Abstract
α-Synuclein is known to play a causative role in Parkinson disease. Although its physiological functions are not fully understood, α-synuclein has been shown to interact with synaptic vesicles and modulate neurotransmitter release. However, the structure of its physiologically relevant membrane-bound state remains unknown. Here we developed a site-directed spin labeling and EPR-based approach for determining the structure of α-synuclein bound to a lipid bilayer. Continuous-wave EPR was used to assign local secondary structure and to determine the membrane immersion depth of lipid-exposed residues, whereas pulsed EPR was used to map long-range distances. The structure of α-synuclein was built and refined by using simulated annealing molecular dynamics restrained by the immersion depths and distances. We found that α-synuclein forms an extended, curved α-helical structure that is over 90 aa in length. The monomeric helix has a superhelical twist similar to that of right-handed coiled-coils which, like α-synuclein, contain 11-aa repeats, but which are soluble, oligomeric proteins (rmsd = 0.82 Å). The α-synuclein helix extends parallel to the curved membrane in a manner that allows conserved Lys and Glu residues to interact with the zwitterionic headgroups, while uncharged residues penetrate into the acyl chain region. This structural arrangement is significantly different from that of α-synuclein in the presence of the commonly used membrane-mimetic detergent SDS, which induces the formation of two antiparallel helices. Our structural analysis emphasizes the importance of studying membrane protein structure in a bilayer environment.
Keywords: EPR, Parkinson's disease, fibril-forming proteins, 11-aa repeats
The interaction of α-synuclein with membranes is thought to be important in its physiologic function in vivo, as well as in its misfolding and aggregation in the pathogenesis of Parkinson disease (1–10). Although the function of α-synuclein in vivo is not fully understood, it has been observed to localize to presynaptic nerve termini, where it modulates presynaptic pool size and neurotransmitter release (11–16). These functions are likely to be mediated by the interaction of α-synuclein with synaptic vesicles, and in vitro studies have shown that α-synuclein interacts strongly with highly curved vesicles that are similar in size to synaptic vesicles (17, 18). The structural characterization of membrane-bound α-synuclein is significant, given the importance of membrane interactions to the pathologic and physiologic roles of α-synuclein.
Previous studies have revealed that the interaction of monomeric α-synuclein with negatively charged vesicles induces a predominantly α-helical structure located in the N-terminal region of the protein (17, 19, 20). This region contains seven 11-aa-repeat regions that share some sequence similarity with apolipoproteins [supporting information (SI) Fig. S1]. Sequence analysis using algorithms for apolipoproteins predicts the formation of five separate helices (17). However, no high-resolution structure is available for α-synuclein in its physiologically relevant membrane-bound form. High-resolution NMR (21) indicates that α-synuclein bound to an SDS detergent micelle forms two antiparallel helices (from Val-3 to Val-37 and Lys-45 to Thr-92) that wrap tightly around the detergent micelle. Recently, it has been suggested (22, 23) that membrane-bound α-synuclein may take up a similar structure, whereas other studies suggest an extended helical structure (20).
In structural biology, detergent micelles are commonly used as membrane-mimetic environments because their small size facilitates high-resolution structural analysis by NMR. However, it is often difficult to test whether the structure of proteins bound to micelles is indeed the same as that of the respective membrane-bound form. We have argued that the much smaller size of SDS micelles might be responsible for the break between the α-synuclein helices and that an extended helical structure might be formed in the presence of membranes (20). Therefore, the main goals of the present study were to develop an approach for refining membrane protein structure in the presence of lipid bilayers and to apply this methodology to determine the three-dimensional structure of membrane-bound α-synuclein.
Results and Discussion
A structural refinement process for membrane-bound proteins was developed based upon site-directed spin labeling, EPR spectroscopy (24–26), and simulated annealing molecular dynamics (SAMD). Continuous-wave EPR of singly-labeled α-synuclein derivatives was used to generate local mobility, accessibility, and membrane immersion depth, whereas pulsed EPR provided intramolecular distances. These data were converted into restraints that were used to refine the structure of membrane-bound α-synuclein.
Local Secondary Structure and Membrane Topology Information from Continuous-Wave EPR Spectroscopy.
To obtain secondary structure and topography information, we generated 26 singly-labeled α-synuclein derivatives and investigated them by continuous-wave EPR spectroscopy. The experimental design for these studies was identical to that of our previous work, which included a nitroxide scan from residue 59 to residue 90 (20). The additional new sites were chosen to generate a contiguous nitroxide scan from residue 25 to residue 90. Collectively, this region contains most of the sequence that forms the two helices and the intervening loop region in the presence of SDS (19, 21, 23, 27–29).
The continuous-wave EPR spectra of all new sites were similar to those obtained previously for residues 59–90 (20), and the mobility information from all spectra is summarized by using the inverse central line width. As illustrated in Fig. S2A, the respective values for residues located within the repeat regions are consistent with the formation of an ordered structure that is devoid of pronounced tertiary or quaternary packing interactions. In contrast, the C-terminal residues exhibit elevated mobility and are unstructured. In an effort to provide more detailed secondary structural information, we determined the membrane topography for each of the labeled sites by measuring their accessibilities to O2 and NiEDDA (ΠO2 and ΠNiEDDA, respectively, EDDA being ethylenediamine-N,N′-diacetic acid). This measurement is based upon the preferential partitioning of O2 into the membrane and NiEDDA into the aqueous environment. As a consequence, membrane-exposed sites show enhanced accessibility to O2, whereas solvent-exposed sites are preferentially accessible to NiEDDA. In agreement with the formation of an extended helical structure, ΠO2 and ΠNiEDDA exhibit continuous periodic oscillations wherein maxima (as well as minima) are spaced 3 to 4 aa apart (Fig. S2B). An important feature of the accessibilities is that they are precisely out-of-phase. Such behavior is typically observed for asymmetrically solvated α-helices that are exposed to solvent on the NiEDDA-accessible side and to the membrane on the O2-accessible side (20, 25).
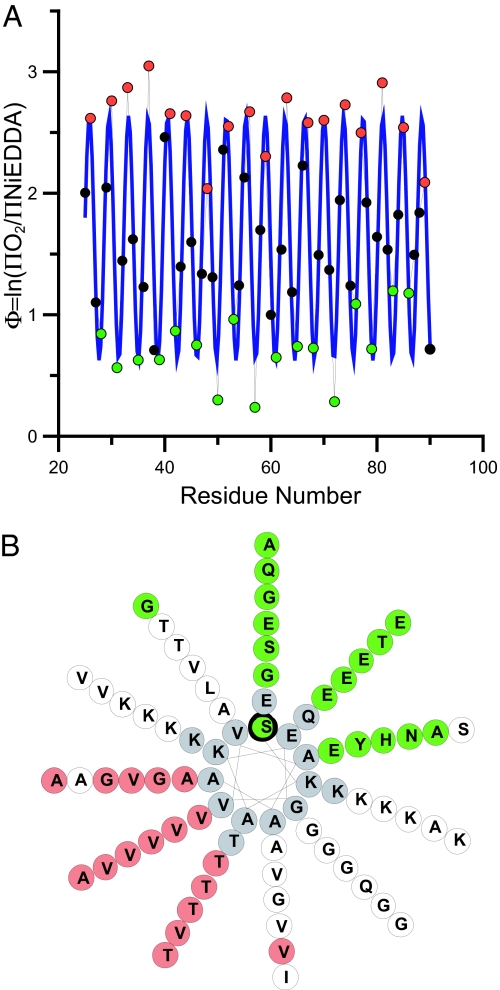
The accessibility data for both colliders can be conveniently summarized by the depth parameter Φ [Φ = ln(ΠO2/ΠNiEDDA)], which increases linearly with increasing immersion depth (see Methods for calibration). As shown in Fig. 1A, Φ also exhibits a pronounced oscillation consistent with an extended α-helical structure. In our prior analysis of residues 59–90, we noted the formation of a helical structure in which each repeat (11 aa) takes up three turns, giving rise to a periodicity of 3.67 aa per turn. This unique feature is retained throughout all other scanned regions (Fig. 1A), as further illustrated by the best fit to a cosine function, which results in a periodicity of ≈3.68 aa per turn (Fig. 1A, blue line). Moreover, when we use a helical wheel representation that is based upon 11 aa taking up exactly 3 turns (Fig. 1B), lipid-exposed (red) and solvent-exposed (green) sites fall onto opposite sides of this helix. Thus, the data indicate the formation of a continuous helical structure that extends parallel to the membrane and that has a slightly unusual helical periodicity.
Fig. 1.
Continuous-wave EPR analysis of singly labeled α-synuclein derivatives indicates the formation of an ordered and continuous helical structure. (A) The ratios of the accessibilities to O2 and NiEDDA for residues 25–90 are summarized by the depth parameter Φ = ln(ΠO2/ΠNiEDDA), with increasing Φ values indicating deeper membrane immersion depth. The blue line indicates the best fit to a cosine function and the resulting periodicity is 3.68 aa per turn (r2 = 0.85), which is close to the theoretically predicted periodicity of 3.67 aa per turn (11 aa per 3 turns = 3.67 aa per turn). (B) The repeat region residues are plotted onto a helical wheel in which 11 aa make up three turns. Lipid-exposed sites (red) fall onto one side, while solvent-exposed sites (green) lie on the opposite side. White circles denote residues with Φ values that are neither maxima nor minima. Residues shown in gray were not tested.
Intramolecular Distances from Four-Pulse DEER Experiments.
We measured interlabel distances in 17 doubly-labeled α-synuclein derivatives to obtain independent verification of an elongated helical structure and to collect additional restraints for the structural refinement. Only sites directly facing the membrane were spin-labeled to ensure that all sites had comparable orientations (chosen from the maxima in Fig. 1A). This strategy minimizes the effect of spin-label orientation and the measured distances are therefore expected to depend mainly on the respective backbone distances.
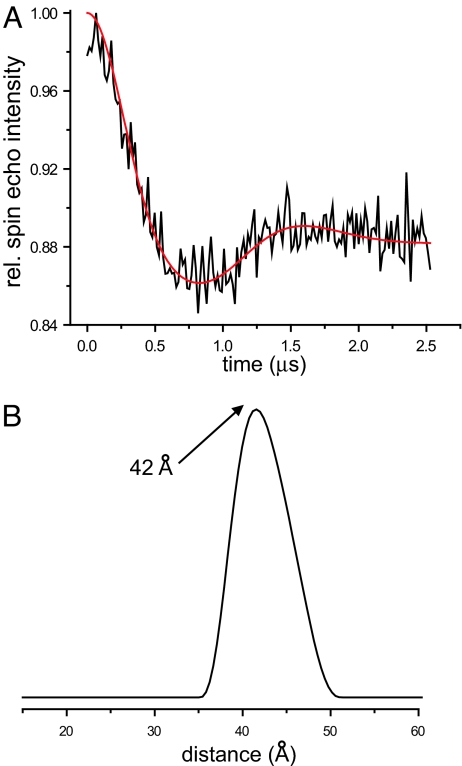
Distances were obtained from a four-pulse DEER experiment, which monitors the time evolution of a spin echo intensity (30). The frequency of the resulting signal is a direct measurement of inter-residue distances, with longer distances giving rise to slower oscillations. This method has been demonstrated to be effective for precise measurement of distances of up to ≈80 Å in model systems and up to ≈60 Å in proteins (30–33). As shown for the 56R1/85R1 derivative (Fig. 2A), the time domain signal exhibits pronounced oscillations, indicating the presence of a specific and ordered structure with a well-defined and relatively narrow distance distribution. The data were fit using Tikhonov regularization (34), and the resulting distance distribution is given in Fig. 2B. The experimentally determined distance (maximum at 42 Å) coincides remarkably well with the backbone distance expected from a rise of 1.5 Å per aa in an idealized α-helix (29 aa × 1.5 Å per aa = 43.5 Å). Similarly good agreement between experimental distances and theoretical estimates for an idealized α-helix was obtained for 13 additional derivatives (Table 1 and Fig. S3). No distances could be determined for the 11R1/70R1, 11R1/81R1, and 41R1/85R1 derivatives, all of which gave slowly decaying signals with oscillations that were too long to fit unambiguously, indicating that the underlying distances are too large (>60 Å as estimated in Fig. S3B) to be resolved using the present method. This result is also consistent with an extended helical structure in which these distances would exceed the upper limit of detection (Table 1).
Fig. 2.
Intramolecular distances from four-pulse DEER experiments. (A) The baseline corrected time evolution data from a four-pulse DEER experiment for the membrane-bound 56R1/85R1 α-synuclein derivative (black line) were fit by using Tikhonov regularization (34) (red line). The resulting distance distribution is given in B. All results, including those from 16 additional membrane-bound doubly labeled derivatives, are summarized in Table 1 (for data, see Fig. S3).
Table 1.
Intramolecular distances for 17 membrane-bound doubly-labeled derivatives from four-pulse DEER experiments
| Derivative | Distance, Å |
||
|---|---|---|---|
| DEER | Ideal helix | SDS (NMR) | |
| 11R1/26R1 | 25 | 22.5 | 22.5 |
| 11R1/41R1 | 48 | 45 | 43.3 |
| 22R1/52R1 | 49 | 45 | 23.3 |
| 26R1/41R1 | 23 | 22.5 | 25.4 |
| 26R1/56R1 | 44 | 45 | 24.4 |
| 37R1/67R1 | 42 | 45 | 39.2 |
| 41R1/56R1 | 23 | 22.5 | 24.0 |
| 41R1/67R1 | 37 | 39 | 38.8 |
| 41R1/70R1 | 41 | 43.5 | 42.0 |
| 44R1/67R1 | 36 | 34.5 | 34.2 |
| 48R1/67R1 | 29 | 28.5 | 27.8 |
| 56R1/70R1 | 22 | 21 | 20.1 |
| 56R1/85R1 | 42 | 43.5 | 42.0 |
| 63R1/81R1 | 26 | 27 | 25.7 |
| 11R1/70R1 | >60 | 88.5 | 22.7 |
| 11R1/81R1 | >60 | 105 | 22.8 |
| 41R1/85R1 | >60 | 66 | 62.6 |
The experimental distances are taken from the peaks of the Tikhonov regularization-based fits (for data, see Fig. S3), but identical maximal distances (within 1 Å or less) were obtained from Gaussian fits. The data are compared to the rise of an ideal helical structure, which is taken to be 1.5 Å per residue. The final column shows the respective α -carbon distances from the high-resolution NMR structure of SDS-bound α -synuclein.
Importantly, the above data are inconsistent with the antiparallel helical structure that forms in the presence of SDS micelles, inasmuch as shorter distances would be expected for the 22R1/52R1, 26R1/56R1, 11R1/70R1, and 11R1/81R1 derivatives (Table 1). To test the structure of the micelle-bound α-synuclein, DEER data were obtained for two of these derivatives (11R1/70R1 and 11R1/81R1) in their SDS micelle-bound state. Indeed, in agreement with previous studies (21, 29), much shorter distances with broad distance distributions were obtained (Fig. S3C). Thus, the membrane-bound helical form of α-synuclein is different from the SDS-bound form in two respects: it adopts a single helical structure and it is better defined.
Based on a small number of distance measurements, it has been suggested (23) that the N- and C-terminal portions of the repeat region have comparable distances in the membrane-bound and SDS-bound states. This conclusion was reached based upon use of very highly charged small unilamellar vesicles (SUVs) containing 100% 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (POPG). Replicating these conditions, we obtained a DEER signal that corresponds to a broad distance distribution for the 11R1/70R1 derivative centered at 26 Å (Fig. S4 A–C). When compared to the conditions of the present study, however, the signal exhibited a very low modulation depth, suggesting that the short distance arises from a small fraction of samples. Moreover, we noted that α-synuclein strongly disrupts the integrity of SUVs containing 100% POPG, as judged by vesicle leakage experiments (Fig. S4D). According to gel filtration, only a subset of α-synuclein binds to intact vesicles, while a large fraction of α-synuclein induces formation of smaller, nonvesicular structures (Fig. S4E). Importantly, after the vesicle-bound α-synuclein had been purified, the 11R1/70R1 derivative no longer gave any clearly detectable short distances (Fig. S4A), suggesting that the shorter distances for this derivative did not arise from vesicle-bound protein.
Structural Refinement.
Collectively, our data indicate that α-synuclein forms an extended helical structure when bound to phospholipid bilayer membranes but that it can take up different structures when bound to nonvesicular lipids or detergents. To generate an atomistic three-dimensional structure of α-synuclein bound to a vesicle, we developed a computational approach for structural refinement based on our EPR data. An in-house algorithm was used to generate the starting structure of spin-labeled α-synuclein as a linear α-helix (see SI Methods and Fig. S5) and to convert the experimental data into three types of structural restraints: (i) distance restraints between spin labels based on the DEER data, (ii) immersion depth restraints (Table S1) modeled as distance restraints from each spin label to the center of the vesicle (Fig. S5), and (iii) backbone dihedral and hydrogen bonding restraints applied to regions found to be α-helical (Table S2). These restraints were used in a SAMD-based refinement using AMBER8 (35). To test the quality of the refinement, some experimental depth and distance data were excluded from the actual refinement process and were used to determine the effectiveness of the refinement.
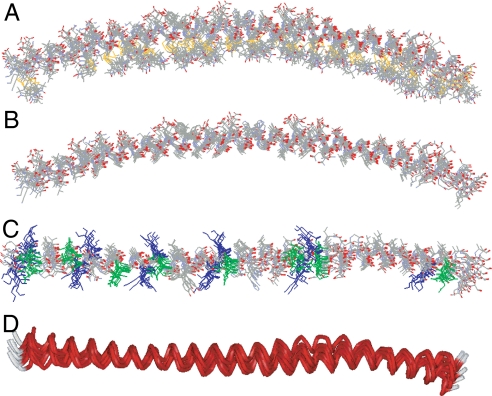
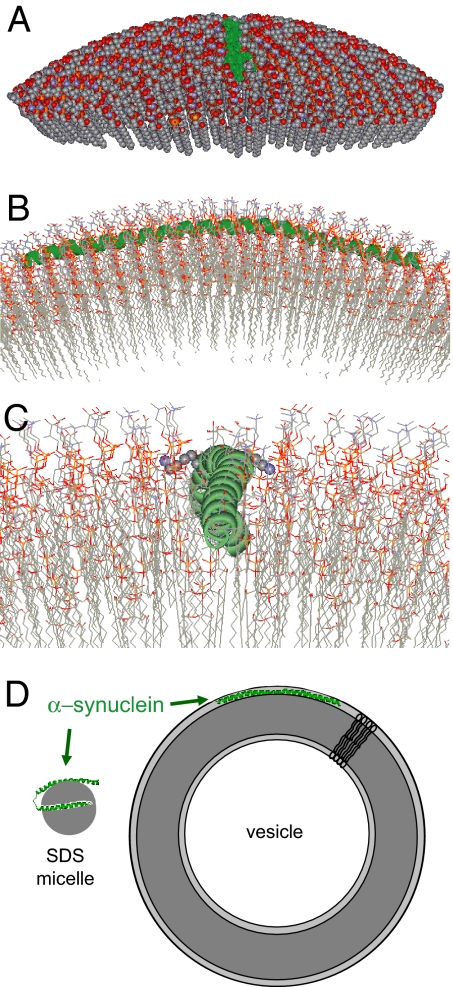
The refinement process produced 10 structures, 9 of which reproduced the omitted experimental data to within experimental error (Table S3 and Fig. S6). These nine structures are overlaid in Fig. 3 and share strong similarities. To generate the final structures, the labeled side chains (Fig. 3A) were replaced by the native side chains (Fig. 3B). Some of the central features of the structure are as follows: α-synuclein forms an extended, continuously curved helical structure (Fig. 3 A and B) with a superhelical twist (Fig. 3D). When viewed from the top, the Lys residues (blue in Fig. 3C) are oriented approximately perpendicular to the helical axis, whereas the Glu residues (green in Fig. 3C) are facing upwards. To illustrate the spatial relationship between this helix and the membrane, we used an in-house algorithm to construct a vesicle of 300 Å in diameter (size of SUVs used in the present study; see ref. 20) around the vesicle center used in the SAMD simulations. The lipids were packed around the SAMD-derived α-synuclein structures and the lipids displaced by α-synuclein were repacked evenly. We emphasize that the protein position was directly obtained from the experimental depth restraints of the SAMD calculations; thus, it is governed by the experimental data. An example of the resulting protein–lipid interaction is shown in Fig. 4. The curved helical structure is aligned in such a way that it follows the curvature of the vesicle surface (Fig. 4A), with the center of the α-helix located just below the phosphate groups of the lipids (Fig. 4B). This alignment permits the lysine residues to interact with the phosphates, as shown for Lys-58 and Lys-60 in Fig. 4C. Negatively charged residues are on the outside surface of the helix (Fig. 3C) and are located at the level of the choline groups of the lipids (Fig. 4). In contrast to these charged residues, all of the lipid-exposed residues are generally hydrophobic or Thr residues.
Fig. 3.
Refinement of the membrane-bound structure of α-synuclein by using SAMD with restraints from EPR data. (A) Overlay of nine structures obtained from SAMD calculations using the amino acids 9–89 fragment of α-synuclein, with spin labels added at 26 sites (Fig. S5). In the refined structures, the labels (identifiable by the S–S bond depicted in yellow) are generally positioned on the concave side of the curved protein structure (i.e., oriented into the membrane). The N terminus is on the left of the figure. (B) The refined labeled structures were converted to the respective α-synuclein structures by replacing each label with the normal amino acid in the α-synuclein sequence, with retention of the Cβ position in the label. (C) View of the overlaid structures from above the lipid surface. The 11 lysine residues (blue) are oriented approximately perpendicular to the helical axis, permitting potential interactions with the lipid phosphates. In contrast, the 8 glutamic acid residues (green) are oriented away from the membrane, on the top surface of the α-helix. (D) Ribbon view of the 9 overlaid structures (perspective similar to that in C), indicating the general goodness of fit of the structures and, most importantly, showing the subtle but reproducible superhelical twist in the α-helix that emerged in the SAMD calculations. We speculate that the superhelicity may allow the helix to adopt a 3-11 conformation that permits the helical axis to follow a curved surface over an extended length.
Fig. 4.
Representations of the interaction of α-synuclein with a curved lipid surface. (A) Space-filled model of α-synuclein (shown in green) binding to the surface of a lipid vesicle 300 Å in diameter; ≈25% of the outer leaflet of the vesicle is shown. The vesicle was fitted around one of the structures derived from the experimentally restrained SAMD calculations. (B) A closer cross-sectional view of the α-synuclein interaction with the lipid surface, with rotation through 90° from the image in A. The protein (green) follows the curved surface of the vesicle, with the helical axis positioned just below the level of the phosphate groups of the lipids. This position of the protein emerged from the SAMD calculations and is a reflection of the immersion depths obtained from the continuous-wave EPR data. (C) A more detailed image of the protein–lipid interaction, viewed from the same angle as the image in A. The N terminus of the α-helix is in the foreground. Lysine residues 58 and 60 are shown in space-filling format (K58 oriented to the right and K60 to the left of the helix). The image indicates the proximity of the positively charged lysine side chains to the negatively charged phosphate groups. (D) Cartoon representations of the structures of α-synuclein on micelles and SUVs. The small and highly curved micelles cannot accommodate the extended helical structure present on the membrane.
A particularly interesting feature of the α-synuclein helix is that it bends around the membrane with a superhelical twist (Figs. 3D and 4C). It is likely that this twist facilitates the formation of a continuously bent helical structure, with the slightly altered periodicity of 3.67 aa per turn. In fact, continually curved helices with superhelical twists have been observed in right-handed coiled-coils (36–38), which also contain 11-aa-repeat regions, but which form soluble oligomers instead of interacting with membranes. To investigate potential structural similarities, we overlaid the α-synuclein structure with that of tetrabrachion, a naturally occurring, right-handed coiled-coil. This overlay resulted in a remarkably good overlap with an rmsd of 0.82 Å (Fig. 5). Thus, the 11-aa-repeat regions in both proteins encode for an α-helical fold that can either mediate interaction of a monomeric protein with curved membranes or mediate protein–protein contacts. In one case, the hydrophobic surface is exposed to the membrane, whereas in the other case it forms the core of a helical bundle. Considering the sequence similarities between α-synuclein and apolipoproteins, it is likely that the latter may use a similar fold to wrap around lipid particles.
Fig. 5.
Comparison of the structure of membrane-bound α-synuclein to that of a right-handed coiled-coil. Overlay of a single helix from tetrabrachion (red, Protein Data Bank ID 1FE6) with that of α-synuclein (green) using Tm-align (41) results in a backbone rmsd of 0.82 Å. The tetrabrachion helices contain 52 aa. Shown are residues 12–62 for α-synuclein (structure 3, which scored highest in the validation (Fig. S6) and residues 2–52 of tetrabrachion. Comparable rmsd values were obtained for all other structures. Overlays with all structures resulted in Tm-scores larger than 0.5, which is an indication of the same fold (41).
Conclusions
Compared with the plethora of structural information available for soluble proteins, relatively little is known about the structures of transmembrane- or membrane-associated proteins in the physiologically important lipid bilayer environment. Here we have presented an approach for refining the structure of membrane-bound α-synuclein based solely upon experimental restraints from continuous-wave and pulsed EPR. The resulting structure of membrane-bound α-synuclein is an extended α-helix and provides detailed molecular insight into the mechanism by which α-synuclein interacts with membranes. Importantly, this structure is different from that obtained in the presence of detergent micelles, whose small diameter may inhibit formation of an extended helix (Fig. 4D). Our data underscore the importance of obtaining direct structural information on membrane proteins in a lipid bilayer environment and show that it is important to consider the lipid composition of a given bilayer because this may have pronounced effects on protein and bilayer structure. The approach presented here should not only be applicable to testing the various modes of α-synuclein interactions with lipids but may also enable structural investigation of other membrane proteins.
Methods
Preparation of Spin-Labeled α-Synuclein Derivatives.
Single and double cysteine mutants of α-synuclein were expressed and purified as described previously (20). Briefly, α-synuclein mutants were expressed in BL21(DE3)pLysS Escherichia coli cells, and the cell pellet was resuspended in lysis buffer [100 mM Tris (pH 8), 300 mM NaCl, 1 mM EDTA]. The cell lysate was boiled and then precipitated with hydrochloric acid. After precipitation, the supernatant was dialyzed against dialysis buffer [20 mM Tris (pH 8), 1 mM EDTA, 1 mM DTT]. Two rounds of anion exchange chromatography were performed, and proteins were eluted with a salt gradient of 0–1 M NaCl. Samples were spin-labeled in 20 mM Hepes (pH 7.4), 100 mM NaCl buffer using 5× molar excess spin label, incubated for 1 h at room temperature, and separated from unreacted spin label by gel filtration using PD10 columns (GE Healthcare).
Vesicle Preparation.
The following synthetic lipids were used: 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), and POPG. All lipids were purchased from Avanti Polar Lipids. Lipids were dried with nitrogen and desiccated overnight. After desiccation, lipids were resuspended in buffer, treated to bath sonication, then sonicated with a tip at 2-W power (Misonix) for 30 min. Lipids were centrifuged for 1 h at 128,400 × g at 22°C, and the supernatant was recovered and used for experiments.
Continuous-Wave EPR.
Continuous-wave EPR spectra were obtained from vesicle-bound α-synuclein derivatives at a molar protein-to-lipid ratio of 1:250 in 20 mM Hepes (pH 7.4), 100 mM NaCl buffer. Spectral scans were collected using a Bruker EMX X-band CW EPR spectrometer, and inverse central line width values were measured from the peak-to-peak distance of the central line as described previously (20). Accessibilities to O2 and NiEDDA (ΠO2 and ΠNiEDDA) were obtained from power saturation experiments using a dielectric resonator (39). The oxygen accessibility was measured in the presence of ambient oxygen and the sample was equilibrated with 3 mM NiEDDA for NiEDDA accessibility. The immersion depth of lipid-exposed sites was determined from the relation d (in Å) = a × Φ + b (39), where the values of a and b were obtained by using calibration with spin-labeled lipids [1-palmitoyl-2-stearoyl-(n-doxyl)-sn-glycero-3-phosphocholine); with n = 5, 7, or 10]. The values of a and b are 5.9 and −4.1, respectively (20).
Pulsed EPR and Distance Analysis.
Samples were prepared at a protein-to-lipid ratio of 1:250 as described above. For all experiments, 25% fully spin-labeled protein, containing two spin labels per protein, was mixed with unlabeled wild-type protein before the addition of vesicles. Unbound protein was washed by using YM-100 concentrators (Amicon). DEER experiments were performed using a Bruker Elexsys E580 X-band pulse EPR spectrometer fitted with a 3-mm split ring (MS-3) resonator, a continuous-flow helium cryostat (CF935, Oxford Instruments), and a temperature controller (ITC503S, Oxford Instruments). Samples (20 μl) were flash-frozen in the presence of 30% sucrose and data were acquired at 78 K. The data were fit using Tikhonov regularization (40) as implemented in DEERAnalysis2006 and DEERAnalysis2008 packages (34). See SI Methods for more detail.
Computational Structural Refinement.
A peptide with 26 spin labels was constructed as a linear α-helix (Fig. S5). Experimental data for 12 interlabel distances (Table 1) and 25 label depths were used to define restraints for SAMD calculations in AMBER8 (35). Interlabel distances were defined between the N atoms of the nitroxide groups of each label pair, and depths were defined between the N atom of each label and an “atom” representing the center of an imaginary vesicle of 300 Å in diameter. The SAMD calculation was performed over 10 heating and cooling cycles of 30 ps each. Full details of the computational procedure and the evaluation of the derived structures are given in SI Methods.
Supplementary Material
Acknowledgments.
We thank M. Kaptein for technical support. This work was supported by grants from the Larry L. Hillblom foundation (to J.C. and R.L.), the National Institutes of Health (P50 AG05142 to R.L.) and the John Douglas French Alzheimer Foundation (to R.L.). C.C.J. is supported by National Institutes of Health Predoctoral Training Grant T32 GM067587.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807826105/DCSupplemental.
References
- 1.Cole NB, et al. Lipid droplet binding and oligomerization properties of the Parkinson's disease protein alpha-synuclein. J Biol Chem. 2002;277:6344–6352. doi: 10.1074/jbc.M108414200. [DOI] [PubMed] [Google Scholar]
- 2.Lee HJ, Choi C, Lee SJ. Membrane-bound alpha-synuclein has a high aggregation propensity and the ability to seed the aggregation of the cytosolic form. J Biol Chem. 2002;277:671–678. doi: 10.1074/jbc.M107045200. [DOI] [PubMed] [Google Scholar]
- 3.Lotharius J, Brundin P. Pathogenesis of Parkinson's disease: Dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci. 2002;3:932–942. doi: 10.1038/nrn983. [DOI] [PubMed] [Google Scholar]
- 4.Zhu M, Li J, Fink AL. The association of alpha-synuclein with membranes affects bilayer structure, stability, and fibril formation. J Biol Chem. 2003;278:40186–40197. doi: 10.1074/jbc.M305326200. [DOI] [PubMed] [Google Scholar]
- 5.Fortin DL, et al. Lipid rafts mediate the synaptic localization of alpha-synuclein. J Neurosci. 2004;24:6715–6723. doi: 10.1523/JNEUROSCI.1594-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vekrellis K, Rideout HJ, Stefanis L. Neurobiology of alpha-synuclein. Mol Neurobiol. 2004;30:1–21. doi: 10.1385/MN:30:1:001. [DOI] [PubMed] [Google Scholar]
- 7.Tofaris GK, Spillantini MG. Physiological and pathological properties of alpha-synuclein. Cell Mol Life Sci. 2007;64:2194–2201. doi: 10.1007/s00018-007-7217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith DP, et al. Formation of a high affinity lipid-binding intermediate during the early aggregation phase of alpha-synuclein. Biochemistry. 2008;47:1425–1434. doi: 10.1021/bi701522m. [DOI] [PubMed] [Google Scholar]
- 9.Soper JH, et al. α-Synuclein-induced aggregation of cytoplasmic vesicles in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:1093–1103. doi: 10.1091/mbc.E07-08-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aisenbrey C, et al. How is protein aggregation in amyloidogenic diseases modulated by biological membranes? Eur Biophys J. 2008;37:247–255. doi: 10.1007/s00249-007-0237-0. [DOI] [PubMed] [Google Scholar]
- 11.Iwai A, et al. The precursor protein of non-A beta component of Alzheimer's disease amyloid is a presynaptic protein of the central nervous system. Neuron. 1995;14:467–475. doi: 10.1016/0896-6273(95)90302-x. [DOI] [PubMed] [Google Scholar]
- 12.Masliah E, Iwai A, Mallory M, Ueda K, Saitoh T. Altered presynaptic protein NACP is associated with plaque formation and neurodegeneration in Alzheimer's disease. Am J Pathol. 1996;148:201–210. [PMC free article] [PubMed] [Google Scholar]
- 13.Abeliovich A, et al. Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron. 2000;25:239–252. doi: 10.1016/s0896-6273(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 14.Murphy DD, Rueter SM, Trojanowski JQ, Lee VM. Synucleins are developmentally expressed, and alpha-synuclein regulates the size of the presynaptic vesicular pool in primary hippocampal neurons. J Neurosci. 2000;20:3214–3220. doi: 10.1523/JNEUROSCI.20-09-03214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabin DE, et al. Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking alpha-synuclein. J Neurosci. 2002;22:8797–8807. doi: 10.1523/JNEUROSCI.22-20-08797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gitler AD, et al. The Parkinson's disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci USA. 2008;105:145–150. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem. 1998;273:9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 18.Kamp F, Beyer K. Binding of alpha-synuclein affects the lipid packing in bilayers of small vesicles. J Biol Chem. 2006;281:9251–9259. doi: 10.1074/jbc.M512292200. [DOI] [PubMed] [Google Scholar]
- 19.Eliezer D, Kutluay E, Bussell R, Jr, Browne G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 20.Jao CC, Der-Sarkissian A, Chen J, Langen R. Structure of membrane-bound alpha-synuclein studied by site-directed spin labeling. Proc Natl Acad Sci USA. 2004;101:8331–8336. doi: 10.1073/pnas.0400553101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 22.Bortolus M, et al. Broken helix in vesicle and micelle-bound alpha-synuclein: Insights from site-directed spin labeling-EPR experiments and MD simulations. J Am Chem Soc. 2008;130:6690–6691. doi: 10.1021/ja8010429. [DOI] [PubMed] [Google Scholar]
- 23.Drescher M, et al. Antiparallel arrangement of the helices of vesicle-bound alpha-synuclein. J Am Chem Soc. 2008;130:7796–7797. doi: 10.1021/ja801594s. [DOI] [PubMed] [Google Scholar]
- 24.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 25.Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 26.Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Chandra S, Chen X, Rizo J, Jahn R, Sudhof TC. A broken alpha-helix in folded alpha-synuclein. J Biol Chem. 2003;278:15313–15318. doi: 10.1074/jbc.M213128200. [DOI] [PubMed] [Google Scholar]
- 28.Bussell R, Jr., Eliezer D. A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J Mol Biol. 2003;329:763–778. doi: 10.1016/s0022-2836(03)00520-5. [DOI] [PubMed] [Google Scholar]
- 29.Borbat P, Ramlall TF, Freed JH, Eliezer D. Inter-helix distances in lysophospholipid micelle-bound alpha-synuclein from pulsed ESR measurements. J Am Chem Soc. 2006;128:10004–10005. doi: 10.1021/ja063122l. [DOI] [PubMed] [Google Scholar]
- 30.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 31.Bhatnagar J, Freed JH, Crane BR. Rigid body refinement of protein complexes with long-range distance restraints from pulsed dipolar ESR. Methods Enzymol. 2007;423:117–133. doi: 10.1016/S0076-6879(07)23004-6. [DOI] [PubMed] [Google Scholar]
- 32.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 34.Jeschke G, et al. DeerAnalysis2006 - a Comprehensive Software Package for Analyzing Pulsed ELDOR Data. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- 35.Pearlman DA, et al. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comp Phys Comm. 1995;91:1–41. 1995. [Google Scholar]
- 36.Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. High-resolution protein design with backbone freedom. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 37.Stetefeld J, et al. Crystal structure of a naturally occurring parallel right-handed coiled coil tetramer. Nat Struct Biol. 2000;7:772–776. doi: 10.1038/79006. [DOI] [PubMed] [Google Scholar]
- 38.Plecs JJ, Harbury PB, Kim PS, Alber T. Structural test of the parameterized-backbone method for protein design. J Mol Biol. 2004;342:289–297. doi: 10.1016/j.jmb.2004.06.051. [DOI] [PubMed] [Google Scholar]
- 39.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: Application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci USA. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang YW, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Skolnick J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.