Abstract
Tyrosine trans-phosphorylation is a key event in receptor tyrosine kinase signaling, yet, the structural basis for this process has eluded definition. Here, we present the crystal structure of the FGF receptor 2 kinases caught in the act of trans-phosphorylation of Y769, the major C-terminal phosphorylation site. The structure reveals that enzyme- and substrate-acting kinases engage each other through elaborate and specific interactions not only in the immediate vicinity of Y769 and the enzyme active site, but also in regions that are as much of 18 Å away from D626, the catalytic base in the enzyme active site. These interactions lead to an unprecedented level of specificity and precision during the trans-phosphorylation on Y769. Time-resolved mass spectrometry analysis supports the observed mechanism of trans-phosphorylation. Our data provide a molecular framework for understanding the mechanism of action of Kallmann syndrome mutations and the order of trans-phosphorylation reactions in FGFRs. We propose that the salient mechanistic features of Y769 trans-phosphorylation are applicable to trans-phosphorylation of the equivalent major phosphorylation sites in many other RTKs.
Keywords: crystal structure, FGF receptor, RTKs
Signaling by receptor tyrosine kinases (RTKs) plays ubiquitous roles throughout the human life cycle commencing at germ cell maturation and continuing throughout embryogenesis into adulthood (1). Ligand binding to the extracellular region of RTKs triggers activation of the intracellular tyrosine kinase domain through the universal process of trans-phosphorylation, whereby one kinase acts as a substrate for another one. Trans-phosphorylation has two major roles in RTK signaling: trans-phosphorylation on A-loop tyrosines elevates enzyme activity while trans-phosphorylation of juxtamembrane and C-terminal tyrosines generate platforms for recruitment and phosphorylation of target substrates (2–6).
Structural studies of tyrosine kinase domains have been instrumental in shaping our current understanding of the mechanisms of tyrosine kinase regulation. Crystal structures of unphosphorylated tyrosine kinase domains have unveiled diverse tactics used by kinases to achieve self-inhibition (7–10). The crystal structures of phosphorylated kinases, however, reveal how tyrosine phosphorylation stabilizes the active conformation of the kinase, and how peptide substrates dock into the enzyme active site (7, 11–13). In contrast, the structural basis for tyrosine trans-phosphorylation has remained elusive. Here, we report the crystal structure of a phosphorylated FGF receptor 2 (FGFR2) kinase domain, which provides the first crystallographic snapshot of trans-phosphorylation in action. Our structural data supported by biochemical data show that trans-phosphorylation proceeds with a much higher degree of specificity than that currently perceived based on the crystal structures of kinases in complexes with short peptide substrates.
Results and Discussion
We recently reported the crystal structure of the A-loop phosphorylated FGFR2 kinase domain comprising residues P458 to Q778 (FGFR2K458–778) in complex with AMP-PCP and a 15-residue FGFR2-derived substrate peptide representing Y769, the major phosphorylation site at the C-tail of FGFR2 (PDB entry 2PVF, referred to as the “kinase-peptide” structure hereafter) (7). Diffraction analysis of a morphologically similar crystal of the same complex grown at slightly higher PEG concentrations showed that it differed from the kinase-peptide structure in the b-axis dimension. The structure of this new crystal form was solved using the kinase-peptide structure as the search model, and has been refined to 2.0 Å resolution (supporting information (SI) Table S1). This new crystal structure, deposited in the RCSB Protein Data Bank under PDB ID 3CLY, contains one kinase molecule in the asymmetric unit consisting of residues L468 to L772, one AMP-PCP molecule, 2 Mg2+ ions, and 107 water molecules (Fig. S1). Superimposition of this new crystal structure onto the kinase-peptide structure, and the crystal structure of unphosphorylated FGFR2 kinase domain comprising residues P458 to E768 (FGFR2K458–768) (PDB entry 2PSQ) (7) shows that the new kinase structure has assumed an active conformation (Figs. S1 and S2). Surprisingly, however, the new kinase structure lacks the substrate peptide.
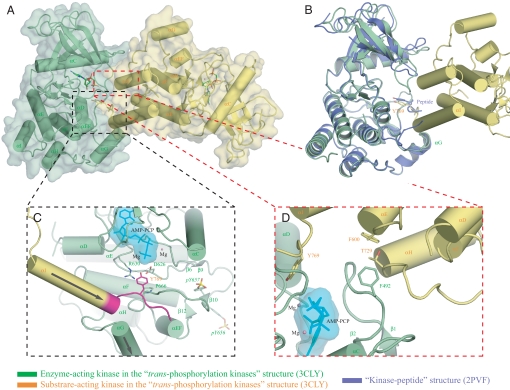
Inspection of the packing of the kinase molecules in the new crystal structure reveals that the lack of substrate peptide is because the kinase molecules are engaged in enzyme-substrate relationship (Fig. 1A; we shall refer to this new structure as the “trans-phosphorylating kinases” structure hereafter). Specifically, the Y769 from one kinase (referred to as the “substrate-acting kinase” hereafter) docks into the active site of the other kinase (referred to as the “enzyme-acting kinase” thereafter), occupying a similar position as Y769 (P0) of the peptide substrate in the kinase-peptide structure (Fig. 1B, also compare Fig. 2 A and B). Reminiscent of the kinase-peptide structure, D626, the catalytic base of the enzyme-acting kinase in the trans-phosphorylating kinases structure, is ready to abstract a proton from Y769 of the substrate-acting kinase (Fig. 1C, also compare Fig. 2 A and B). In the trans-phosphorylating kinases structure, the enzyme-acting kinase interacts with residues T765 (P-4) through L772 (P+3) of the substrate-acting kinase (Fig. 2 A and C) whereas in the kinase-peptide structure, besides Y769 (P0), only L770 (P+1) of the substrate peptide is involved in enzyme-substrate recognition (Fig. 2B). Moreover, in the trans-phosphorylating kinases structure, the enzyme-acting kinase interacts with other regions in the C-lobe of the substrate-acting kinase remote from Y769 (Figs. 1D and 2D). These extensive interactions between the enzyme- and substrate-acting kinases apparently compete with the binding of the substrate peptide into the active site, accounting for the lack of substrate in the structure.
Fig. 1.
Crystallographic snapshot of the trans-phosphorylation reaction at Y769, a major phosphorylation site in FGFR2K. (A) The substrate-acting kinase (in yellow) interacts with both N- and C-lobe of the enzyme-acting kinase (in green) during the trans-phosphorylation on Y769. (B) The tyrosine of the peptide substrate in the kinase-peptide structure (in blue) occupies a similar position as the Y769 in the substrate-acting kinase. (C) The trans-phosphorylation reaction on Y769 phosphorylation site. The near parallel arrangement of the αI helix from the substrate-acting kinase and the αG helix from the enzyme-acting kinase is denoted by the two arrows. The extra ordered residues at the C-tail of the trans-phosphorylating kinases structure compared with the kinase-peptide structure are highlighted in magenta. (D) The interaction between the C-lobe of the substrate-acting kinase and the N-lobe of the enzyme-acting kinase. Selected residues are show in stick diagrams. Atom colorings are as follows: red, oxygens; blue, nitrogens; yellow, phosphorus; carbons are colored according to the kinase molecule to which they belong. Hydrogen bonds are shown as black dashed lines. The ATP analogue (in cyan) is shown in stick representation, and its molecular surface is also shown as a solid semitransparent surface. Mg2+ ions are in pink.
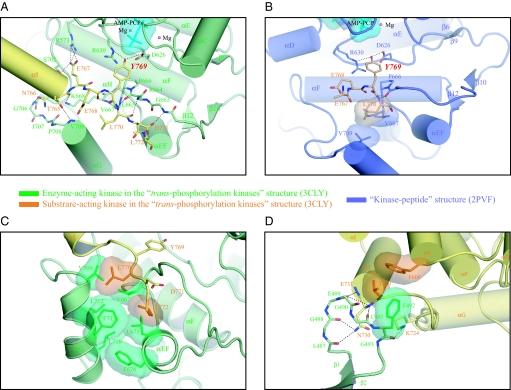
Fig. 2.
Structural basis for the trans-phosphorylation on Y769 of FGFR2K. (A) Detailed view of the interactions between the substrate-acting kinase and the enzyme-acting kinase in the vicinity of active site. (B) In the kinase-peptide structure, the peptide substrate makes limited contacts with the enzyme. (C) Detailed view of the hydrophobic interactions between the L770 (P+1) and L772 (P+3) residues of the substrate-acting kinase and the residues from the A-loop and the αG and αEF helices of the enzyme-acting kinase. (D) Detailed view of the interaction between the C-lobe of the substrate-acting kinase and the nucleotide-binding loop of the enzyme-acting kinase. Yellow, substrate-acting kinase; green, enzyme-acting kinase; blue, kinase in the kinase-peptide structure; wheat, peptide substrate. Atom colorings are as in Fig. 1. Hydrogen bonds, the ATP analogue and Mg2+ ions are rendered as in Fig. 1. Hydrophobic interactions are rendered as solid semitransparent surfaces.
Enzyme-Substrate Interface.
Our trans-phosphorylating kinases structure reveals an unprecedented level of specificity during the trans-phosphorylation on Y769. The enzyme-substrate relationship between the two FGFR2 kinases in the trans-phosphorylating kinases structure is achieved by contacts between the bottom corner of the C-lobe of the substrate-acting kinase and the front side of the enzyme-acting kinase (Fig. 1A). A total of 1,852.57 Å2 surface area is buried between the enzyme- and substrate-acting kinases, of which ∼70% is with the C-lobe of the enzyme-acting kinase and 30% with the N-lobe of the enzyme-acting kinase. The enzyme-substrate interface has a shape complementarity of 0.724 and a hydrophobic/polar fraction of 50%, values consistent with a physiological interface (14, 15). Specifically, the αH and αE helices from the substrate-acting kinase lean via their N termini tips against the N-lobe of the enzyme-acting kinase (Figs. 1D and 2D), accounting for the closer disposition of the kinase lobes in the trans-phosphorylating kinases structure relative to that observed in the kinase-peptide structure (Fig. S1C). The αI helix of the substrate-acting kinase descends in near parallel fashion via its C-terminal tip onto the N-terminal tip of the αG helix of the enzyme-acting kinase (Fig. 1C and 2A). This near parallel arrangement of the αI helix from the substrate-acting kinase and αG helix from the enzyme-acting kinase brings about close contacts between the negative pole of the αI helix and the positive pole of the αG helix and optimally presents residues following the αI helix including the phosphorylation site Y769 into the active site.
In the kinase-peptide structure, the αI helix terminates at residue T765 and the following residues are disordered. In the trans-phosphorylating kinases structure, however, the αI helix is elongated by one helical turn incorporating residues T765, N766 and E767 into the αI helix (Fig. S1B). Moreover, residues E768 to L772 after the αI helix are ordered as well (Fig. S1B). These restructurings are clearly due to the key role this region plays in enzyme binding. Residues N-terminal to Y769 (P0) make numerous contacts with the enzyme-acting kinase, none of which are seen in the kinase-peptide structure (compare Fig. 2 A and B). The most prominent contact is the salt bridge between E767 (P-2), the last residue of the αI helix, and R573 of the enzyme-acting kinase (Fig. 2A). R573 is located in the αD helix and has not been previously implicated in substrate recognition. Also at the negative pole of the αI helix dipole, the backbone carbonyl oxygens of T765 (P-4) and N766 (P-3), and the side chain of N766 make hydrogen bonds with residues at the positive pole of the αG helix of the enzyme-acting kinase (Fig. 2A). Notably, the loop leading to the αG helix and the N-terminal end of the αG helix are two regions in the C-lobe, which deviate structurally from those in the kinase-peptide structure (Fig. S1E). In the kinase-peptide structure, this region has high temperature factors consistent with the presence of fewer contacts between the peptide substrate and this region.
In the trans-phosphorylating kinases structure, contacts between residues following Y769 (P0) of the substrate-acting kinase and the enzyme-acting kinase are also more extensive than those observed in the kinase-peptide structure. In the trans-phosphorylating kinases structure, residues L770 (P+1), D771 (P+2) and L772 (P+3) of the substrate-acting kinase participate in enzyme binding whereas in the kinase-peptide structure only the P+1 residue of peptide substrate is engaged (compare Fig. 2 A and B). In the trans-phosphorylating kinases structure, three hydrogen bonds are made between the backbone atoms of L770 (P+1) and the side chain of D771 (P+2) of the substrate-acting kinase and the backbone atoms of the A-loop of the enzyme-acting kinase (Fig. 2A). In addition, L770 (P+1) and L772 (P+3) engage a shallow hydrophobic depression in the enzyme-acting kinase composed of residues from the A-loop, and the αG and αEF helices (Fig. 2C).
The most striking feature of our trans-phosphorylating kinases structure is that it also implicates the N-terminal lobe of the enzyme in conferring specificity during trans-phosphorylation. At the heart of the interface between the substrate-acting kinase and the N-lobe of the enzyme-acting kinase, the aromatic ring of F600 of the substrate-acting kinase packs perpendicularly against the aromatic ring of F492 in the β1-β2 loop of the enzyme-acting kinase (Figs. 1D and 2D). This type of perpendicular aromatic-aromatic interaction is energetically favorable and has been observed in many protein–protein interfaces (16). Several hydrogen bonds fortify this interface as well. The carbonyl oxygens in the β1-β2 loop of the enzyme-acting kinase make hydrogen bonds with the backbone nitrogens of N730 and E731 and the side chain of N730 at the positive pole of the αH helix dipole from the substrate-acting kinase (Fig. 2D). Moreover, the side chain of K724 from the substrate-acting kinase makes a hydrogen bond with the backbone carbonyl oxygen of F492 in the β1-β2 loop of the enzyme-acting kinase (Fig. 2D). As a consequence of these extensive contacts, the nucleotide binding β1-β2 loop is well-ordered and deviates from that of the kinase-peptide structure, which has high temperature factors (Fig. S1C). Taken together, the trans-phosphorylating kinases structure reveals that the trans-phosphorylation on Y769 proceeds with an exceptional degree of specificity, and that the kinase-peptide structure provides a partial picture of trans-phosphorylation process on Y769.
Analysis of Structure-Based Mutations Support the Physiological Relevance of the Mode of Transphosphorylation on Y769.
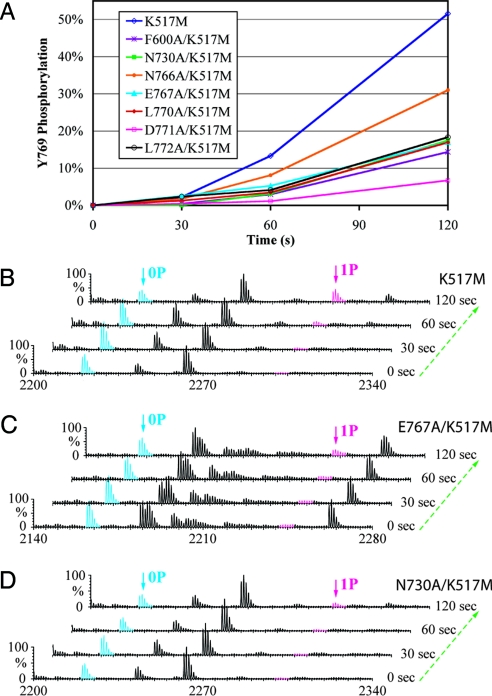
To test the functional relevance of the specific contacts between the substrate- and enzyme-acting kinases in our trans-phosphorylating kinases structure, we individually substituted F600, N730, N766, E767, L770, D771, and L772 with alanine in the kinase-dead (K517M) version of FGFR2K458–778 protein (referred to as the “WT kinase substrate” and “mutant kinase substrates” hereafter) rather than in the wild type version. These WT and mutant kinase substrates were then allowed to be trans-phosphorylated by a wild type kinase lacking Y769 (FGFR2K458–768). We chose this experimental design for two reasons: (i) to exclude the influence of the mutations on the kinase activity (Fig. S3A) and (ii) to rule out the contribution of the enzyme-acting kinase to the Y769 phosphorylation. The status of phosphorylation on Y769 in the WT and mutant kinase substrates as a function of time was monitored by matrix-assisted laser desorption/ionization quadrupole-time of flight mass spectrometry (MALDI Q-TOF MS). Trans-phosphorylation of Y769 in F600A, N730A, N766A, E767A, L770A, D771A, and L772A mutant kinase substrates was reduced relative to the parent WT kinase substrate, supporting the functional relevance of the observed specific contacts in the trans-phosphorylating kinases structure (Fig. 3). To test whether the observed contacts between the enzyme- and substrate-acting kinases are specific for trans-phosphorylation on Y769, we also studied the impact of some of the interface mutations on the phosphorylation of the A-loop and kinase insert tyrosines (Fig. 3 B and C and Fig. S3B). As shown in Fig. 3 B and C and Fig. S3B, the E767A mutation slows down Y769 trans-phosphorylation but has little effect on the phosphorylation of the A-loop or kinase insert tyrosines indicating that the observed mode of trans-phosphorylation in the crystal is specific for Y769.
Fig. 3.
Comparison of the time course of trans-phosphorylation on Y769 in WT and mutant kinase substrates by time-resolved MALDI Q-TOF MS. (A) Relative to the parent WT kinase substrate (K517M), trans-phosphorylation on Y769 is reduced in the 7 mutant kinase substrates harboring structure-based mutations. Ion counts of the phosphopeptide and its unphosphorylated version are used to estimate the percentage of phosphorylation of Y769. (B–D) Time-resolved MALDI Q-TOF MS spectra show the progression of the trans-phosphorylation reaction on Y769 peptide in K517M (B), E767A/K517M (C), and N730/K517M (D) mutant substrates. 0P and 1P denote the FGFR2K peptide in unphosphorylated and single-phosphorylated states, respectively.
The physiological significance of the mechanism of trans-phosphorylation on Y769 revealed by our trans-phosphorylating kinases structure is also corroborated by a double pathogenic mutation (Q764H/D768Y) in the FGFR1 gene in a patient with isolated GnRH deficiency (idiopathic hypogonadotropic hypogonadism, IHH) (N.P., unpublished data). Approximately 10% of the Kallmann syndrome (IHH with anosmia) patients harbor FGFR1c mutations and structural and functional analysis of >15 of these mutations has documented that they impair FGFR1 function and signaling (17–19). Based on our structural data, the double mutation should affect trans-phosphorylation on Y766, which corresponds to Y769 of FGFR2, thus manifesting in reduced recruitment and phosphorylation of PLCγ by FGFR1.
The enzyme-substrate interface mediating the trans-phosphorylation of Y769 in FGFR2 is not fully conserved among the other three human FGFRs. Nevertheless, we propose that overall structural model for trans-phosphorylation of Y769 will be applicable to the other three FGFRs but the details/extent of interactions may well be different. We propose that the sequence divergence among FGFRKs is probably responsible for the differences that exist in the speed/efficiency of phosphorylation of the equivalent C-terminal phosphorylation sites among these 4 FGFRs. This hypothesis is currently being investigated in our laboratory.
Order of Transphosphorylation Reactions.
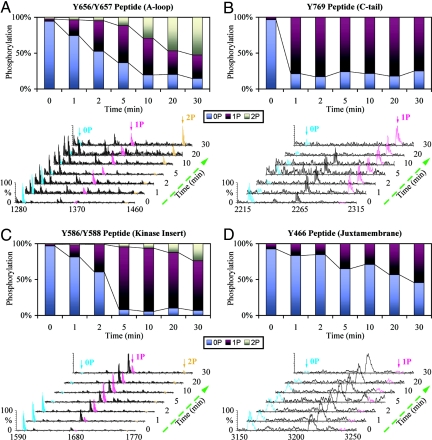
A comparison of the primary sequences surrounding the different phosphorylation sites of FGFR2 kinase shows little sequence similarity, implying the existence of mechanistic differences in the trans-phosphorylation of different sites. These sequence differences may also be responsible for different efficacies at which these different sites are phosphorylated, and thus dictate the order of trans-phosphorylation events. Interestingly, modeling studies based on the trans-phosphorylating kinases structure predicts fewer enzyme-substrate contacts during trans-phosphorylation of kinase insert tyrosines, Y586 and Y588, and the juxtamembrane site, Y466 (data not shown). This implies that trans-phosphorylation on Y769 is preferred over other trans-phosphorylation reactions suggesting that it may kinetically precede other phosphorylation reactions. To test this hypothesis we allowed wild-type kinase (FGFR2K458–778) to trans-phosphorylate itself in the presence of ATP and MgCl2 over time, and monitored the phosphorylation status of each phosphorylation site as a function of time by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS). This analysis shows that Y769 is phosphorylated much faster than the kinase insert sites and the juxtamembrane phosphorylation sites (Fig. 4). After only 1 min, phosphorylation of Y769 is nearly complete whereas at this time only 20% of the A-loop peptide is singly phosphorylated. Hence, only a catalytic amount of mandatory A-loop phosphorylation is sufficient to initiate and complete the phosphorylation of Y769, which reaffirms the structural finding that Y769 is a highly preferred site (Fig. 4 A and B). It is noteworthy that the A-loop tyrosine in Eph RTKs is also a suboptimal phosphorylation site compared with juxtamembrane sites (13). Our data differ from the data on sequential phosphorylation of the FGFR1 kinase showing that phosphorylation on the A-loop Y653 is completed before the kinase insert and juxtamembrane sites are phosphorylated (20). This discrepancy could be because the FGFR1 kinase domain used by Furdui et al. lacks the C-terminal Y766, the equivalent of Y769 in FGFR2.
Fig. 4.
Analysis of order of tyrosine autophosphorylation by time-resolved MALDI-TOF MS in wild type FGFR2K458–778. Y769 is highly preferred tyrosine phosphorylation site in FGFR2K. Time-resolved MALDI-TOF MS spectra and ion counts estimations at different time points show the progression of the autophosphorylation reaction at the A-loop [Y656/Y657 peptide (A)], the C-tail [Y769 peptide (B)], the kinase insert [Y586/Y588 peptide (C)] and the juxtamembrane [Y466 peptide (D)], respectively. 0P, 1P, and 2P represent peptides in unphosphorylated, single-phosphorylated, and double-phosphorylated states, respectively.
Conclusion
Tyrosine trans-phosphorylation is an ordered event that is initiated by phosphorylation on the A-loop tyrosines and is pursued by trans-phosphorylation on tyrosines in the C-tail, juxtamembrane region and kinase insert. A-loop phosphorylation up-regulates the kinase activity of the receptor, whereas the latter phosphorylations create platforms for signaling molecules to facilitate their phosphorylation. In this report, we provided the molecular basis for the trans-phosphorylation reaction of Y769, which upon phosphorylation serves as a docking site for the SH2 domains of PLCγ-1, a key mediator of FGFR signaling. Our data reveal that during trans-phosphorylation, kinases engage each other through elaborate and extensive interactions involving regions outside of the immediate vicinity of the tyrosine phosphorylation sites and the active site resulting in an exceptional level of specificity. Additional crystal structures of kinases captured in autophosphorylation reactions should further enrich our understanding of the order of receptor trans-phosphorylation and signaling events down stream of RTKs.
A structure-based sequence alignment of RTKs reveals that besides FGFRs, many other RTKs possess autophosphorylation sites that are similarly positioned relative to the αI helix as Y769 in FGFR2 (Fig. S4) (21–25). However, many of the FGFR2 residues that mediate the enzyme-substrate interface in our structure are not conserved in these other RTKs. Nevertheless, we propose that the overall structural mode of enzyme-substrate engagement, particularly the αI-αG dipole-dipole contacts, is probably applicable to trans-phosphorylation of these equivalent sites in other RTKs, even though the details of the respective enzyme-substrate interfaces will be different.
An asymmetric dimer of EGF receptor (EGFR) kinase was recently shown to play a role in the initial activation of the receptor by homodimerization (26). The asymmetric arrangement of enzyme- and substrate-acting kinases during Y769 trans-phosphorylation is, however, very different from the one observed between the kinase monomers in the asymmetric EGFR kinase dimer (PDB entry 2GS2) (Fig. S5). The asymmetric EGFR dimer reveals a mechanism by which EGFRK is activated by homodimerization, an event that precedes tyrosine trans-phosphorylation. The asymmetric FGFR2K enzyme-acting kinase:substrate-acting kinase dimer in our study purely depicts an enzyme-substrate relationship between already activated FGFR2 kinases, and as such gives insights into how trans-phosphorylation can occur once kinases are activated through A-loop phosphorylation. However, it is formally possible that FGFR kinases are also initially activated by forming asymmetric dimers as seen for the EGFR kinases.
Experimental Procedures
Protein Expression, Purification, and Crystallization.
The wild type kinases (FGFR2K458–778 and FGFR2K458–768), and the 7 kinase “dead” versions of FGFR2K458–778 (K517M, F600A/K517M, N730A/K517M, N766A/K517M, E767A/K517M, L770A/K517M, D771A/K517M, L772A/K517M) were expressed in E. coli with an N-terminal 6XHis-tag to aid in protein purification. The kinases were purified using sequential Ni2+-chelating, anion exchange and size exclusion chromatography. The purity of the proteins was estimated to be >98% based on SDS/PAGE analysis. The purified wild type FGFR2K458–778 kinase was then mixed with ATP and MgCl2 and the completion of tyrosine autophosphorylation was monitored by native PAGE analysis. Phosphorylated kinase was then separated from excess of ATP on a size-exclusion column, followed by anion exchange chromatography to yield a kinase that was homogenously phosphorylated on both A-loop tyrosines, Y656 and Y657, and on the C-terminal Y769 (confirmed by sequencing the phosphopeptides, using MALDI Q-TOF MS. Tyrosine phosphorylated FGFR2K458–778 kinase protein was concentrated to ∼30 mg/ml using a Centricon-10. Before crystallization, the protein was mixed with peptide substrate, ATP-analogue (AMP-PCP) and MgCl2 at a molar ratio of 1:1:3:15. The peptide substrate comprises residues 764 to 778, and contains the major tyrosine phosphorylation site which upon phosphorylation serves as the recruitment site for the SH2 domain(s) of PLCγ-1 (27). Crystals were grown by hanging drop vapor diffusion at 20 °C using crystallization buffer composed of 0.1 M Hepes (pH 7.5), 26% PEG 4000 and 0.2 M ammonium sulfate. After the completion of data collection, the crystal was subjected to SDS/PAGE and analyzed by MS, which showed that Y769 was not phosphorylated, consistent with the crystal structure.
Data Collection and Structure Determination.
Diffraction data were collected on a single cryocooled crystal at beamline X-4A at the National Synchrotron Light Source, Brookhaven National Laboratory. The crystal was stabilized in mother liquor by stepwise increasing the glycerol concentration to 20%, and then flash-frozen in dry nitrogen stream. All diffraction data were processed using HKL2000 Suite (28). Molecular replacement solution was found using the related phosphorylated wild type FGFR2K structure in complex with peptide substrate (PDB entry 2PVF) as the search model using the program AMoRe (29). Rigid-body refinements were performed using CNS (30) by treating the N-lobe and C-lobe of the kinases as two separate entities. Model building was carried out using O (31) and iterative positional and B-factor refinements were done using CNS (30). The refined structure displays good geometry and Ramachandran statistics. Data collection and structure refinement statistics are listed in Table S1. Atomic superimpositions and calculations of lobe rotations were performed using program lsqkab (32) in CCP4 Suite (33) and structural representations were prepared using PyMol (34). Shape complementarity was calculated using program sc in CCP4 Suite (33) and hydrophobic/polar fraction was calculated using CNS (30).
Analysis of the Time Course of Trans-phosphorylation on Y769 by Mass Spectrometry.
Trans-phosphorylation on Y769 was started by adding a mixture of the WT or mutant kinase substrate (K517M, F600A/K517M, N730A/K517M, N766A/K517M, E767A/K517M, L770A/K517M, D771A/K517M, or L772A/K517M), ATP and MgCl2 to the active kinase (FGFR2K458–768) in equal volumes at room temperature. The reaction concentrations for the active enzyme, kinase substrate, ATP, and MgCl2 were 1 mg/ml, 10 mg/ml, 10 mM, and 20 mM, respectively. The reactions were quenched at different time points by adding an equal volume of 100 mM EDTA (final concentration 33.33 mM) to the reaction mix. After SDS/PAGE and Coomassie-Blue staining, gel bands corresponding to the WT or mutant kinase substrate were excised and digested with trypsin according to a published protocol (35). Approximately 1/8 of the protein digests were analyzed by positive ion MALDI Q-TOF MS (MALDI Q-TOF Ultima, Waters–Micromass). 50 mg/ml 2,5-Dihydroxybenzoic acid (DHB) in 0.1% trifluoroacetic acid (TFA) and 50% Acetonitrile was used as MALDI matrix. A total of 1.0 μL of protein digests in 0.1% TFA were mixed with equal volume of matrix solution and dried in air. For each sample, ion signals from 310 laser shots were combined into one mass spectrum. After spectrum smoothing and background subtraction, the ion counts from the phosphopeptide and its unphosphorylated version were used to estimate the ratio of phosphorylation on tyrosines at different time points.
Analysis of the order of tyrosine autophosphorylation by Mass Spectrometry.
Autophosphorylation reaction was initiated by adding equal volumes of the kinase (FGFR2K458–778) to a mixture of ATP and MgCl2 at room temperature. The reaction concentrations for the enzyme, ATP and MgCl2 were 5 mg/ml, 25 mM and 50 mM, respectively. Preparation of the samples and MALDI-TOF MS (Tof Spec 2E, Waters–Micromass) analysis were carried out as above.
Kinase Assays.
A continuous spectrophotometric assay (36) was used to measure the kinase activity of wild type and mutant FGFR2Ks. The FGFR2K autophosphorylation assays were carried out at 30 °C using 1 μM enzyme and 1 mM ATP with 100 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 1 mM phosphoenolpyruvate, 0.28 mM NADH, 89 units/ml pyruvate kinase, 124 units/ml lactate dehydrogenase, in a total volume of 50 μL. Data were recorded every 6 s. The kinetic parameters were determined by fitting initial rate data to the Michaelis–Menten equation.
Supplementary Material
Acknowledgments.
We thank Drs. R. Abramowitz and J. Schwanof for synchrotron beamline assistance and Dr. X-P. Kong for comments on the manuscript. Beamlines X-4A and X-4C at the National Synchrotron Light Source, Brookhaven National Laboratory, a DOE facility, are supported by New York Structural Biology Consortium. This work was supported by National Institutes of Health Grants DE13686 (to M.M.) and CA122091 (to W.T.M.). The New York University Protein Analysis Facility is supported by National Institutes of Health Shared Instrumentation Grants S10 RR14662 and S10 RR017990, National Institute of Neurological Disorders and Stroke Grant P30 NS050276, and National Cancer Institute Core Grant P30 CA016087 (to T.A.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in Protein Data Bank, www.pdb.org (PDB ID code 3CLY).
This article contains supporting information online at www.pnas.org/cgi/content/full/0807752105/DCSupplemental.
References
- 1.Hunter T. Signaling—2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T. Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 4.Jiang G, Hunter T. Receptor signaling: when dimerization is not enough. Curr Biol. 1999;9:R568–R571. doi: 10.1016/s0960-9822(99)80357-1. [DOI] [PubMed] [Google Scholar]
- 5.Wilson IA, Jolliffe LK. The structure, organization, activation and plasticity of the erythropoietin receptor. Curr Opin Struct Biol. 1999;9:696–704. doi: 10.1016/s0959-440x(99)00032-9. [DOI] [PubMed] [Google Scholar]
- 6.Stroud RM, Wells JA. Mechanistic diversity of cytokine receptor signaling across cell membranes. Sci STKE. 2004;2004:re7. doi: 10.1126/stke.2312004re7. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, et al. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell. 2007;27:717–730. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellicena P, Kuriyan J. Protein–protein interactions in the allosteric regulation of protein kinases. Curr Opin Struct Biol. 2006;16:702–709. doi: 10.1016/j.sbi.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Lietha D, et al. Structural basis for the autoinhibition of focal adhesion kinase. Cell. 2007;129:1177–1187. doi: 10.1016/j.cell.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubbard SR. Autoinhibitory mechanisms in receptor tyrosine kinases. Front Biosci. 2002;7:d330–d340. doi: 10.2741/A778. [DOI] [PubMed] [Google Scholar]
- 11.Favelyukis S, Till JH, Hubbard SR, Miller WT. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat Struct Biol. 2001;8:1058–1063. doi: 10.1038/nsb721. [DOI] [PubMed] [Google Scholar]
- 12.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mol CD, et al. Structure of a c-kit product complex reveals the basis for kinase transactivation. J Biol Chem. 2003;278:31461–31464. doi: 10.1074/jbc.C300186200. [DOI] [PubMed] [Google Scholar]
- 14.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 15.Dill KA. Theory for the folding and stability of globular proteins. Biochemistry. 1985;24:1501–1509. doi: 10.1021/bi00327a032. [DOI] [PubMed] [Google Scholar]
- 16.Deindl S, et al. Structural basis for the inhibition of tyrosine kinase activity of ZAP-70. Cell. 2007;129:735–746. doi: 10.1016/j.cell.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 17.Albuisson J, et al. Kallmann syndrome: 14 novel mutations in KAL1 and FGFR1 (KAL2) Hum Mutat. 2005;25:98–99. doi: 10.1002/humu.9298. [DOI] [PubMed] [Google Scholar]
- 18.Zenaty D, et al. Paediatric phenotype of Kallmann syndrome due to mutations of fibroblast growth factor receptor 1 (FGFR1) Mol Cell Endocrinol. 2006;254–255:78–83. doi: 10.1016/j.mce.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Pitteloud N, et al. Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Mol Cell Endocrinol. 2006;254–255:60–69. doi: 10.1016/j.mce.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 20.Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 21.Mohammadi M, Schlessinger J, Hubbard SR. Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell. 1996;86:577–587. doi: 10.1016/s0092-8674(00)80131-2. [DOI] [PubMed] [Google Scholar]
- 22.Knowles PP, et al. Structure and chemical inhibition of the RET tyrosine kinase domain. J Biol Chem. 2006;281:33577–33587. doi: 10.1074/jbc.M605604200. [DOI] [PubMed] [Google Scholar]
- 23.Shewchuk LM, et al. Structure of the Tie2 RTK domain: Self-inhibition by the nucleotide binding loop, activation loop, and C-terminal tail. Structure. 2000;8:1105–1113. doi: 10.1016/s0969-2126(00)00516-5. [DOI] [PubMed] [Google Scholar]
- 24.Schiering N, et al. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc Natl Acad Sci USA. 2003;100:12654–12659. doi: 10.1073/pnas.1734128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mol CD, et al. Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J Biol Chem. 2004;279:31655–31663. doi: 10.1074/jbc.M403319200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Mohammadi M, et al. Point mutation in FGF receptor eliminates phosphatidylinositol hydrolysis without affecting mitogenesis. Nature. 1992;358(6388):681–684. doi: 10.1038/358681a0. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 29.Navaza J. AMoRe: An automated package for molecular replacement. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 30.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr A. 1976;32:922–923. [Google Scholar]
- 33.Collaborative. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.DeLano WL. The PyMOL User's Manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 35.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 36.Barker SC, et al. Characterization of pp60c-src tyrosine kinase activities using a continuous assay: Autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry. 1995;34:14843–14851. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.