosomes were aligned.
To estimate a “neutral” DNA mutation rate, we identified 2,653 putative gene orthologs of C. albicans in C. dubliniensis (see Materials and Methods). We aligned these genes with T-Coffee (21) and measured the synonymous mutation rates, using seven codons that are “fully degenerate” in the third position (the first 2 bases determine the coded amino acid). A naïve count of the third-position mutation rate yields 27%. Correcting for genomewide codon biases yields 42%, an upper-boundary estimate for the neutral rate of DNA mutation between these two yeasts (see Materials and Methods). This rate corresponds to a pairwise conservation rate (“proximity”) q = 0.58 or a proximity to a common ancestor of 0.76. Tests on synthetic DNA sequence (as reported in ref. 21) suggest that Sigma would easily align such sequence; therefore, it appears that CaCse4p-binding sequences (but not pericentric regions) have diverged faster than expected from the neutral point-mutation rate in these yeasts.
We also identified 309 homologous intergenic regions in these species that were between 1,000 and 5,000 bp long (comparable in length with the Cse4p-binding regions). We aligned these regions with Sigma and DIALIGN2 and measured mutation rates in aligned regions only (ignoring insertions and deletions). Sigma aligned 56% of the input intergenic sequence, with a mutation rate of 17%; DIALIGN2 aligned 89% of the input sequence, with a mutation rate of 29%. This rate is less than our estimated neutral mutation rate of 42%, suggesting constraints on the evolution of intergenic DNA sequences. Although pericentric regions evolve slower than the neutral rate determined above, they have a smaller fraction of conserved blocks and a greater mutation rate than intergenic sequences.
Interestingly, despite the rapid divergence of CEN DNA sequences, the relative position of the CEN on each chromosome is conserved in all cases (Fig. S5). The relative location of the Cse4p-rich centromeric chromatin in the ORF-free region is also similar in both species (Fig. 1). Although we find no homology among Cse4p-binding regions in matching chromosomes, some of the ORF-free pericentric regions have repeated segments, both within the same species and across the two species (Fig. S6 and Table S4). These repeats are mostly singles and in some cases flank a core region; mostly these repeats are not conserved across chromosomes in C. dubliniensis but sometimes they are conserved across species (e.g., chromosome 5 repeats). However, these repeats are mostly chromosome specific and not restricted to only core centromeric or pericentric regions. These results strongly suggest that mechanisms other than the DNA sequence of Cse4p-bound regions, such as specific chromatin architecture, determine centromere identity in these species. The role of pericentric regions in determining centromere identity remains unclear.
Discussion
We have identified and characterized the core CdCse4p-rich centromeric DNA sequences of all eight chromosomes of C. dubliniensis. Two important evolutionarily conserved kinetochore proteins, CdCse4p and CdMif2p are shown to be bound to these regions. Each of these CEN regions has unique and different DNA sequence composition without any strong sequence motifs or centromere-specific repeats that are common to all of the eight centromeres and has A-T content similar to that of the overall genome. In these respects they are remarkably similar to CEN regions of C. albicans (11, 12). Although genes flanking corresponding CENs in these species are syntenous, the Cse4p-binding regions show no significant sequence homology. They appear to have diverged faster than other intergenic sequences of similar length and even faster than our best estimated neutral mutation rate for ORFs.
A study, based on computational analysis of centromere DNA sequences and kinetochore proteins of several organisms, indicates that point centromeres have probably derived from regional centromeres and appeared only once during evolution (22). The core Cse4p-rich regions of C. albicans and C. dubliniensis are intermediate in length between the point S. cerevisiae-like centromeres and the regional S. pombe centromeres. The characteristic features of point and regional yeast centromeres are the presence of consensus DNA sequence elements and repeats, respectively, organized around a nonhomologous core CenH3-rich region (CDEII and the central core of S. cerevisiae and S. pombe, respectively). Both C. albicans and C. dubliniensis centromeres lack such conserved elements or repeats around their nonconserved core centromere regions in each chromosome. On the basis of these features, we propose that these Candida species possess centromeres of an “intermediate” type between point and regional centromeres.
On rare occasions, functional neocentromeres form at nonnative loci in some organisms. However, neocentromere activation occurs only when the native centromere locus becomes nonfunctional. Therefore, native centromere sequences may have components that cause them to be preferred in forming functional centromeres. Despite sequence divergence, the location of the Cse4p-rich regions in orthologous regions of C. albicans and C. dubliniensis has been maintained for millions of years. We also observe homology in orthologous pericentric regions in a pairwise chromosome-specific analysis in these two species. Moreover, several short stretches of DNA sequences are found to be common in pericentric regions of some, but not all, C. albicans and C. dubliniensis chromosomes. Both in budding and in fission yeasts, pericentric regions contain conserved elements that are important for CEN function. In the absence of any highly specific sequence motifs or repeats in these regions, it is possible that specific histone modifications at more conserved pericentric regions facilitate the formation of a specialized three-dimensional common structural scaffold that favors centromere formation in these Candida species.
It is an enigma that, despite their conserved function and conserved neighboring orthologous regions, core centromeres evolve so rapidly in these closely related species. Satellite repeats, which constitute most of the Arabidopsis centromeres, have been shown to be evolving rapidly (23). However, because of their repetitive nature, these centromeres are subject to several events such as mutation, recombination, deletion, and translocation that may contribute to rapid change in centromere sequence. In the absence of any such highly repetitive sequences at core centromere regions of C. albicans and C. dubliniensis, such accelerated evolution is particularly striking. It is important to mention that a very recent report based on comparison of chromosome III of three closely related species of S. paradoxus suggests that the centromere seems to be the fastest evolving part in the chromosome (24).
Several studies reveal that centromeres function in a highly species-specific manner. Henikoff and colleagues proposed that rapid evolution of centromeric DNA and associated proteins may act as a driving force for speciation (1, 25). The consequence of the rapid change in centromere sequence we observed in these two closely related Candida species may contribute to generation of functional incompatibility of centromeres to facilitate speciation. These two Candida species are both parasitic and clonally propagated. It is possible that the lack of recombination at the centromere and the more constant environment that a parasite finds itself in relative to a free-living organism may contribute to differential sequence evolution observed between centromeres and the rest of the genome. It is still unclear how centromeres are packaged, and it is possible that the presence of CenH3-containing chromatin with very different biochemical properties from bulk chromatin (26) provide less protection from random mutation. To understand the mechanisms of centromere formation in the absence of specific DNA sequence cues, it will be important to identify more genetic and epigenetic factors that may contribute to the formation of specialized centromeric chromatin architecture.
Materials and Methods
Strains, Media, and Transformation Procedures.
The Candida dubliniensis and C. albicans strains used in this study are listed in Table S5 and the strain construction strategies are mentioned in the SI Text. These strains were grown in yeast extract/peptone/dextrose (YPD), yeast extract/peptone/succinate (YPS), or supplemented synthetic/dextrose (SD) minimal media at 30 °C as described. C. albicans and C. dubliniensis cells were transformed by standard techniques (27, 28).
Identification of CdCse4p and CdMif2p.
The C. dubliniensis Cse4p was identified by a BLAST search (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/c_dubliniensis) with C. albicans Cse4p (CaCse4p) as the query sequence against the C. dubliniensis genome sequence database. This sequence analysis revealed three protein sequences with high homology to CaCse4p: two are the C. dubliniensis putative histone H3 proteins (Chr R- Cd36_32350 and Chr1- Cd36_04010 with BLAST scores of 333 each) and the other is CdCse4p (Chr 3- Cd36_80790 with BLAST score of 661). The CdCSE4 gene encodes a putative 212-aa-long protein with 100% identity in the C-terminal histone-fold domain of CaCse4p. A pairwise comparison of the CaCse4p and CdCse4p sequences revealed that they share 97% identity over a 212-aa overlap (Fig. S2). Using CaMif2p as the query sequence in the BLAST search against the C. dubliniensis genome database, we retrieved a single hit that was identified as the CENP-C homolog (Cd36_63360) in C. dubliniensis, showing 77% identity in a 516-aa overlap with CaMif2p. The CdMIF2 gene codes for a putative 520-aa-long protein with a conserved CENP-C box required for centromere targeting (29, 30, 11) that is identical in C. albicans and C. dubliniensis.
Complementation Assay, Indirect Immunofluorescence, ChIP assay, and Sequence Analysis.
The construction of C. albicans strain CAKS3b, the pAB1-based plasmids carrying CSE4 genes of C. albicans and C. dubliniensis, and the procedure for the complementation assay are described in the SI Text. Intracellular CdCse4p or CdMif2p was visualized by indirect immunofluorescence microscopy as described previously (16). ChIP assays were performed as described before (11). Details of the indirect immunofluorescence, ChIP procedure and WU-BLAST 2.0 analysis to identify CdCENs and flanking ORFs are available in the SI Text.
Homology Detection and Mutation Rate Measurement.
We used Sigma (version 1.1.3) and DIALIGN 2 (version 2.2.1) to align ORF-free DNA sequences. Default parameters were used for both programs, but Sigma was given an auxiliary file of intergenic sequences from which to estimate a background model. Orthologous genes were aligned (at the amino acid level) with T-Coffee. We examined instances of the following seven codons where the first two positions were conserved in both species: GTn (valine), TCn (serine), CCn (proline), ACn (threonine), GCn (alanine), CGn (arginine), and GGn (glycine) (n, any nucleotide). Third-position mutations here do not change the amino acid. (Leucine was ignored because of a variant codon in these species.) A naïve count of mutation rates in the third position yields 0.27. Taking into consideration genomewide bias for each codon (details are in SI Text), an upper-bound mutation rate of 0.42 was obtained.
Data Availability.
The coordinates of the ORFs of C. dubliniensis mentioned in Table S2 are obtained by the BLAST analysis from the C. dubliniensis genome database (www.sanger.ac.uk/cgi-bin/blast/submitblast/c_dubliniensis) as of May 16, 2007, and the coordinates apply to the 031907 release of the contigs. Subsequent to this work, an independent annotation and new nomenclature of ORFs in the C. dubliniensis genome have been made available from the GeneDB database (www.genedb.org/genedb/). The CdCse4p-rich centromere sequences of C. dubliniensis can be obtained from www.jncasr.ac.in/sanyal/Cdsequences.txt.
Supplementary Material
Acknowledgments.
We thank P. Magee, J. Morschhaeuser, and P. Koetter for the strains and reagents, M. A. Lone for plasmid constructs, Suma for confocal images, and Mary Baum for critical comments on the manuscript. Sequence data for C. dubliniensis were obtained from the Wellcome Trust Sanger Institute website at http://www.sanger.ac.uk/cgi-bin/blast/submitblast/c_dubliniensis. This work was supported by a research grant from the Department of Science and Technology, Government of India (SR/SO/BB-24/2007) and by Jawaharlal Nehru Centre for Advanced Scientific Research (JNCASR) (K.S.). J.T. is a junior research fellow funded by the Department of Biotechnology, Government of India. R.S. was supported by the PRISM project at the Institute of Mathematical Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 3.Clarke L. Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr Opin Genet Dev. 1998;8:212–218. doi: 10.1016/s0959-437x(98)80143-3. [DOI] [PubMed] [Google Scholar]
- 4.Cumberledge S, Carbon J. Mutational analysis of meiotic and mitotic centromere function in Saccharomyces cerevisiae. Genetics. 1987;117:203–212. doi: 10.1093/genetics/117.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaudet A, Fitzgerald-Hayes M. Alterations in the adenine-plus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:68–75. doi: 10.1128/mcb.7.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 8.Marschall LG, Clarke L. A novel cis-acting centromeric DNA element affects S. pombe centromeric chromatin structure at a distance. J Cell Biol. 1995;128:445–454. doi: 10.1083/jcb.128.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baum M, Ngan VK, Clarke L. The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol Biol Cell. 1994;5:747–761. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 11.Sanyal K, Baum M, Carbon J. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci USA. 2004;101:11374–11379. doi: 10.1073/pnas.0404318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra PK, Baum M, Carbon J. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol Genet Genomics. 2007;278:455–465. doi: 10.1007/s00438-007-0263-8. [DOI] [PubMed] [Google Scholar]
- 13.Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc Natl Acad Sci USA. 2006;103:14877–14882. doi: 10.1073/pnas.0606958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 15.Talbert PB, Bryson TD, Henikoff S. Adaptive evolution of centromere proteins in plants and animals. J Biol. 2004;3:18.1–18.17. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanyal K, Carbon J. The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc Natl Acad Sci USA. 2002;99:12969–12974. doi: 10.1073/pnas.162488299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Copenhaver GP. Who's driving the centromere? J Biol. 2004;3:17. doi: 10.1186/jbiol15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corvey C, et al. Carbon source-dependent assembly of the Snf1p kinase complex in Candida albicans. J Biol Chem. 2005;280:25323–25330. doi: 10.1074/jbc.M503719200. [DOI] [PubMed] [Google Scholar]
- 19.Siddharthan R. Sigma: multiple alignment of weakly-conserved non-coding DNA sequence. BMC Bioinformatics. 2006;7:143. doi: 10.1186/1471-2105-7-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgenstern B. DIALIGN2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- 21.Notredame C, Higgins D, Heringa J. T-Coffee: a novel method for multiple sequence alignments. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 22.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23.1–R23.21. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall SE, Kettler G, Preuss D. Centromere satellites from Arabidopsis populations: maintenance of conserved and variable domains. Genome Res. 2003;13:195–205. doi: 10.1101/gr.593403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bensasson D, Zarowiecki M, Burt A, Koufopanou V. Rapid evolution of yeast centromeres in the absence of drive. Genetics. 2008;178:2161–2167. doi: 10.1534/genetics.107.083980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malik HS, Henikoff S. Conflict begets complexity: the evolution of centromeres. Curr Opin Genet Dev. 2002;12:711–718. doi: 10.1016/s0959-437x(02)00351-9. [DOI] [PubMed] [Google Scholar]
- 26.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:1798–1809. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgers PM, Percival KJ. Transformation of yeast spheroplasts without cell fusion. Anal Biochem. 1987;163:391–397. doi: 10.1016/0003-2697(87)90240-5. [DOI] [PubMed] [Google Scholar]
- 28.Hull CM, Johnson AD. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science. 1999;285:1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- 29.Yu HG, Hiatt EN, Dawe RK. The plant kinetochore. Trends Plant Sci. 2000;5:543–547. doi: 10.1016/s1360-1385(00)01789-1. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki N, et al. CENP-B interacts with CENP-C domains containing Mif2 regions responsible for centromere localization. J Biol Chem. 2004;279:5934–5946. doi: 10.1074/jbc.M306477200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of the ORFs of C. dubliniensis mentioned in Table S2 are obtained by the BLAST analysis from the C. dubliniensis genome database (www.sanger.ac.uk/cgi-bin/blast/submitblast/c_dubliniensis) as of May 16, 2007, and the coordinates apply to the 031907 release of the contigs. Subsequent to this work, an independent annotation and new nomenclature of ORFs in the C. dubliniensis genome have been made available from the GeneDB database (www.genedb.org/genedb/). The CdCse4p-rich centromere sequences of C. dubliniensis can be obtained from www.jncasr.ac.in/sanyal/Cdsequences.txt.