Abstract
Aquaporins (AQPs) are a family of ubiquitous membrane channels that conduct water across cell membranes. AQPs form homo-tetramers containing four functional and independent water pores. Aquaporin-0 (AQP0) is expressed in the eye lens where its water permeability is regulated by calmodulin (CaM). Here we use a combination of biochemical methods and NMR spectroscopy to probe the interaction between AQP0 and CaM. We show CaM binds the AQP0 C-terminal domain in a calcium dependent manner. We demonstrate that only two CaM molecules bind a single AQP0 tetramer in a non-canonical fashion, suggesting a form of co-operativity between AQP0 monomers. Based on these results, we derive a structural model of the AQP0/CaM complex, which suggests CaM may be inhibitory to channel permeability by capping the vestibules of two monomers within the AQP0 tetramer. Finally, phosphorylation within AQP0’s CaM binding domain inhibits the AQP0/CaM interaction suggesting a temporal regulatory mechanism for complex formation.
Aquaporins (AQPs) are a family of ubiquitous membrane channels that conduct water and small solutes across membranes (Agre et al., 1993). At least 13 differentially expressed AQPs have been identified in humans (reviewed in (Gonen and Walz, 2006)). These AQPs differ in their permeability rates and in their selectivity: some allowing only water to permeate the pore (for example AQP1 and AQP0) while others also allow small solutes such as glycerol to permeate (for example AQP7 and AQP9) (Agre and Kozono, 2003; Yang and Verkman, 1997). The atomic structure of a number of AQPs have been determined to date (Fu et al., 2000; Gonen et al., 2005; Gonen et al., 2004a; Harries et al., 2004; Hiroaki et al., 2005; Lee et al., 2005; Murata et al., 2000; Ren et al., 2001; Savage et al., 2003; Sui et al., 2001; Tajkhorshid et al., 2002; Tornroth-Horsefield et al., 2005) confirming that aquaporins are tetrameric assemblies in which each monomer forms a functional pore (Shi et al., 1994). Each monomer in the AQP tetramer consists of 6 transmembrane α-helices that pack against each other to form a barrel-like structure with a hydrophilic pore at the center. Both the N- and C- termini localize to the cell cytoplasm. These structural studies in combination with molecular dynamic simulations have provided insight into the structural basis for channel specificity and proton exclusion mechanism (reviewed in (Gonen and Walz, 2006)). While the core transmembrane domains of aquaporins are largely conserved, the cytoplasmic amino- and carboxyl-termini vary greatly in primary sequence and have been implicated in regulation of several aquaporins (Nielsen et al., 2007). Current structural studies of AQPs aim at understanding their regulatory mechanisms.
Aquaporin-0 (AQP0), also known as the Major Intrinsic Polypeptide (MIP), is only expressed in the eye lens, where it constitutes more than 60% of the total membrane protein content of fiber cells (Alcala et al., 1975; Bloemendal et al., 1972) and forms a channel for water permeation. In mature lens fiber cells, AQP0 functions as an adhesive protein forming the 11 nm “thin” membrane junctions between apposing fiber cells (Bok et al., 1982; Costello et al., 1989; Gonen et al., 2005). Normal function of AQP0 is therefore required for lens homeostasis and transparency. Not surprisingly, mutation or malfunction of this protein causes severe developmental lesions and cataracts (Francis et al., 2000a; Gu et al., 2007). AQP0 permeability is tightly regulated in the lens by at least 3 separable bona fide mechanisms: C-terminal cleavage (Gonen et al., 2004b), pH and Ca2+/calmodulin (CaM) (Nemeth-Cahalan and Hall, 2000; Nemeth-Cahalan et al., 2004; Varadaraj et al., 2005). We have previously demonstrated that C-terminal cleavage of AQP0 leads to junction formation and pore closure (Gonen et al., 2004b; Gonen et al., 2005; Gonen et al., 2004a). In contrast, AQP0 regulation by pH and Ca2+/CaM dynamically modulate AQP0 permeability. The pH regulation has been linked to histidine residues on the extracellular loops of AQP0 (Gonen et al., 2004a; Nemeth-Cahalan and Hall, 2000) where a drop in pH from 7.2 to 6.5 increases the water permeability by 2 times. The regulation of AQP0 by Ca2+/CaM may be inhibitory or excitatory (Nemeth-Cahalan et al., 2004; Varadaraj et al., 2005) and appears to involve the intracellular C-terminal helix of AQP0 (Girsch and Peracchia, 1991).
Calmodulin is a 17 kDa bi-lobed Ca2+ binding protein that functions as an ubiquitous secondary messenger in several Ca2+ signaling pathways and has been shown to interact with, and modulate the function of, a number of channels and transporters including aquaporins, connexins, tetraspanins and voltage gated ion channels (Arneson et al., 1995; Pitt, 2007; Zuhlke et al., 1999). Under conditions of high intracellular calcium levels (>100 nM) CaM binds two Ca2+ ions at each of its N- and C-terminal EF-hand domains, resulting in a dramatic conformational rearrangement that exposes hydrophobic binding pockets in CaM (Babu et al., 1988; Vogel, 1994). CaM uses these hydrophobic domains to bind a variety of unique target proteins to evoke a regulatory response in a Ca2+ dependent fashion. Structural data on CaM-membrane protein complexes is sparse despite the ubiquitous nature of CaM and its critical role in the modulation of a number of channels and transporter activity.
Here we use a combination of biochemical methods and nuclear magnetic resonance (NMR) spectroscopy to characterize the AQP0/CaM interaction with the aim of obtaining structural insight into the mechanism of water channel regulation by CaM. Our results reveal a non-canonical Ca2+ dependent CaM interaction with AQP0, where 2 CaM molecules interact with a single AQP0 tetramer, suggesting possible structural co-operativity between monomers within the tetramer. This interaction is greatly inhibited by phosphorylation of AQP0 at a conserved serine position. Finally, we used our experimental results to generate an architectural model of the AQP0/CaM complex that offers new insights into the mechanism of water channel regulation by CaM.
RESULTS
CaM binds to lens membrane proteins in a Ca2+ dependent manner
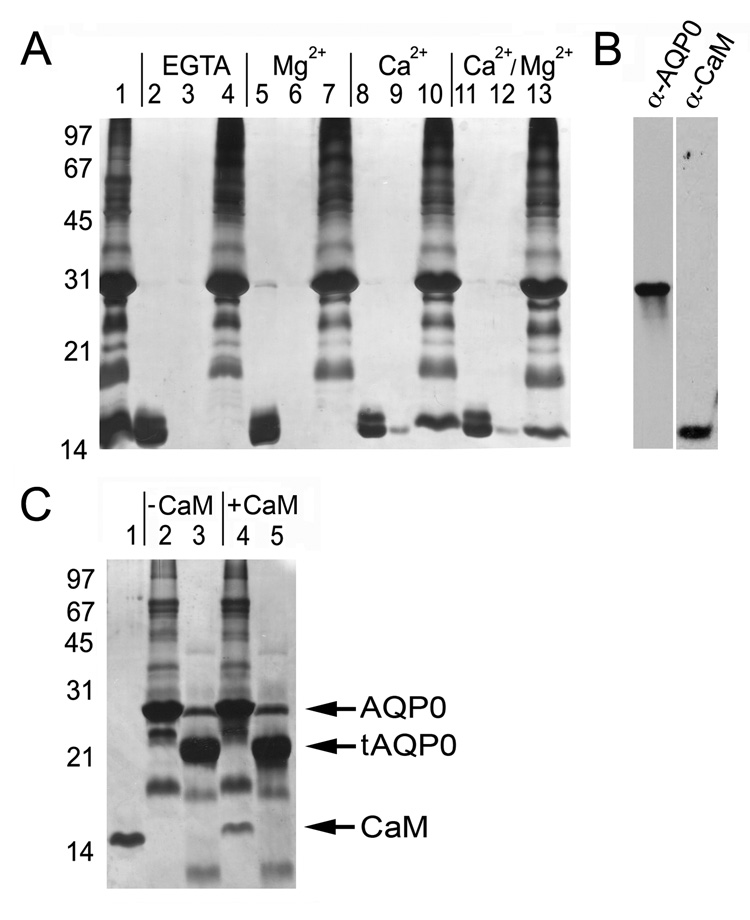
We have developed a lens membrane pull-down assay as a first approach to detect CaM binding to lens membrane proteins, and investigate the Ca2+ dependence of these interactions. Lens membranes were prepared as described previously (Gonen et al., 2000) and incubated with recombinant vertebrate CaM in a binding buffer that contained EGTA, 10 mM Ca2+ and/or 50 mM Mg2+. The binding mixtures were incubated at 37°C for 30 minutes. Unbound CaM was separated from membrane-bound CaM by centrifugation followed by several washes with appropriate buffers. Lens membranes (pellet) or unbound material (supernatant) were then suspended in Laemmli buffer and assayed for the presence of CaM by SDS-PAGE and silver staining and western blotting (Figure 1A and B). In the absence of Ca2+, membrane pellets contained no detectable amounts of CaM (Figure 1A, Lanes 4 and 7), while significant amounts of CaM were routinely observed in conditions containing Ca2+ (Figure 1A, Lanes 10 and 13). Mg2+ alone or in combination with Ca2+ showed no significant effect on CaM binding (Figure 1A, Lanes 7 and 13, respectively). The supernatants from the final wash of conditions containing Ca2+ show trace amounts of CaM (Figure 1A, Lanes 9 and 12), probably a result of leaching of weakly bound Ca2+/CaM during the wash steps. CaM was not observed in the final wash steps in the absence of Ca2+ (Figure 1A, Lanes 3 and 6). Membrane-bound CaM could be efficiently removed from Ca2+ treated lens pellets by the addition of molar excess EGTA (data not shown). CaM has been shown to bind lipid vesicles rich in phosphatidylcholine (PC) (Johnson and Wittenauer, 1983); and indeed lens membarnes are PC rich (Borchman et al., 2004). We therefore treated lens membrane with a protease cocktail to assay CaM binding. In the case of AQP0, treatment with protease results in the truncation of AQP0 into its 21kDa species (Figure 1C, Lane 3), which lacks its entire carboxyl terminus (Gonen et al., 2004b). CaM did not bind to these proteolytically cleaved preparations (Figure 1C Lane 5) highlighting that CaM interaction is to lens membrane protein targets and does not involve binding to lens lipids directly. We note, however, that some of these “lens membrane protein targets” may also include connexins and MP20 (Swamy-Mruthinti, 2001; Zhou et al., 2007).
Figure 1. Calmodulin binds AQP0 carboxyl terminus in a Ca2+ dependent manner.
A, CaM pull-down assay with lens membranes. Molecular weight markers are indicated on the left in kDa. Lane 1: lens membranes incubated with calmodulin. Lanes 2, 5, 8 and 11: supernatants containing unbound CaM. Lanes 3, 6, 9 and 12: supernatants following 4 rounds of pellet washing. Lanes 4, 7, 10 and 13: pellets containing lens membranes. CaM is associated with lens membranes only when Ca2+ is present (Lanes 10 and 13). B, Western blot detection identifying AQP0 (left) as a 26kDa species and CaM (right) as a 17kDa species. C, Pull-down assay with proteolytically cleaved lens membrane proteins. Lanes 1, 2, and 3: CaM, lens membranes and cleaved lens membranes; respectively. “tAQP0” is C-terminally truncated AQP0 (Gonen et al., 2004b). Lanes 4 and 5: pellets containing untreated membranes and cleaved lens membrane proteins, respectively. CaM is found only in the untreated, un-cleaved preparation (Lane 4).
CaM binds the C-terminal α-helix of AQP0 and is inhibited by serine phosphorylation
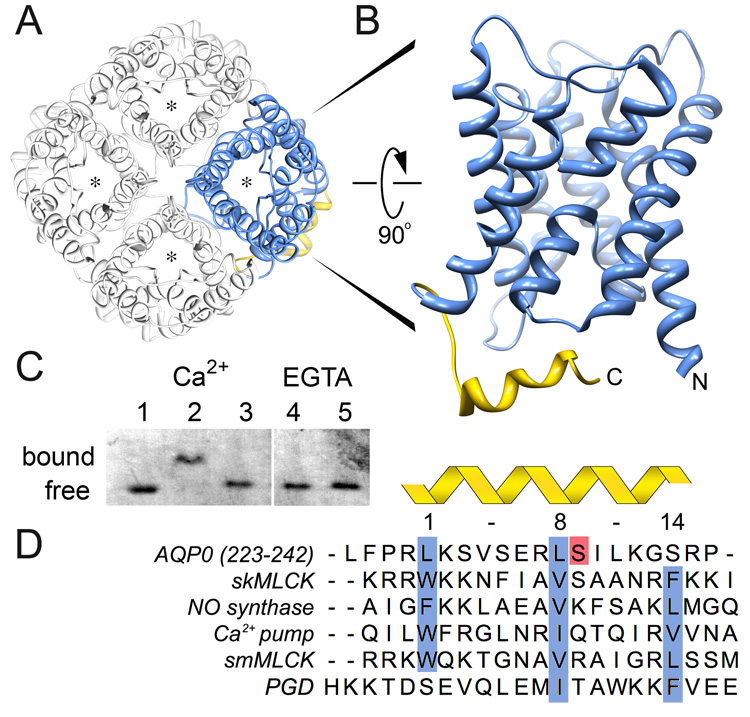
Aquaporins assemble into tetramers in which each monomer forms a functional and independent water pore (Shi et al., 1994) (Figure 2A). Although the core transmembrane domain of different AQPs is well conserved, the cytoplasmic C-terminal domains vary significantly both in sequence and post-translational modifications suggesting that this domain could play an important role in AQP regulation (Gonen and Walz, 2006). The C-terminal domain of AQP0 begins at the end of transmembrane helix 6, at residue Leu 222, and ends at residue Leu 263. The distal part of the AQP0 carboxyl terminus, residues Leu 223 - Pro 242, contains a short α-helix, which is a putative CaM binding site (Girsch and Peracchia, 1991) (Figure 2B, yellow). We synthesized this peptide (referred to in this manuscript as the AQP0 Calmodulin Binding Domain or “AQP0CBD”) and tested for CaM binding activity by native gel electrophoresis. In the presence of Ca2+, we obtain a clear CaM band-shift relative to free CaM (Figure 2C, Lane 1 versus 2). Substituting Ca2+ with EGTA inhibited this interaction even in the presence of 5 times molar excess of the peptide (Figure 2C, Lanes 4 and 5, respectively). Therefore, CaM interacts with AQP0CBD in a Ca2+ dependent manner consistent with a recent report using fluorescent binding assays of CaM to human AQP0CBD (Rose et al., 2008).
Figure 2. Identification of the AQP0-Calmodulin binding domain and effect of serine phosphorylation.
A, Aquaporins form tetrames in which each monomer forms a functional and independent water pore (asterisk) (Shi et al., 1994). The AQP0 tetramer is viewed from the extracellular side of the cell membrane (Protein Data Bank Accession number 2B6O). B, Side view of an AQP0 monomer highlighting the cytoplasmic C-terminal α-helix as the calmodulin binding domain (AQP0CBD, yellow). C, Native gel electrophoresis of calmodulin bound to AQP0CBD. A clear shift in the calmodulin band is seen in the presence of Ca2+ (Lanes 1 versus 2) but not when Ser 235 is phosphorylated (Lane 3) or in the presence of EGTA even with excess peptide (Lanes 4 and 5, respectively). D, Sequence alignment of AQP0CBD and 4 other 1-8-14 calmodulin binding motifs (highlighted in blue) from different proteins as indicated in the figure. The AQP0CBD is missing the large hydrophobic residue at position 14. Ser 235 is highlighted in red and is the site of phosphorylation assayed in this study. The sequence of the non-cononical plant glutamate decarboxylase (PGD) CaM binding site is at the bottom.
The C-terminus of AQP0 is heavily post-translationally modified in the lens as fiber cells mature (Ball et al., 2004). Some of the well-characterized modifications include a conserved phosphorylation at Ser 235 (Figure 2D, red highlight). The biological significance of this phosphorylation is unknown, and it does not seem to impede the water conductance of AQP0 (Ball et al., 2003). However, the localization of this phophorylation site within the AQP0CBD suggests that it could play a role in regulating AQP0’s interaction with CaM, a mechanism used by other proteins to regulate their interaction with CaM (Enyedi et al., 1997; Turner et al., 2004). We synthesized an AQP0CBD peptide containing a phosphoserine at residue 235 (S235-P) and assayed for CaM binding by native gel electropheresis. The S235-P peptide did not cause a band shift, even at 5 times molar excess of the peptide (Figure 2C, Lane 3). This result clearly demonstrates that phosphorylation at this conserved serine position in the AQP0CBD greatly inhibits the CaM interaction.
Close inspection of the AQP0CBD amino acid sequence reveals a significant distinction from classical CaM binding domains. CaM can bind a large number of different substrates, most sharing certain consensus sequences and/or structural motifs (Rhoads and Friedberg, 1997). A comparison of the primary sequence of AQP0CBD with different CaM ligands shows that AQP0 most closely resembles a common CaM binding domain type termed the “1-8-14 motif” (Rhoads and Friedberg, 1997) (Figure 2D). These domains are usually ~20 amino acids long, and form a positively charged ampiphillic helix containing three large hydrophobic residues that typically fit snuggly into the CaM hydrophobic binding pockets. Usually the C- and N-terminal lobes of CaM bind the residues at positions 1 and 14, respectively (Yamniuk and Vogel, 2004) (Figure 2D, blue highlights). Although the AQP0CBD most closely mimics this 1-8-14 motif, it lacks the third hydrophobic residue at position 14.
Non-canonical binding of CaM to the AQP0 C-terminal domain
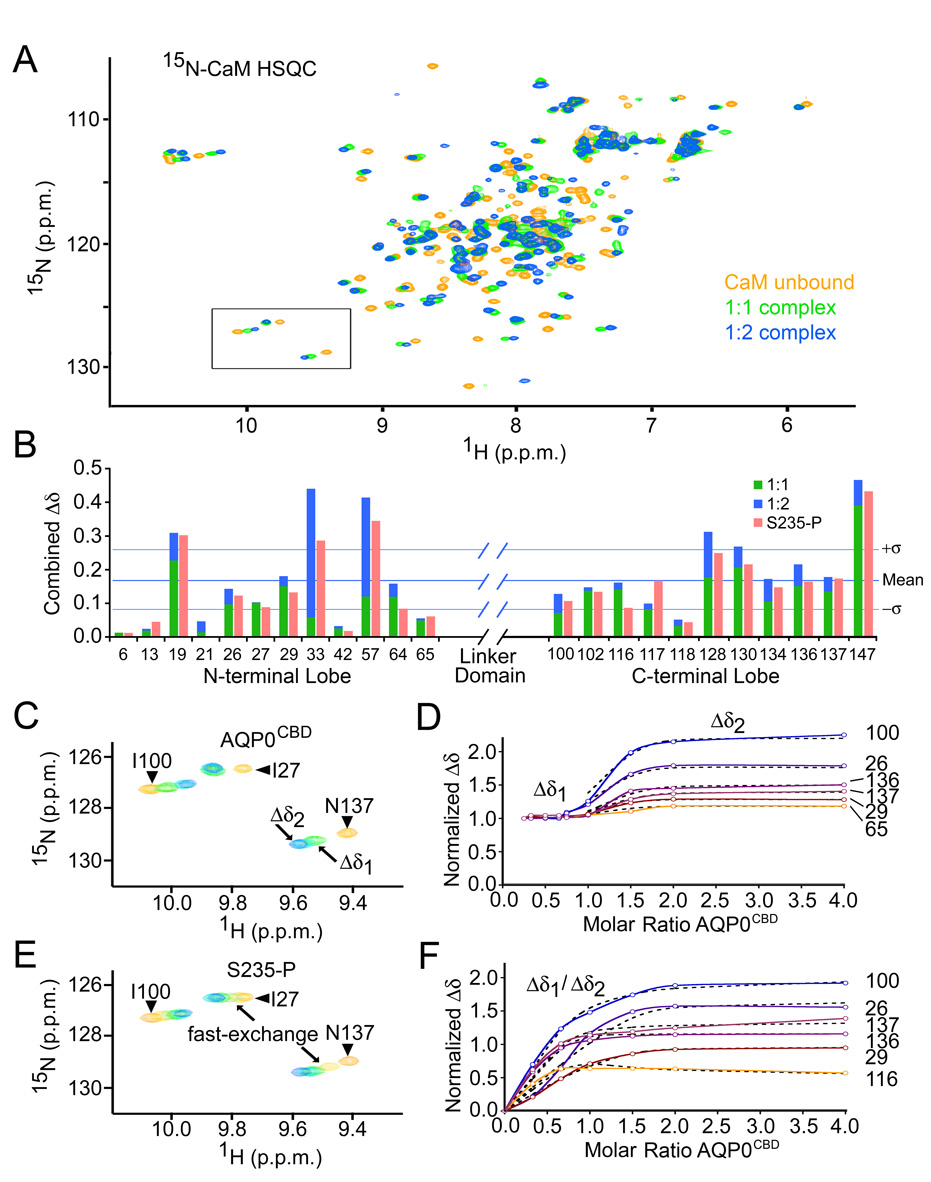
We employed heteronuclear NMR spectroscopy with uniformly 15N-labelled CaM (15N-CaM) to perform titration experiments using the AQP0CBD and an S235-P modified peptide to further characterize the CaM/AQP0CBD interaction and the effect of serine phosphorylation on CaM binding, respectively. As negative control we synthesized and tested a peptide corresponding to a cytosolic loop of AQP0 (AQP0loop residues Thr 148 – Gly 158). The NMR chemical shift offers a highly sensitive and residue specific probe for monitoring protein-protein interactions. NMR titration experiments were conducted at 25°C and in the presence of 5mM Ca2+. A reference heteronuclear single quantum coherence (HSQC) spectrum was recorded for the 15N-CaM (Figure 3A, yellow). Chemical shifts were assigned for several of the well-resolved resonances from both the N- and C-lobes of CaM by matching them to previously published data obtained by others under similar conditions (Ikura et al., 1990) (assigned HSQC presented in Supplementary Figure S1). In this way, we were able to obtain site-specific information about the CaM/AQP0CBD interaction, as well as binding exchange rates by monitoring the chemical shift changes to 15N-CaM HSQC spectra that occurred during the titration experiments.
Figure 3. Calmodulin binds two AQP0CBD peptides in a step-wise manner.
A, Overlay of 15N-calmodulin HSQC spectra of unbound (yellow), singly bound (green) and doubly bound (blue) species in the presence of Ca2+. B, Combined 1H/15N chemical shift changes (Δδ) for several amino acids from the N- and C-lobes of calmodulin. Green and blue bars correspond to Δδ values from the 1:1 and 1:2 complexes, respectively. Red bars correspond to the overall Δδ values from the S235-P titration. Horizontal lines indicate the mean and standard deviation of the overall Δδ values from the AQP0CBD titration. C, Close-up view of the boxed area from panel A, for AQP0CBD. Residue assignments are identified in the figure. Arrows indicate different points in the titration experiment, corresponding to singly bound (Δδ1) and doubly bound (Δδ2) 15N-calmodulin species. D, Titration curves of normalized Δδ values for several 15N-calmodulin residues versus mole ratio of AQP0CBD peptide (colored and labeled). The flat portion of the trace “Δδ1” indicates slow-exchange. Theoretical fits to the fast exchange Δδ2 used to obtain binding constants are overlaid in black hatch lines. E and F, are the same as C and D, but were obtained for the S235-P modified AQP0CBD. In E, arrows indicate S235-P titration points that resulted in fast-exchange. In F, the titration data for both binding events were fit to a two-state binding isotherm.
Titrating 15N-CaM up to equimolar amounts of the unmodified AQP0CBD peptide resulted in the appearance of a new set of resonances in the HSQC spectra, corresponding to a singly bound species of 15N-CaM (Figure 3A, green). The relative intensities for the bound and unbound 15N-CaM resonances reflected the expected molar ratios for a 1:1 binding stoichiometry. These characteristics are classical for molecules interacting with rate of exchange (Kex) that is slow on the NMR time-scale (Kex ≪ 1ms−1), and indicative of specific high-affinity complex formation (Klevit et al., 1985). Significant combined 1H/15N chemical shift changes “Δδ” were observed for nearly every resonance in the 15N-CaM HSQC spectra, consistent with the large conformational changes that occur in CaM upon substrate binding. Bound resonances could be mapped for several residues from the N- and C-lobe of CaM (Figure 3B, green bars). The most significant chemical shift changes mapped to residues located within the hydrophobic binding domains of CaM (for example, Phe 19 and Ala 147), while residues in regions of secondary structure distant from the CaM binding site showed the smallest perturbations (for example, Glu 6 and Lys 13). The drastic chemical shift changes in 15N-CaM HSQC involving residues from both the N- and C-terminal lobes, and slow-exchange binding behavior for the CaM/AQP0CBD, is similar to what has been reported for several high-affinity CaM-peptide interactions (Klevit et al., 1985). We observe no interaction between our control peptide AQP0loop and CaM (data not shown).
In addition to the classical binding behavior exhibited by the AQP0CBD peptide, a non-classical secondary binding event was observed when the titration was continued beyond a 1:1 molar ratio of CaM:AQP0CBD peptide (Figure 3A, blue). The bound 15N-CaM resonances in the HSQC exhibited secondary perturbations that became fully saturated near the 1:2 stoichiometric point in the titration, indicating that CaM is able to bind a second AQP0CBD peptide. These secondary chemical shift perturbations involved many of the same residues from both the N- and C-lobe of CaM that were perturbed during the first titration step (Figure 3B, blue bars). As with the primary binding event, the most pronounced perturbations belonged to residues within and near both CaM binding pockets (for example, Gly 33, Ala 57 and Ala 128). The secondary perturbations followed a unique chemical shift trajectory and displayed intermediate to fast-exchange binding characteristics (Figure 3C). Under conditions of fast-exchange in NMR, Kex ≫ 1/Δδ, a single resonance is observed during the titration, and is located at a chemical shift corresponding to the weighted average of the free and bound CaM species. These characteristics allowed fitting of the data to a binding isotherm for several resonances perturbed by this secondary binding event (Figure 3D) using the program XCRVFIT v4.11 (Sykes Laboratory, University of Alberta, Canada). Analysis of the data yielded apparent dissociation constants for this secondary binding event (Kd2) of about 6.5 ± 5.5 × 10−6 M for resonances from both N- and C-lobes of CaM. The large variance reflects the limited number of data points used for this calculation. However, this low micro-molar affinity is consistent with the appearance of intermediate exchange broadening for resonances with very large Δδ2 values (for example, Gly 33 and Ala 57). The appreciable affinity for forming the 1:2 complex, the large secondary perturbations involving residues in the CaM hydrophobic binding pocket, and the clear saturation end-point in the titration data, all indicate CaM forms a specific ternary complex in which CaM is bound to two AQP0CBD peptides. Significant secondary perturbations in chemical shift (Δδ2 > 0.5) observed for several residues from both lobes of CaM suggest a concerted structural rearrangement occurring over the entire CaM molecule to accommodate the second AQP0CBD peptide.
We next used NMR perturbation analysis to monitor the effects of AQP0CBD phosphorylation on CaM binding. Titrating 15N-CaM with the S235-P modified AQP0CBD peptide also resulted in significant chemical shift perturbations to the same resonances in the 15N-CaM HSQC affected by the unmodified AQP0CBD (Figure 3B, red bars). However, in sharp contrast, the NMR titration data for the S235-P peptide was exclusively characterized with intermediate to fast-exchange binding (Figure 3E), signifying a greatly increased exchange rate for this complex compared to the unmodified CaM/AQP0CBD complex. The S235-P titration data suggest that CaM retains the ability to bind two equivalents of the S235-P peptide, although the interaction is greatly inhibited. This is demonstrated by the observation of several resonances from both N- and C-lobes of CaM that follow two unique chemical shift trajectories during the titration (for example, Asn 117 in Figure 3E). Nearly every perturbed 15N-CaM resonance was shifted beyond the Δδ1 values obtained for the AQP0CBD 1:1 complex (Figure 3B, green versus red bars). Several resonances exhibiting fast-exchange binding behavior during the S235-P peptide titration were fit to a two-state binding isotherm using least-squared minimization (Figure 3F). This analysis yielded similar dissociation constants for the primary binding event for both N- and C-terminal residues, Kd1 = 3.5 ± 2.2 × 10−6M. The dissociation constants for the secondary binding event for the S235-P peptide converged to different values for the N- and C-lobe residues, Kd2 = 33 ± 9 × 10−6 M and 175 ± 30 × 10−6 M, respectively, suggesting this post-translational modification may have a larger effect on the C-lobe of CaM during the secondary binding event. Although the slow-exchange binding characteristics of the unmodified AQP0CBD prohibited similar Kd calculations, this behavior is typically associated with Kd values less than ~1×10−7 M (reviewed in (Pellecchia, 2005)). This means 35 fold reduction (or even greater) in affinity for the S235-P versus the AQP0CBD peptide for 1:1 complex; and a 5-25 fold reduction in affinity for forming the 1:2 complex.
CaM binding to native AQP0 tetramers supports a non-canonical interaction
Aquaporins are tetramers in cell membranes in which each monomer forms a water pore (Figure 2A). An AQP0 tetramer could potentially bind up to four molecules of CaM under a classical 1:1 stoichiometry. However, our NMR titration studies using AQP0CBD indicate that a single CaM is capable of binding two such peptides simultaneously. If this is indeed the case also for the full-length AQP0 then only two CaM molecules would saturate a single AQP0 tetramer. We therefore conducted a second set of NMR titration experiments using natively purified AQP0 to delineate the binding stoichiometry of CaM to AQP0 tetramers. The phosphorylation of S235 in vivo occurs in an age-dependent manner (Ball et al., 2004) so extracting and purifying AQP0 from young tissue guarantees that no significant phosphorylation occurred. Lenses from young lambs were therefore used for the following binding experiments.
Stripped membranes were prepared as described previously (Gonen et al., 2000) and solublized with 1% decyl-maltopyranoside (DM) (Anatrace). AQP0 was purified to homogeneity using protocols we published previously (Gonen et al., 2004b; Gonen et al., 2004a). DM is a mild, non-ionic detergent known to preserve the tetrameric quaternary structure of AQP0 (Gonen et al., 2004a). Typically AQP0 purifies on a size exclusion S-200 chromatography column (Pharmacia Biotech) as a ~220 kDa species, accounting for ~120 kDa for the AQP0 tetramer, plus ~100 kDa in detergent micelle (Gonen et al., 2004b). The large size of this protein therefore renders it “invisible” to standard 15N-NMR studies. Similarly, therefore, the NMR signal for any 15N-CaM bound to the AQP0 tetramer will be severely broadened and is well below the detection limit of a standard HSQC experiment. This allowed us to monitor the loss of free 15N-CaM HSQC signal during the AQP0 titration experiment, and extract the binding stoichiometry for the CaM association with AQP0 tetramers in solution. The 1H dimension of the 15N-CaM HSQC titration series with AQP0 tetramers is presented in figure 4A. In the presence of 5mM Ca2+, the intensity of the 15N-CaM NMR signal progressively vanished in a linear fashion, until it was completely abolished at a 1:2 stoichiometric ratio of CaM:AQP0 (Figure 4B). No such association was observed in the absence of Ca2+ (Figure 4C). This result clearly illustrates that the AQP0 tetramer is capable of binding only two copies of CaM in a Ca2+ dependent fashion and demonstrates that the non-canonical complex detected using the AQP0CBD peptide is likely relevant to the native AQP0/CaM complex.
Figure 4. Two calmodulin molecules bind to a single AQP0 tetramer.
A, 1H dimension of 15N-calmodulin HSQC titration series using full-length AQP0 tetramers in the presence of Ca2+. The molar stoichiometric ratio of CaM:AQP0 is indicated. B, Plot of signal intensity for several resonances from 15N-calmodulin during the titration. Standard deviations are indicated. The NMR signal for 15N-calmodulin is almost completely attenuated at a 1:2 molar ratio, indicating that two calmodulin molecules bind to a single AQP0 tetramer. C, As in panel A, but in the presence of EGTA. No AQP0/CaM interaction is detected in the absence of Ca2+.
Modeling the AQP0/CaM Complex
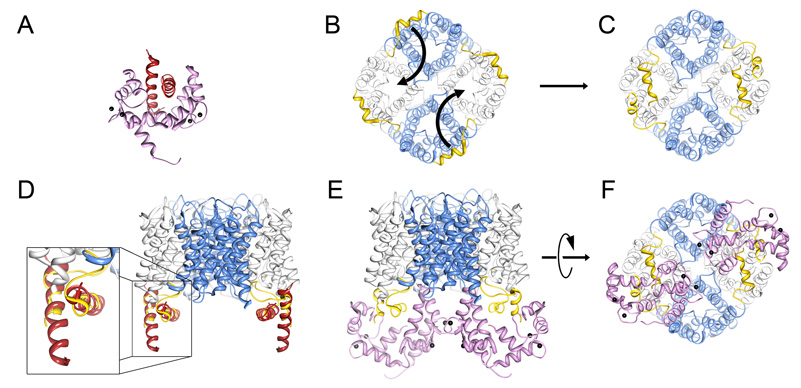
We built an architectural model of AQP0/CaM complex based on our biochemical and NMR data, as well as knowledge of known structures available at the protein data bank (http://www.rcsb.org/pdb/home/home.do). This model provides insights towards a possible mechanism of AQP0 channel regulation by Ca2+/CaM (Figure 5). Our results show that two AQP0CBD bind simultaneously to a single CaM. The reported petunia glutamate decarboxylase (PGD) structure is the only previously known example where Ca2+/CaM is bound to two α-helical target peptides (Yap et al., 2003) (Figure 5A). Notably, the PGD CaM binding domain also deviates from the 1-8-14 CaM motif, in that it lacks the first hydrophobic position (see sequence alignment in Figure 2D). In the reported structure, the two PGD α-helical peptides pack against each other in an anti-parallel fashion. The central domain of CaM wraps around the vertex of the interacting peptides placing its N- and C-lobes on either side.
Figure 5. Model of the AQP0/calmodulin complex.
A, Structure of the petunia glutamate decarboxylase (PGD) calmodulin complex (Protein Data Bank Accession number 1NWD). Calmodulin wraps itself around two PGD α-helices oriented in an anti-parallel fashion. Ca2+ indicated by black speheres. B, AQP0 tetramer viewed from the cytoplasmic side of the cell membrane (bottom view). The carboxyl terminal tails are highlighted in yellow. Curved arrows indicate the proposed movement of the carboxyl tails. C, Rotation of the AQP0 carboxyl terminal tails places neighboring AQP0CBD in close proximity in an orientation similar to the PGD calmodulin structure (Yap et al., 2003). D, Overlay of the PGD α-helices with AQP0CBD. E and F, Side and bottom views of the modeled AQP0/CaM complex, respectively. In this model, calmodulin obstructs only two of the water pores out of the AQP0 tetramer (grey versus blue). Structure fitting and modeling was performed in UCSF chimera (Pettersen et al., 2004).
In the crystallographic structure of full-length AQP0, the cytoplasmic C-terminal helices are arranged in an anti-clockwise orientation along the outside edge of the AQP0 tetramer (Gonen et al., 2004a; Harries et al., 2004) (Figure 5B, yellow). In order to mimic the CaM binding of PGD (Yap et al., 2003) and translate it to our NMR studies, we changed the phi-psi angles between residues 223–225 located at the start of the AQP0 C-terminus. More specifically, we rotated the C-termini to bring the CaM binding domains from two neighboring AQP0 monomers in close proximity to each other (Figure 5B, motion indicated by arrows). The high crystallographic temperature factors observed for the AQP0 C-terminus (Gonen et al., 2005) and the rapid digestion of this domain by proteases (Figure 1B), suggest that it is conformationally dynamic and likely capable of such structural rearrangement. The snug fit between the PGD α-helices and the rotated AQP0CBD allowed us to place the CaM molecule in the same orientation as presented in the PGD structure (Figure 5D, zoom overlay). Visual inspection of the AQP0/CaM model from the cytoplasmic side shows no gross steric overlap between the two CaM molecules bound to the AQP0 tetramer (Figure 5E). In this orientation, the two CaM molecules are positioned directly over the water pores of two AQP0 monomers within the tetramer, while the other two water pores remain are unobstructed (Figure 5F).
DISCUSSION
In this study we employed a combination of biochemical methods and heteronuclear NMR spectroscopy to probe the interaction of CaM with AQP0. Our studies show that CaM binds to AQP0 in lens fiber cell membranes in a Ca2+ dependent manner. This interaction involves the carboxyl tail of AQP0, as removal of this domain by proteolysis abolishes the interaction. Moreover, we show that CaM binds in vitro to synthetic peptides corresponding to the AQP0CBD; and to detergent-extracted full-length AQP0 tetramers purified from lens fiber cells.
Our NMR analysis revealed a stepwise binding mechanism between AQP0 and CaM, where CaM binds two AQP0CBD peptides sequentially. Only the high affinity 1:1 CaM/AQP0CBD peptide complex is detectable using native gel electrophoresis, but both the 1:1 and 1:2 complexes are detectable by NMR. NMR is ideally suited for detecting weak interactions such as the 1:2 CaM/AQP0CBD peptide complex seen in this study due to the high protein concentrations used (~1mM). We observe the same 1:2 binding stoichiometry when CaM is incubated with full-length AQP0 tetramers. CaM is “invisible” in our NMR experiment when bound to AQP0 tetramers because of the large size of the AQP0/CaM complex. It is therefore uncertain if CaM follows a similar step-wise binding mechanism to full-length AQP0 as for the AQP0CBD peptides. However, the probability that two AQP0 C-termini would by chance arrange themselves in an orientation that allows CaM to bind is low. Therefore a step-wise mechanism could eliminate this stochastic issue if CaM first binds to one AQP0 C-terminus, and could then more easily facilitate the second binding event.
The regulation of AQP0 water permeability by Ca2+/CaM has been demonstrated by several laboratories (Nemeth-Cahalan and Hall, 2000; Nemeth-Cahalan et al., 2004; Peracchia and Girsch, 1985; Varadaraj et al., 2005). Our structural model of the AQP0/CaM complex suggests that in the presence of Ca2+, CaM binds the C-terminus from two neighboring AQP0 monomers, locking these domains in a orientation that seemingly blocks the pores of two AQP0 monomers within the tetramer (Figure 5E and F). A similar channel capping mechanism has been proposed for the plant aquaporin SoPiP2;1, where a cadmium cation bound to the N-terminus and cytoplasmic loop of the protein induced pore closure (Gonen and Walz, 2006; Hedfalk et al., 2006; Tornroth-Horsefield et al., 2005). In AQP0, therefore, CaM may act as an inhibitor of channel permeability, and predictably should result in a 2-fold reduction in the overall observed permeability for AQP0. Indeed a 2-fold reduction in channel activity was observed in AQP0 permeability studies conducted in Xenopus oocytes in response to increased Ca2+ levels (Nemeth-Cahalan et al., 2004). This effect was CaM dependent and is in agreement with our proposed structural model; however, high-resolution structural analysis of the AQP0/CaM complex is required to unambiguously decipher this gating mechanism. The structural model presented here provides an architectural framework to guide further biochemical and functional studies aimed at elucidating the water channel gating mechanism of AQP0.
What is the possible role of AQP0 serine phosphorylation in the eye lens? Phosphorylation is recognized as an important mechanism used to regulate both the trafficking and water permeability of some AQPs (reviewed in (Gonen and Walz, 2006)). Serine residues on AQP0’s carboxyl terminus, including Ser 235, are phosphorylated in the lens in an age dependent manner (Ball et al., 2004). Ser 235 is located within a consensus site for protein kinase A, suggesting that the AQP0/CaM complex formation could be dynamically regulated by kinase activity in vivo. Our NMR data show that phosphorylation of AQP0 Ser 235 inhibits the AQP0/CaM interaction. A recent study reported a similar inhibitory affect on CaM binding affinity to AQP0CBD Ser 235 and/or Ser 231 phospho-peptides (Rose et al., 2008). Thus it seems that these serine modifications may be used to temporally modulate the Ca2+ regulatory mechanism of AQP0 within the eye lens. Other post-translational modifications of AQP0 include an age dependent partial proteolysis of its C-terminus, which triggers AQP0 junction formation (Gonen et al., 2004b). Our data shows that CaM can bind to unmodified full-length AQP0. This binding event could shield the AQP0 carboxyl terminus from digestion by proteases thereby regulating junction formation. CaM cannot bind to phosphorylated AQP0 and therefore will not be able to protect the protein from partial proteolysis. It is possible that in this way the lens is able to dynamically regulate junction formation as well as channel permeability. We do note, however, that 1. the protease responsible for cleaving the AQP0 C-tail is unknown; and 2. the kinase responsible for AQP0 phosphorylation is also unkown. Although definitive biochemical studies are needed, it appears that serine phosphorylation within the AQP0CBD is emerging as a conserved and dynamic mechanism for providing control over the function of AQP0.
EXPERIMENTAL PROCEDURES
Lens membrane preparation, AQP0 purification and CaM expression
Lamb lenses were obtained from the Wolverine Packers slaughterhouse (Detroit, MI). Fiber cell membranes were prepared as described previously (Gonen et al., 2000) and stored at a total membrane protein concentration of 2mg/ml at −20°C as determined by BCA (Pierce). AQP0 was solubilized with 1% decyl maltoside (DM) (Anatrace) and purified with a combination of ion exchange and Superdex 200 size exclusion chromatography as described before (Gonen et al., 2004b). Purified AQP0 was exchanged to NMR buffer containing 10mM BisTris pH 6.5, 50mM NaCl, 0.3% DM during the gel-filtration, and concentrated using a centrifugal filter device to ~25mg/ml (Vivaspin). pET expression vectors containing the full-length sequence for vertabrate calmodulin (CaM) and an N-terminal 6xHis tagged version of CaM were kindly donated to us by Rachel Klevit (University of Washington). CaM was expressed and purified similarly according to well-established protocols (Li et al., 2001). For NMR studies, 15N isotopically labeled CaM (15NCaM) was prepared by growing cells in minimal M9 media, supplemented with 15N-ammonium sulfate (Spectra Stable Isotopes).
AQP0 peptide constructs
Peptides used in this study were obtained at >95% purity from Anaspec (San Jose, CA). Lyophilized peptides were dissolved in milliQ water at 8–10mg/ml as determined by the BCA method (Pierce) and stored at −80°C until needed. Both unmodified and phosphorylated peptides were chosen to correspond to a putative calmodulin binding domain from the C-terminus of AQP0 residues 223–242 (AQP0CBD 223LFPRLKSVSERLSILKGSRP242). The phosphorylated peptide contained a phosphoserine at residue 235 (S235-P). An additional peptide corresponding to a cytosolic AQP0 loop, residues 148–158 (AQP0loop) was used as a negative control for calmodulin binding studies.
CaM pull-down assay and native gel electrophoresis
Direct binding of CaM to lens cortex membranes was assayed by preparing a 100µl binding mixture containing purified lens membranes (2mg/ml) and CaM (0.67mg/ml), in 10mM Tris pH 8.0 and 10mM CaCl2. To assay calcium dependence of the CaM interaction, CaCl2 was replaced with either 10mM EGTA or 50mM MgCl2. The effect of combining Mg2+ and Ca2+ was also assayed by supplementing 50mM MgCl2 to the original binding mixture. The ability of CaM to bind lens membranes treated with protease was also assayed. For this experiment, lens membranes were treated with protease cocktail (α-chymotrypsin and thrombin at 1:250 w/w) at 37°C for 1 hour and the reaction stopped with PMSF. The preparations was then washed several times with 10mM Tris pH 8.0 and 10mM CaCl2 to remove the proteases prior to the addition of CaM. CaM binding was then assayed as described above at 37°C for 30 minutes. The binding reaction was stopped on ice, free CaM was removed by centrifugation at 10,000xg and membrane pellets washed 4 times in this way. Samples were re-suspended in Laemmli buffer and the CaM content at each step was assessed by SDS-PAGE followed by silver staining. For native gel electrophoresis, CaM binding to AQP0 peptides was assessed by monitoring retardation of CaM mobility on a 12% nondenaturing PAGE. Binding mixtures were prepared with 5mg/ml CaM and a 1.5 molar excess of the AQP0CBD or S235-P peptide in the presence of 50mM Tris pH-8.0 and 5mM CaCl2. The binding mixtures were incubated at 37°C for 30 minutes and loaded to the native gel in 20% glycerol. The calcium dependence of binding was assayed by replacing CaCl2 with 1mM EGTA in both the binding mixture and loading buffer. Protein bands were visualized by silver stain. The identities of AQP0 and CaM were confirmed by western blot analysis using a monoclonal antibody directed against the carboxyl terminus of AQP0 (Alpha Diagnostic International, San Antonio, Texas, USA) or to a 6xHis tag on CaM (His-Probe, Pierce). Antibody detection was performed using chemiluminescence (ECL kit, Amersham Life Science) following manufacturer’s protocol.
NMR titration experiments
NMR titrations with uniformly labeled 15N-CaM were all conducted at 25°C. For the titration of 15N-CaM with AQP0 peptides (AQP0CBD, S235-P or the control AQP0loop), 0.33mM 15N-CaM was prepared in 10mM BisTris pH 6.5, 10mM CaCl2 and 8% (v/v) D20. 15N-CaM was titrated with the desired molar equivalent of AQP0 peptide from highly concentrated stocks dissolved in water. The concentration of 15N-CaM was corrected for dilutions >5% of the sample volume. 15N-HSQC assignments for free calmodulin were obtained for well-resolved resonances by matching them to previously published data (Biomagnetic Research Data Bank accession number 547 (Ikura et al., 1990)). Bound calmodulin resonances were obtained by mapping results from the fast-exchange titration data. For NMR titrations using full-length purified lens AQP0 tetramers, individual samples containing increasing amount of AQP0 were prepared with 0.50mM 15N-CaM in 10mM BisTris pH 6.5, 5mM CaCl2, 50mM NaCl, 0.3% DM and 0.8% (v/v) D20. The Ca2+ dependence for the AQP0/15N-CaM interaction was tested by replacing CaCl2 with 1mM EGTA. For the AQP0CBD peptide titrations, 15N-HSQC spectra (spectral widths 14 p.p.m. (1H)×28 p.p.m (15N)) were collected on a Bruker 600MHz spectrometer equipped with a cryo-probe. For the full-length AQP0 tetramer titrations, 15N HSQC spectra were collected on a Bruker 750MHz spectrometer. The NMR data were processed similarly using NMRPipe (Delaglio et al., 1995) and analyzed with Sparky (T. D. Goddard and D. G. Kneller, University of California, San Francisco). Combined 1H and 15N chemical shift changes (Δδ=√((1Hbound−1Hfree)2 + (15Nbound−15Nfree/6.51)2) were calculated for the NMR titration data and plotted using Excel and dissociation constants were obtained by least-squared fitting in XCRVFIT v4.11 (Sykes Laboratory, University of Alberta, Canada). The program UCSF Chimera (Pettersen et al., 2004) was used for visualizing protein structures and building the AQP0/CaM model.
Supplementary Material
A reference heteronuclear single quantum coherence (HSQC) spectrum was recorded for the 15N-CaM and chemical shifts were assigned for well-resolved resonances by matching them to previously published results obtained under similar conditions (Biomagnetic Research Data Bank accession number 547 (Ikura et al., 1990)).
Acknowledgements
The authors would like to thank Rachel Klevit (University of Washington) for helpful discussions and providing us with the calmodulin clone. We would also like to thank Gabriele Varani (University of Washington) for helpful discussions; for critically reading this manuscript and for giving us access to the 600MHz and 750MHz NMR. This work was supported by NIH grant R01 GM079233. The authors declare that none have financial interests related to this work.
The abbreviations used are
- AQP0
aquaporin-0
- tAQP0
C-terminally truncated AQP0
- CaM
calmodulin
- AQP0CBD
aquaporin-0 calmodulin binding domain
- NMR
nuclear magnetic resonance
- DM
decyl maltopyranoside
- PC
phosphatidylcholine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agre P, Kozono D. Aquaporin water channels: molecular mechanisms for human diseases. FEBS Lett. 2003;555:72–78. doi: 10.1016/s0014-5793(03)01083-4. [DOI] [PubMed] [Google Scholar]
- Agre P, Sasaki S, Chrispeels MJ. Aquaporins: a family of water channel proteins. Am J Physiol. 1993;265:F461. doi: 10.1152/ajprenal.1993.265.3.F461. [DOI] [PubMed] [Google Scholar]
- Alcala J, Lieska N, Maisel H. Protein composition of bovine lens cortical fiber cell membranes. Exp Eye Res. 1975;21:581–595. doi: 10.1016/0014-4835(75)90040-8. [DOI] [PubMed] [Google Scholar]
- Arneson ML, Cheng HL, Louis CF. Characterization of the ovine-lens plasma-membrane protein-kinase substrates. Eur J Biochem. 1995;234:670–679. doi: 10.1111/j.1432-1033.1995.670_b.x. [DOI] [PubMed] [Google Scholar]
- Babu YS, Bugg CE, Cook WJ. Structure of calmodulin refined at 2.2 A resolution. J Mol Biol. 1988;204:191–204. doi: 10.1016/0022-2836(88)90608-0. [DOI] [PubMed] [Google Scholar]
- Ball LE, Garland DL, Crouch RK, Schey KL. Post-translational modifications of aquaporin 0 (AQP0) in the normal human lens: spatial and temporal occurrence. Biochemistry. 2004;43:9856–9865. doi: 10.1021/bi0496034. [DOI] [PubMed] [Google Scholar]
- Ball LE, Little M, Nowak MW, Garland DL, Crouch RK, Schey KL. Water permeability of C-terminally truncated aquaporin 0 (AQP0 1–243) observed in the aging human lens. Invest Ophthalmol Vis Sci. 2003;44:4820–4828. doi: 10.1167/iovs.02-1317. [DOI] [PubMed] [Google Scholar]
- Bloemendal H, Zweers A, Vermorken F, Dunia I, Benedetti EL. The plasma membranes of eye lens fibres. Biochemical and structural characterization. Cell Differ. 1972;1:91–106. doi: 10.1016/0045-6039(72)90032-2. [DOI] [PubMed] [Google Scholar]
- Bok D, Dockstader J, Horwitz J. Immunocytochemical localization of the lens main intrinsic polypeptide (MIP26) in communicating junctions. J Cell Biol. 1982;92:213–220. doi: 10.1083/jcb.92.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchman D, Yappert MC, Afzal M. Lens lipids and maximum lifespan. Exp Eye Res. 2004;79:761–768. doi: 10.1016/j.exer.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Costello MJ, McIntosh TJ, Robertson JD. Distribution of gap junctions and square array junctions in the mammalian lens. Invest Ophthalmol Vis Sci. 1989;30:975–989. [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Enyedi A, Elwess NL, Filoteo AG, Verma AK, Paszty K, Penniston JT. Protein kinase C phosphorylates the "a" forms of plasma membrane Ca2+ pump isoforms 2 and 3 and prevents binding of calmodulin. J Biol Chem. 1997;272:27525–27528. doi: 10.1074/jbc.272.44.27525. [DOI] [PubMed] [Google Scholar]
- Francis P, Berry V, Bhattacharya S, Moore A. Congenital progressive polymorphic cataract caused by a mutation in the major intrinsic protein of the lens, MIP (AQP0) Br J Ophthalmol. 2000a;84:1376–1379. doi: 10.1136/bjo.84.12.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Libson A, Miercke LJ, Weitzman C, Nollert P, Krucinski J, Stroud RM. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- Girsch SJ, Peracchia C. Calmodulin interacts with a C-terminus peptide from the lens membrane protein MIP26. Curr Eye Res. 1991;10:839–849. doi: 10.3109/02713689109013880. [DOI] [PubMed] [Google Scholar]
- Gonen T, Cheng Y, Kistler J, Walz T. Aquaporin-0 membrane junctions form upon proteolytic cleavage. J Mol Biol. 2004b;342:1337–1345. doi: 10.1016/j.jmb.2004.07.076. [DOI] [PubMed] [Google Scholar]
- Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison S, Walz T. Lipid-protein interactions in double-layered two-dimensional crystals of aquaporin-0. Nature. 2005 doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen T, Donaldson P, Kistler J. Galectin-3 is associated with the plasma membrane of lens fiber cells. Invest Ophthalmol Vis Sci. 2000;41:199–203. [PMC free article] [PubMed] [Google Scholar]
- Gonen T, Sliz P, Kistler J, Cheng Y, Walz T. Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature. 2004a;429:193–197. doi: 10.1038/nature02503. [DOI] [PubMed] [Google Scholar]
- Gonen T, Walz T. The structure of aquaporins. Q Rev Biophys. 2006;39:361–396. doi: 10.1017/S0033583506004458. [DOI] [PubMed] [Google Scholar]
- Gu F, Zhai H, Li D, Zhao L, Li C, Huang S, Ma X. A novel mutation in major intrinsic protein of the lens gene (MIP) underlies autosomal dominant cataract in a Chinese family. Mol Vis. 2007;13:1651–1656. [PubMed] [Google Scholar]
- Harries WE, Akhavan D, Miercke LJ, Khademi S, Stroud RM. The channel architecture of aquaporin 0 at a 2.2-A resolution. Proc Natl Acad Sci U S A. 2004;101:14045–14050. doi: 10.1073/pnas.0405274101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedfalk K, Tornroth-Horsefield S, Nyblom M, Johanson U, Kjellbom P, Neutze R. Aquaporin gating. Curr Opin Struct Biol. 2006;16:447–456. doi: 10.1016/j.sbi.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Hiroaki Y, Tani K, Kamegawa A, Gyobu N, Nishikawa K, Suzuki H, Walz T, Sasaki S, Mitsuoka K, Kimura K, et al. Implications of the Aquaporin-4 Structure on Array Formation and Cell Adhesion. J Mol Biol. 2005;355:628–639. doi: 10.1016/j.jmb.2005.10.081. [DOI] [PubMed] [Google Scholar]
- Ikura M, Kay LE, Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990;29:4659–4667. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- Johnson JD, Wittenauer LA. A fluorescent calmodulin that reports the binding of hydrophobic inhibitory ligands. Biochem J. 1983;211:473–479. doi: 10.1042/bj2110473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevit RE, Blumenthal DK, Wemmer DE, Krebs EG. Interaction of calmodulin and a calmodulin-binding peptide from myosin light chain kinase: major spectral changes in both occur as the result of complex formation. Biochemistry. 1985;24:8152–8157. doi: 10.1021/bi00348a047. [DOI] [PubMed] [Google Scholar]
- Lee JK, Kozono D, Remis J, Kitagawa Y, Agre P, Stroud RM. Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A. Proc Natl Acad Sci U S A. 2005;102:18932–18937. doi: 10.1073/pnas.0509469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Wu JG, Si JL, Guo DW, Xu JP. High-level expression of human calmodulin in E. coli and its effects on cell proliferation. Shi Yan Sheng Wu Xue Bao. 2001;34:131–135. [PubMed] [Google Scholar]
- Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- Nemeth-Cahalan KL, Hall JE. pH and calcium regulate the water permeability of aquaporin 0. J Biol Chem. 2000;275:6777–6782. doi: 10.1074/jbc.275.10.6777. [DOI] [PubMed] [Google Scholar]
- Nemeth-Cahalan KL, Kalman K, Hall JE. Molecular basis of pH and Ca2+ regulation of aquaporin water permeability. J Gen Physiol. 2004;123:573–580. doi: 10.1085/jgp.200308990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S, Kwon TH, Frokiaer J, Agre P. Regulation and dysregulation of aquaporins in water balance disorders. J Intern Med. 2007;261:53–64. doi: 10.1111/j.1365-2796.2006.01760.x. [DOI] [PubMed] [Google Scholar]
- Pellecchia M. Solution nuclear magnetic resonance spectroscopy techniques for probing intermolecular interactions. Chem Biol. 2005;12:961–971. doi: 10.1016/j.chembiol.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Peracchia C, Girsch SJ. Permeability and gating of lens gap junction channels incorporated into liposomes. Curr Eye Res. 1985;4:431–439. doi: 10.3109/02713688509025157. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pitt GS. Calmodulin and CaMKII as molecular switches for cardiac ion channels. Cardiovasc Res. 2007;73:641–647. doi: 10.1016/j.cardiores.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Ren G, Reddy VS, Cheng A, Melnyk P, Mitra AK. Visualization of a water-selective pore by electron crystallography in vitreous ice. Proc Natl Acad Sci U S A. 2001;98:1398–1403. doi: 10.1073/pnas.041489198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. Faseb J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- Rose KM, Wang Z, Magrath GN, Hazard ES, Hildebrandt JD, Schey KL. Aquaporin 0-calmodulin interaction and the effect of aquaporin 0 phosphorylation. Biochemistry. 2008;47:339–347. doi: 10.1021/bi701980t. [DOI] [PubMed] [Google Scholar]
- Savage DF, Egea PF, Robles-Colmenares Y, O'Connell JD, 3rd, Stroud RM. Architecture and selectivity in aquaporins: 2.5 a X-ray structure of aquaporin Z. PLoS Biol. 2003;1:E72. doi: 10.1371/journal.pbio.0000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LB, Skach WR, Verkman AS. Functional independence of monomeric CHIP28 water channels revealed by expression of wild-type mutant heterodimers. J Biol Chem. 1994;269:10417–10422. [PubMed] [Google Scholar]
- Sui H, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- Swamy-Mruthinti S. Glycation decreases calmodulin binding to lens transmembrane protein, MIP. Biochim Biophys Acta. 2001;1536:64–72. doi: 10.1016/s0925-4439(01)00031-x. [DOI] [PubMed] [Google Scholar]
- Tajkhorshid E, Nollert P, Jensen MO, Miercke LJ, O'Connell J, Stroud RM, Schulten K. Control of the selectivity of the aquaporin water channel family by global orientational tuning. Science. 2002;296:525–530. doi: 10.1126/science.1067778. [DOI] [PubMed] [Google Scholar]
- Tornroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, Neutze R, Kjellbom P. Structural mechanism of plant aquaporin gating. Nature. 2005 doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- Turner JH, Gelasco AK, Raymond JR. Calmodulin interacts with the third intracellular loop of the serotonin 5-hydroxytryptamine1A receptor at two distinct sites: putative role in receptor phosphorylation by protein kinase C. J Biol Chem. 2004;279:17027–17037. doi: 10.1074/jbc.M313919200. [DOI] [PubMed] [Google Scholar]
- Varadaraj K, Kumari S, Shiels A, Mathias RT. Regulation of aquaporin water permeability in the lens. Invest Ophthalmol Vis Sci. 2005;46:1393–1402. doi: 10.1167/iovs.04-1217. [DOI] [PubMed] [Google Scholar]
- Vogel HJ. The Merck Frosst Award Lecture 1994. Calmodulin: a versatile calcium mediator protein. Biochem Cell Biol. 1994;72:357–376. [PubMed] [Google Scholar]
- Yamniuk AP, Vogel HJ. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- Yang B, Verkman AS. Water and glycerol permeabilities of aquaporins 1–5 and MIP determined quantitatively by expression of epitope-tagged constructs in Xenopus oocytes. J Biol Chem. 1997;272:16140–16146. doi: 10.1074/jbc.272.26.16140. [DOI] [PubMed] [Google Scholar]
- Yap KL, Yuan T, Mal TK, Vogel HJ, Ikura M. Structural basis for simultaneous binding of two carboxy-terminal peptides of plant glutamate decarboxylase to calmodulin. J Mol Biol. 2003;328:193–204. doi: 10.1016/s0022-2836(03)00271-7. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yang W, Lurtz MM, Ye Y, Huang Y, Lee HW, Chen Y, Louis CF, Yang JJ. Identification of the calmodulin binding domain of connexin 43. J Biol Chem. 2007;282:35005–35017. doi: 10.1074/jbc.M707728200. [DOI] [PubMed] [Google Scholar]
- Zuhlke RD, Pitt GS, Deisseroth K, Tsien RW, Reuter H. Calmodulin supports both inactivation and facilitation of L-type calcium channels. Nature. 1999;399:159–162. doi: 10.1038/20200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A reference heteronuclear single quantum coherence (HSQC) spectrum was recorded for the 15N-CaM and chemical shifts were assigned for well-resolved resonances by matching them to previously published results obtained under similar conditions (Biomagnetic Research Data Bank accession number 547 (Ikura et al., 1990)).