Abstract
Fragile X syndrome is the most common form of inherited mental retardation caused by loss of the fragile X mental retardation protein 1 (FMRP). The detailed molecular pathways underlying the pathogenesis of this disorder remain incompletely understood. Here, we show that miR-124a, a nervous-system-specific miRNA, is associated with the Drosophila homolog of FMRP (dFMR1) in vivo. Ectopic expression of wild-type but not mutant miR-124a precursors decreased dendritic branching of dendritic arborization sensory neurons, which was partially rescued by the loss of dFMR1 activity, suggesting that the biogenesis and/or function of miR-124a are partially dependent on dFMR1. Indeed, in contrast with the complete loss of mature miR-124a in Dicer-1 mutants, steady-state levels of endogenous or ectopically expressed mature miR-124a were partially reduced in dfmr1 mutants, whereas the level of pre-miR-124a increased. This effect could be explained in part by the reduced abundance of the Dicer-1-Ago1 complex in the absence of dFMR1. These findings suggest a modulatory role for dFMR1 to maintain proper levels of miRNAs during neuronal development.
Keywords: miR-124a, processing, Drosophila, fragile X syndrome, dendrites, RNA
Introduction
The loss of fragile X mental retardation protein 1 (FMRP) activity causes fragile X syndrome, the most common form of inherited mental retardation in humans (Verkerk et al., 1991). FMRP is an evolutionarily conserved RNA-binding protein with two ribonucleoprotein (RNP) K homology domains and an arginine- and glycine-rich domain (RGG box). It has been shown that FMRP preferentially binds to tertiary RNA structures named the “kissing complex” and the “G quartet” (Darnell et al., 2001, 2005). Although hundreds of mRNAs associate preferentially with FMRP-containing complexes (Brown et al., 2001; Miyashiro et al., 2003), the detailed mechanisms to account for FMRP action remain to be illustrated.
Drosophila has been used successfully as a model system to dissect the genetic pathways implicated in fragile X syndrome. The Drosophila fragile X mental retardation protein 1 (dFMR1) is involved in multiple aspects of neuronal development, including synapse formation, axonal growth and dendritic branching (Zhang et al., 2001; Dockendorff et al., 2002; Morales et al., 2002; A. Lee et al., 2003; Michel et al., 2004). dFMR1 is also implicated in the microRNA (miRNA) pathway (Caudy et al., 2002; Ishizuka et al., 2002), although the exact in vivo function of dFMR1 in this pathway is largely unknown.
During animal development, multiple genes must be expressed coordinately at precise levels both spatially and temporally. miRNAs are an important class of regulatory molecules that ensure the accuracy of gene expression. Primary miRNAs (pri-miRNAs) are usually transcribed by RNA polymerase II in the nucleus and processed mostly by the RNase III Drosha to generate 70–80 nt hairpin structures called precursor miRNAs (pre-miRNAs) (Y. Lee et al., 2003). Pre-miRNAs are transported to the cytoplasm by exportin-5 (Yi et al., 2003) and processed by Dicer into ∼22 nt mature miRNAs (Bernstein et al., 2001; Grishok et al., 2001; Hutvágner et al., 2001). These small, noncoding RNAs destabilize target mRNAs or suppress their translation by binding to complementary sequences in the 3′ untranslated regions (3′UTRs) (Filipowicz et al., 2008).
Hundreds of miRNAs have been identified in worms, flies, and humans, and many are evolutionarily conserved at the nucleotide level and regulate various aspects of animal development (Bushati and Cohen, 2007; Hobert, 2008). Some miRNAs are specifically expressed in developing and mature nervous systems (Lagos-Quintana et al., 2002; Aboobaker et al., 2005; Wienholds et al., 2005), and their roles in neuronal development and function have begun to be unraveled in different model systems (Gao, 2008). One of them is miR-124a, an evolutionarily conserved and nervous-system-specific miRNA (Lagos-Quintana et al., 2002). Transcriptional activation of miR-124a expression during neuronal specification seems to require derepression by the RE1 silencing transcription factor (Conaco et al., 2006). Although its function has not been analyzed using genetic null mutations in any model system, miR-124a may be involved in downregulating target gene expression during neuronal specification (Cao et al., 2007; Makeyev et al., 2007; Visvanathan et al., 2007). In this study, we used the Drosophila dendritic arborization (DA) sensory neurons as an assay system and examined the interactions between miR-124a and dFMR1, providing novel mechanistic insights into the exact role of dFMR1 in the miRNA pathway and neuronal development.
Materials and Methods
Fly strains and genetics.
All the flies were raised at 25°C on standard food medium. Gal4221 was used to visualize ddaE and ddaF neurons, and Gal4477 to visualize ddaC neurons and drive ectopic expression of pre-miR-124a in these cells. The UAS-pre-miR-9a fly line was generated in a previous study (Li et al., 2006). The dfmr13 mutant fly line was obtained from Dr. T. Jongens (University of Pennsylvania, Philadelphia, PA) and the dicer-1 mutants were from Dr. R. Carthrew (Northwestern University, Chicago, IL). The UAS-pre-miR-124a fly lines were generated by cloning a 92 bp genomic DNA fragment containing the 77 nt wild-type pre-miR-124a into the UAST vector between EcoRI and XbaI restriction enzyme sites. For UAS-mutant pre-miR-124a, a 6 nt long deletion was introduced into pre-miR-124a. UAS-pre-miR-124a, UAS-mutant pre-miR-124a, and UAS-pre-miR-9a were expressed either in the wild type background or in the dfmr13/dfmr14 mutant background. dcr-1 mutant alleles dcr-1d102 or dcr-1Q1147X were recombined onto the same chromosome as FRT82B and used for MARCM analysis as described previously (Li et al., 2004). The quantitative analysis of dendritic ends and dendritic fields was done also as described previously (Li et al., 2004).
In situ hybridization.
In situ hybridization was performed as described previously (Li et al., 2006). Briefly, Drosophila embryos at different developmental stages were collected and fixed. Drosophila miR-124a locked nucleic acid (LNA) probe was purchased from Exiqon, end-labeled with a DIG oligonucleotide 3′-end labeling kit (Roche), and purified on a G-25 Microspin column (Amersham Biosciences). The labeling with the LNA probe was detected by AP-conjugated anti-Dig antibody (Li et al., 2006).
miR-124a-Gal4 fly line.
Primers 5′-GGGCGGCCGCGGACCTAGCTTTGT GCGTG-3′ and 5′-GGAGATCTCGTGCCTTATGGTGGAAATAC-3′ were used to obtain 3.2 kb PCR fragments corresponding to the miR-124a promoter region, which was subsequently cloned into the pPTGAL vector (Sharma et al., 2002) between the NotI and BglII restriction enzyme sites. The resulting vector was used to generate transgenic flies.
Quantification of dendritic ends.
The number of dendritic ends of different DA neurons was quantified as previously described (Li et al., 2004). Briefly, dendritic ends of DA neuron images were identified visually and highlighted with dots. The number of dots was counted using the Illustrator software.
Immunoprecipitation and Western blot analysis.
The brains and ventral nerve cords from hundreds of third instar larvae were dissected out in PBS on ice and homogenized in cold RNase-free lyses buffer (20 mm Hepes, pH 7.4, 150 mm NaCl, 2 mm MgCl2, 0.5% Nonidet P-40, 1 mm dithiothreitol, and protease inhibitors). In some case, EDTA was used at a final concentration of 10 mm. The protein extracts was prepared by centrifugation for 15 min at 4°C. The supernatant was incubated with anti-hemagglutinin (HA) antibody (Roche) and protein-G-agarose beads at 4°C for 4 h. The precipitated complex was washed with PBS three times. The associated RNA was isolated with Trizol (Invitrogen).
Western blot analysis was performed according to the standard protocol provided by Bio-Rad. Briefly, protein extracts was prepared from three instar larvae as above. Protein (25 μg), as measured by the Bio-Rad reagent, was separated on 8% SDS-PAGE followed by transferring onto PVDF membranes and immunoblotting with appropriate antibodies. The primary antibodies, anti-dFMR1 (5A11) and anti-tubulin (E7) were from the Developmental Studies Hybridoma Bank at the University of Iowa. Anti-Ago1 antibody (1B8) is a gift from Dr. H. Siomi (Keio University, Tokyo, Japan). Anti-Dcr-1 antibody was purchased from the Abcam. The HRP-conjugated anti-mouse IgG antibody was used as secondary antibody (Jackson Laboratory).
Northern blot.
Northern blot was performed as described previously (Li et al., 2006) with some minor modifications. Total RNA (30 μg) extracted from third instar larvae with Trizol were resolved on 12.5% polyacrylamide gels (Sequagel; National Diagnostics) and transferred to Nytran Super Charge Signal membrane (Whatman Schleicher and Schuell) or Zeta-Probe GT membrane (Bio-Rad). Antisense oligonucleotides complementary to mature microRNA sequences were 5′-labeled with 32P (GE Healthcare) and purified with Quick Spin columns (Sigma). The following oligonucleotides were used to detect mature microRNAs: 5′-CTTGGCATTCACCGCGTGCCTTA-3′ for miR-124a, 5′-CTCCATACTTCTTTACATTCCA-3′ for miR-1, and 5′-ACAACAAAATCACTAGTCTTCCA-3′ for miR-7. In some cases, the membrane was stripped by boiling in 0.1% SDS for 3 min and then re-probed with different 32P-labeled oligonucleotides. The intensity of miRNA bands on Northern blot was quantified on a scanning densitometer and averaged from multiple independent experiments.
Quantitative real-time PCR analysis.
Total RNA was extracted from third instar larvae as described above, treated with DNase I and purified with RNeasy mini kit (Qiagen). The purified RNA was used in reverse transcription reaction using Taqman reverse transcription reagent (Applied Biosystems). The first-strand cDNA was used as template for quantitative real-time PCR (qRT-PCR) in a final volume of 25 μl containing primers and SYBR Green PCR master mix (Applied Biosystems). The reaction was done using the ABI7700 sequence detection system. For mature microRNAs, expression levels were measured by qRT-PCR analysis with TaqMan miRNA reverse transcription kit and TaqMan microRNA assays containing specific primers for mature miR-124a (Applied Biosystems). A standard curve was run in each PCR. Individual values were normalized with the value of the gene encoding the ribosomal protein RP-49. For qRT-PCR analysis of the primary and precursor transcripts of miR-124a, the following primers were used: forward primer for pri-miR-124a: 5′-CACTTTCGGTGACCTCA-3′; reverse primer for pri-miR-124a: 5′-CCAATGGCGAGAATATCCTTG-3′; forward primer for pre-miR-124a: 5′-ACGTTTTTCTCCTGGTATCCACTG-3′; reverse primer for pre-miR-124a: 5′-CACCGCGTGCCTTATGG-3′; forward primer for rp-49: 5′-AGATCGTGAAGAAGCGCACCAAG-3′; and reverse primer for rp-49: 5′-CACCAGGAACTTCTTGAATCCGG-3′. All reactions were done three times in triplicate, and relative expression of RNAs was calculated by using the standard curve method and the Delta-Delta Ct method.
Results
miR-124a is associated with dFMR1 in vivo
To examine the role of miR-124a in neuronal development and its biogenesis in Drosophila, we first confirmed its expression pattern during development. miR-124a is specifically expressed in the developing nervous system, mostly in the brain and the ventral nerve cord, as shown by in situ hybridization (Fig. 1A). Other laboratories reported similar findings during the course of our work (Aboobaker et al., 2005; Stark et al., 2005). To further examine this expression pattern at a high resolution, we generated transgenic flies in which Gal4 expression from the pPTGAL vector (Sharma et al., 2002) was directly under the control of a minimal promoter and a 3.2-kb genomic DNA fragment upstream of the pre-miR-124a sequence. When the miR-124a-Gal4 line was crossed with the UAS-GFP line, GFP was specifically expressed in the developing embryonic nervous system (Fig. 1B), and this expression pattern persisted to adulthood (data not shown). In Drosophila embryos and larvae, GFP expression driven by miR-124a promoter is high in the brain, ventral nerve cord and motor neurons but lower in DA neurons in the peripheral nervous system.
Figure 1.
miR-124a and dFMR1 are present in the same RNP complex in the Drosophila nervous system. A, In situ analysis shows prominent miR-124a expression in the brain and ventral nerve cord in a stage 13 Drosophila embryo. B, miR-124a-Gal4 drives GFP expression specifically in the embryonic nervous system, and the expression persists to adulthood. The axon bundles from motor neurons are visible in this image. C, Immunoprecipitation (IP) experiments demonstrate that miR-124a and dFMR1 are present in the same RNP complex independent of polyribosomes. dFMR1-containing RNP complexes were pulled down with HA antibody, and miR-124a in the immunoisolates was detected with a 32P-labeled ribonucleotide probe. A portion of total RNA from control larvae before IP was loaded as a positive control. D, Western blot analysis of immunoisolates with HA antibody. HA-tagged dFMR1 was readily detectable in lysates isolated from larval brains or in immunoisolates after IP. The absence of tubulin in immunoisolates indicates the specificity of the IP experiment with HA antibody. In C and D, control larvae express elav-Gal4 alone, and other larvae express dFMR1 under the control of elav-Gal4.
The exact in vivo function of dFMR1 in the miRNA pathway is largely unknown. We set out to determine whether dFMR1 and miR-124a are present in the same RNP complex. We generated UAS transgenic flies that express dFMR1 with C-terminally tagged HA epitope (dFMR1-HA), expressed dFMR1-HA in all neurons with Elav-Gal4, and dissected out the brain and ventral nerve cord from a large number of third instar larvae. Dissected brain tissues provide “cleaner” lysates for immunoprecipitation experiments than whole larvae, in which contents from guts are problematic. dFMR1-containing RNP complexes were pulled down with the HA antibody, and RNA was extracted from immunoisolates for analysis by Northern blot.
miR-124a was found in immunoisolates from flies expressing both Elav-Gal4 and dFMR1-HA but not in those from Elav-Gal4 control flies (Fig. 1C). The specific isolation of dFMR1-containing RNP complexes was confirmed by Western blot analysis, which revealed dFMR1 but not tubulin in the immunoisolates (Fig. 1D). To confirm that the presence of both miR-124a and dFMR1 in the same RNP complex did not depend on polyribosomes, we also treated larval brain lysates with 10 mm EDTA before immunoprecipitation and obtained the same result (Fig. 1C). Thus, it is unlikely that miR-124a and dFMR1 are associated with polyribosomes through separate RNP complexes, instead, they are present in the same RNP complex. This result raises the possibility that dFMR1 affects the biogenesis and/or function of miR-124a in vivo.
miR-124a suppresses dendritic branching
Before we examine the biological significance of the association of miR-124a and dFMR1 in the same RNP complex, we first set out to examine the effects of miR-124a on dendritic morphology and take this as the readout for our following studies on the functional interactions between the two genes. To this end, we ectopically expressed pre-miR-124a in Drosophila DA sensory neurons, a model system we have used extensively to investigate mechanisms of dendritic morphogenesis. The pre-miR-124a sequence was cloned into the UAST vector, and transgenic flies were generated. In ddaE and ddaF neurons in the dorsal cluster (Sweeney et al., 2002), expression of pre-miR-124a with Gal4221 resulted in significantly fewer dendritic ends than in wild-type neurons [ddaE: 17.0 ± 0.6 (n = 24) vs 25.9 ± 1.0 (n = 20), p < 0.001; ddaF: 17.0 ± 0.7 (n = 24) vs 20.3 ± 0.6 (n = 20), p < 0.001, quantified as we did before in other studies (Li et al., 2004)] (Fig. 2B,D). A similar phenotype was observed in some other DA neurons (supplemental Fig. S1, available at www.jneurosci.org as supplemental material).
Figure 2.
miR-124a suppresses dendritic branching of DA neurons in Drosophila. A, Wild-type ddaE and ddaF neurons in the dorsal cluster were labeled with mCD8-GFP driven by Gal4221. B, Ectopic expression of pre-miR-124a reduced dendritic branching of DA neurons. C, Ectopic expression of pre-miR-9a increased dendritic branching. D, Quantification of dendritic ends in wild-type (WT) ddaE or ddaF neurons and in neurons expressing pre-miR-124a, pre-miR-9a, and mutant pre-miR-124a. The values are mean ± SEM. ***p < 0.001 versus wild type.
To exclude the possibility that ectopic expression of any small RNA was responsible for this effect, we generated a mutant pre-miR-124a molecule with a 6 nt deletion (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Expression of this mutant construct did not affect the dendritic morphology of DA neurons (Fig. 2D). Moreover, ectopic expression of pre-miR-9a significantly increased the number of dendritic ends of both ddaE neurons [56.8 ± 1.9 (n = 18) vs 25.9 ± 1.0 (n = 20), p < 0.001) and ddaF neurons (40.0 ± 3.1 (n = 10) vs 20.3 ± 0.6 (n = 10), p < 0.001] (Fig. 2C,D). Ectopic expression of pre-miR-124a and pre-miR-9a also had opposite effects on dendritic branching in vpda sensory neurons (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). These findings indicate that miR-124a specifically affects dendritic branching and that different miRNAs may exert opposite effects on neuronal morphology, probably by modulating the expression of different sets of downstream target mRNAs.
Genetic interactions between dfmr1 and pre-miR-124a
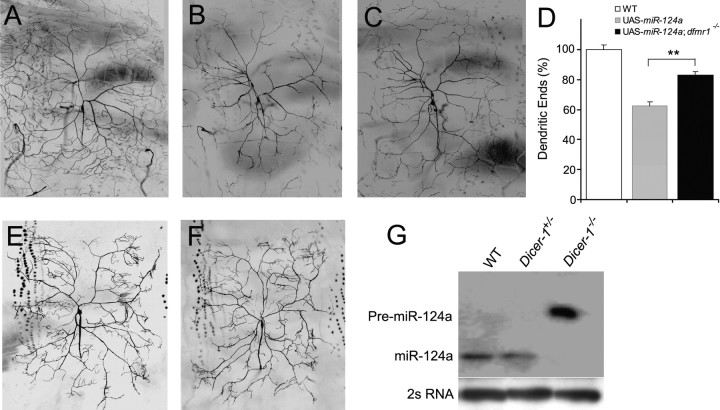
Based on our finding that dFMR1 and miR-124a are present in the same RNP complex (Fig. 1C,D), we performed genetic interaction experiments to determine if miR-124a and dfmr1 indeed interact to control the dendritic branching of DA neurons. We did not perform similar experiments for miR-9a because unlike miR-124a, miR-9a is not detectable in postmitotic neurons. For this experiment, we used ddaC neurons that elaborate extensive dendritic arbors to innervate a large area of the epidermis in the hemisegment (Sweeney et al., 2002). Ectopic expression of pre-miR-124a in ddaC neurons decreased the number of dendritic ends compared with wild-type neurons [342.5 ± 21.2 (n = 18) vs 548.2 ± 13.1 (n = 18), p < 0.001] and the size of the dendritic field [0.27 ± 0.01 mm2 (n = 10) vs 0.42 ± 0.01 mm2 (n = 10) vs p < 0.001] (Fig. 3B,D), consistent with dendritic phenotypes exerted by miR-124a in other DA neurons (Fig. 2B; supplemental Fig. S1, available at www.jneurosci.org as supplemental material). To examine whether dFMR1 mediates the effects of pre-miR-124a, we expressed pre-miR-124a in DA neurons in mutant flies containing two independently generated dfmr1 loss-of-function alleles: dfmr13 (Dockendorff et al., 2002) and dfmr14 (A. Lee et al., 2003). The effects of pre-miR-124a expression in ddaC neurons were significantly attenuated by loss of dfmr1 activity (Fig. 3C,D): the number of dendritic ends was partially rescued [437.2 ± 21.0 (n = 18) vs 342.5 ± 21.2 (n = 18), p < 0.01], although not to the wild type level [437.2 ± 21.0 (n = 18) vs 548.2 ± 13.1 (n = 20), p < 0.001] (Fig. 3D). Interestingly, the dendritic fields of ddaC neurons were similar in size to those of wild-type neurons [0.40 ± 0.02 mm2 (n = 10) vs 0.42 ± 0.01 mm2 (n = 10), p > 0.4]. These results indicate that dFMR1 is required in vivo, at least in part, for pre-miR-124a to exert its effects on dendritic morphology. Our earlier published work indicated that loss of dfmr1 led to increased dendritic branching of DA neurons (A. Lee et al., 2003), raising the possibility that attenuated effects of endogenous miRNAs on dendritic branching could be a potential underlying mechanism.
Figure 3.
dFMR1 is required for pre-miR-124a to exert its effect on dendritic morphogenesis of DA neurons. A, A wild-type (WT) ddaC neuron is labeled with mCD8-GFP whose expression is driven by Gal4477. B, Reduced dendritic branching and dendritic field size in a ddaC neuron expressing pre-miR-124a. C, Expression of pre-miR-124a in ddaC neurons in dfmr1 mutant background (dfmr14/dfmr14 or dfmr13/dfmr14) resulted in a dendritic field of nearly normal size. D, Quantification of dendritic ends of wild-type (WT) ddaC neurons and neurons expressing pre-miR-124a in normal or dfmr1 mutant backgrounds. The values are mean ± SEM. **p < 0.01. E, A MARCM-generated dcr-1 mutant ddaC neuron. F, A dcr-1 mutant ddaC neuron expressing pre-miR-124a. G, Northern blot analysis of miR-124a processing in WT or dcr-1 mutant larvae demonstrates that Dcr-1 is absolutely required for the production of mature miR-124a in vivo.
As a positive control for dFMR1, we performed a similar analysis with dicer-1 mutants. Genetic analysis of zebrafish suggested that Dicer is essential for proper brain morphogenesis (Giraldez et al., 2005). In Drosophila, Dicer-1 (Dcr-1), but not Dicer-2 (Dcr-2), is required for the processing of pre-miRNAs into mature miRNAs (Lee et al., 2004). Because dcr-1Q1147X and dcr-1d102 mutants die at the first or second instar larval stage, the dendritic morphology of DA neurons cannot be analyzed in detail in mutant animals. To examine the cell-autonomous effects of Dcr-1 and miR-124a on dendritic morphology of DA neurons in third instar larvae, we used either allele to generate GFP-labeled single dcr-1 mutant ddaC neurons in an otherwise wild-type animal with mosaic analysis with a repressible cell marker (MARCM) (Lee and Luo, 1999).
Expression of pre-miR-124a in dcr-1 mutant neurons did not affect the number of dendritic ends [613.0 ± 14.3 (n = 7) vs 586 ± 44.6 (n = 9), p > 0.5] (Fig. 3E,F), indicating that Dcr-1 is absolutely essential for pre-miR-124a to exert its biological function in neurons, as one would expect. Indeed, Northern blot analysis demonstrates the absence of endogenous mature miR-124a in dcr-1Q1147X or dcr-1d102 mutant first instar larvae and a corresponding accumulation of endogenous pre-miR-124a (Fig. 3G). It is interesting to note that loss of Dcr-1 activity by itself in MARCM clones did not significantly reduce the number of dendritic ends of ddaC neurons; however, terminal branches fail to elaborate properly, suggesting an essential role for Dcr-1 and the miRNA pathway in some aspects of dendritic morphogenesis.
dFMR1 is required to maintain the steady-state level of miR-124a in Drosophila
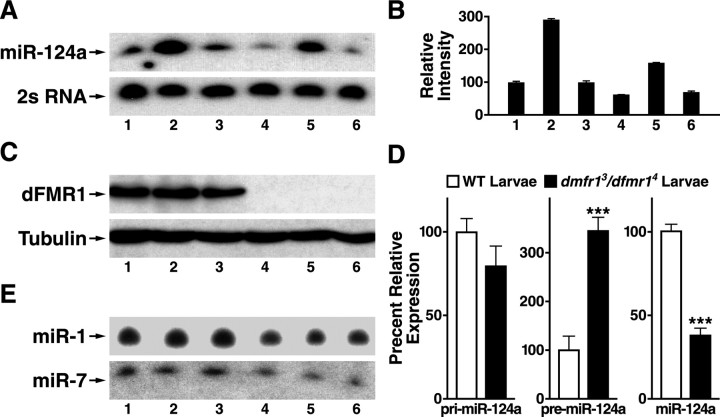
The similarity between the absolute requirement for Dcr-1 (Fig. 3E,F) and the partial requirement for dFMR1 (Fig. 3A–D) in the effect of pre-miR-124a on dendritic branching prompted us to examine the exact role of dFMR1 in miR-124a biogenesis and the biochemical basis for the genetic interaction we observed between miR-124a and dfmr1. Because Dcr-1 is absolutely required for pre-miR-124a processing (Fig. 3G), we tested the hypothesis that dFMR1 might affect the steady-state levels of mature miR-124a in vivo. Ectopic expression of pre-miR-124a in a subset of PNS sensory neurons with 109(2)80-Gal4 led to increased level of mature miR-124a as shown by Northern blot (Fig. 4A, lane 2 vs lane 1 as the control). As expected, ectopic expression of mutant pre-miR-124a did not increase steady-state level of mature miR-124a in vivo (Fig. 4A, lane 3), consistent with the finding that mutant pre-miR-124a did not cause any dendritic phenotype (Fig. 2D). However, in dfmr13/dfmr14 mutant larvae, endogenous mature miR-124a was decreased by 42% (Fig. 4A,B, lane 4), and mature miR-124a processed from ectopically expressed pre-miR-124a was also decreased similarly by 47% (Fig. 4A,B, lane 5). The absence of dFMR1 in mutant larvae was confirmed by Western blot analysis (Fig. 4C, lanes 4–6). These results suggest that the steady-state level of miR-124a in the Drosophila nervous system is modulated by dFMR1. Interestingly, overexpression of dFMR1 did not lead to increased steady-state level of mature miR-124a in vivo (supplemental Fig. S3, available at www.jneurosci.org as supplemental material), suggesting that dFMR1 is not a rate-limiting factor in miRNA biogenesis.
Figure 4.
dFMR1 is required to ensure normal steady-state levels of mature miR-124a in the Drosophila nervous system. A, Northern blot analysis of miR-124a levels in Drosophila third instar larvae. Lane 1: Gal4 109(2)80, UAS-mCD8-GFP/± larvae (controls). Lane 2: Gal4 109(2)80, UAS-mCD8-GFP+; UAS-pre-miR-124a/+ larvae that express miR-124a in all MD neurons and a small number of CNS neurons. Lane 3: Gal4 109(2)80, UAS-mCD8-GFP/+; UAS-mutant pre-miR-124a/+ larvae. Lane 4: Gal4 109(2)80, UAS-mCD8-GFP/+; dfmr13/dfmr14 larvae. Lane 5: Gal4 109(2)80, UAS-mCD8-GFP/UAS-pre-miR-124a; dfmr13/dfmr14 larvae. Lane 6: Gal4 109(2)80, UAS-mCD8-GFP/UAS-mutant pre-miR-124a; dfmr13/dfmr14 larvae. Equal amounts of total RNAs were loaded for each genotype, and 2 s RNA was used as the internal control. B, Relative expression levels of miR-124a in larvae with different genetic backgrounds and transgene expression. The data (mean ± SEM) were derived from three independent experiments and normalized against the levels of 2s RNA. C, Western blot analysis verified the absence of dFMR1 in dfmr1 mutant larvae as described in A. D, Quantitative RT-PCR analysis of relative expression levels of pri-miR-124a, pre-miR-124a, and mature miR-124a in wild-type (WT) and dfmr1 mutant larvae. E, The Northern blot membrane used in A was stripped and reprobed for miR-1 and miR-7 which showed that the in vivo expression levels of miR-1 and miR-7 were also reduced in dfmr1 mutant larvae. The values are mean ± SEM. ***p < 0.001 versus wild type.
To further support the notion that dFMR1 modulates miR-124a biogenesis, we analyzed the steady-state levels of miR-124a and its precursors in wild-type and dfmr1 mutant larvae by qRT-PCR. Consistent with the Northern blot analysis, dfmr13/dfmr14 mutant larvae had a lower level of mature miR-124a than wild-type larvae and a correspondingly higher steady-state level of pre-miR-124a (Fig. 4D). Thus, it appears that dFMR1 modulates the processing efficiency and the steady-state levels of miR-124a in vivo.
To determine if dFMR1 is required for the proper biogenesis of other miRNAs, we examined by Northern blot analysis muscle-specific miR-1 and miR-7 that is highly expressed in the nervous system. The steady-state levels of both miRNAs were reduced by ∼40–45% (Fig. 4E, lanes 4–6). Moreover, the levels of pre- and mature but not pri-miR-7 and miR-1 as measured by qRT-PCR were also decreased (supplemental Fig. S4, available at www.jneurosci.org as supplemental material), suggesting that loss of dFMR1 affects the levels of multiple miRNAs in Drosophila. Because each miRNA may target hundreds of mRNAs, these results suggest that the expression of a very large number of genes may be affected to some extent by the loss of dFMR1.
The Dcr-1–Ago1 complex is less abundant in the absence of dFMR1
In Drosophila, Dcr-1 is primarily responsible for converting pre-miRNAs to mature miRNAs, whereas Dcr-2 is required for processing siRNA precursors (Lee et al., 2004). Although different miRNAs can associate predominantly with either Ago1- or Ago2-containing RNA-induced silencing complex (RISC) (Förstemann et al., 2007), it seems that Ago1 is more essential for miRNA biogenesis (Okamura et al., 2004). To provide further mechanistic insight for the action of dFMR1 in the miRNA pathway, we performed co-immunoprecipitation experiments to examine the abundance of the Dcr-1-Ago1 complex in the presence or absence of dFMR1. We prepared lysates from a large number of dissected larval brains and used Ago1 antibody to pull down the complex. We found that Ago1 was associated with Dcr-1 and dFMR1 in vivo (Fig. 5A). Interestingly, based on three independent experiments, there was an ∼44% decrease in the abundance of the Dcr-1-Ago1 complex attributable to the absence of dFMR1 (Fig. 5A,B). This novel finding suggests a functional role for dFMR1 in facilitating the formation of the Dcr-1-Ago1 complex. This difference is not attributable to reduced Dcr-1 and Ago1 expression levels in dfmr1 mutants (Fig. 5C,D). These findings provide a molecular explanation for the observed lower levels of miR-124a in the developing Drosophila nervous system.
Figure 5.
The Dcr-1-Ago1 complex is less abundant in the absence of dFMR1. A, Control mouse IgG or Ago1 antibody were used to immunoprecipitate Ago1-containing complexes from lysates of dissected wild-type (WT) or dfmr1 mutant larval brains. The presence or absence of Dcr-1 or dFMR1 was detected by Western blot. This experiment was repeated three times. B, Quantification of the abundance of Dicer-1 in Ago1 immunoprecipitates based on three independent experiments. The values are mean ± SEM. p < 0.01 versus wild type. C, The protein band recognized by the Dcr-1 antibody obtained from Abcam is absent in the dcr-1 mutants. D, An Ago1 antibody (Okamura et al., 2004) identifies Ago1 isoforms that are absent in ago1 mutants. Note that the expression levels of Dcr-1 and Ago1 are the same in dfmr1 mutants and wild-type larvae. IP, Immunoprecipation.
Discussion
The major novel findings in this study are that elevated miR-124a expression decreases dendritic branching and dFMR1, the fly homolog of the human protein responsible for fragile X syndrome, modulates the biogenesis and function of miRNAs in the Drosophila nervous system. This conclusion is supported by the presence of dFMR1 and miR-124a in the same RNP complex, the genetic interaction between the two genes, and biochemical analysis of steady-state levels of miRNAs in the absence of dFMR1. Moreover, we found that the Dcr-1-Ago1 complex was less abundant in the nervous system of dfmr1 mutants, providing a mechanistic explanation for the observed effect of loss of dFMR1 activity on miRNA biogenesis.
miR-124a is one of the most abundant miRNAs in the brain and whose nucleotide sequence is 100% conserved across species yet it functions in the brain remain incompletely understood (Lagos-Quintana et al., 2002; Gao, 2008). Our findings suggest that elevated pre-miR-124a level, such as in pathological conditions, can lead to decreased dendritic branching, which may be detrimental to neuronal function and connectivity. The effect of pre-miR-124a on dendritic branching in vivo seems to require dFMR1. Although dFMR1 is implicated in neuronal development through translational control, the exact mechanism remains unclear. Our findings shed new light on the precise roles of dFMR1 in the developing nervous system. dFMR1 is not an absolute essential factor for the biogenesis of mature miRNAs in vivo, consistent with earlier findings in cultured S2 cells (Caudy et al., 2002; Ishizuka et al., 2002). Nonetheless, the finding that dFMR1 modulates the steady-state levels of miR-124a and other miRNAs is of considerable significance, because multiple genes must be expressed coordinately at precise levels both spatially and temporally during brain development. Because miRNAs fine-tune protein synthesis of many genes (Baek et al., 2008; Selbach et al., 2008), the developmental and functional defects seen in dfmr1 mutants are likely the consequence of changes in multiple proteins and pathways.
As an RNA-binding protein, dFMR1 is present in multiple distinct RNP complexes that are likely involved in many aspects of RNA metabolism. For instance, dFMR1 forms a complex with PIWI in germ cells and play a role in the biogenesis and function of piRNAs (Megosh et al., 2006). dFMR1 is also associated with Ago2 and likely plays a role in the RNAi pathway (Caudy et al., 2002; Ishizuka et al., 2002). Here, we show that endogenous dFMR1 and Ago1 are associated with each other in the developing Drosophila nervous system, consistent with a recent finding in oocytes (Yang et al., 2007). More importantly, we found that the Dcr-1-Ago1 complex in the nervous system is less abundant in the absence of dFMR1, providing novel mechanistic insight into the exact role of dFMR1 in the miRNA pathway. Considering the essential roles of the Dcr-1-Ago1 complex in miRNA biogenesis (Okamura et al., 2004; Förstemann et al., 2007), impaired association between Dcr-1 and Ago1 attributable to the absence of dFMR1 seems to be responsible, at least in part, for the observed lower steady-state levels of the nervous-system-specific miR-124a and other miRNAs in vivo. Global regulation of miRNA levels by RNA-binding proteins was also reported in the case of Rbm3, a glycine-rich RNA-binding protein that regulates global protein synthesis under cold-stress conditions (Dresios et al., 2005).
In a human B-cell line, FMRP was associated with small 20 nt RNAs in a common RNP complex (Jin et al., 2004). This small RNA is unlikely to be miR-124a, which is nervous-system-specific and 23 nt long as many other miRNAs. dFMR1 has three mammalian homologs, FMRP, FXR1, and FXR2. Their exact functions in the miRNA pathway remain largely unknown, although one recent report suggested that FXR1 was involved in some aspects of miRNA function in HeLa cells (Vasudevan et al., 2007). It is possible that these three mammalian proteins may have redundant functions in the miRNA pathway. Alternatively, different paralogs may have evolved to carry out at least some distinct molecular functions in postmitotic neurons in mammals. It will be interesting to determine the extent to which dFMR1 and FMRP are functionally conserved and whether the miRNA pathway is also misregulated in mouse models of fragile X syndrome.
Footnotes
This work was supported by the FRAXA Research Foundation and the National Institutes of Health (F.-B.G.). We thank the Bloomington Stock Center, R. Carthrew and T. Jongens for fly lines, S. Ordway for editorial assistance, and L.-P. Chang for help with miR-124a constructs. We are grateful to lab members for discussions and comments during the course of this work.
References
- Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci U S A. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorff TC, Su HS, McBride SM, Yang Z, Choi CH, Siwicki KK, Sehgal A, Jongens TA. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34:973–984. doi: 10.1016/s0896-6273(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Dresios J, Aschrafi A, Owens GC, Vanderklish PW, Edelman GM, Mauro VP. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc Natl Acad Sci U S A. 2005;102:1865–1870. doi: 10.1073/pnas.0409764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci. 2008;31:20–26. doi: 10.1016/j.tins.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Li W, Wang F, Menut L, Gao FB. BTB/POZ-zinc finger protein abrupt suppresses dendritic branching in a neuronal subtype-specific and dosage-dependent manner. Neuron. 2004;43:823–834. doi: 10.1016/j.neuron.2004.08.040. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Michel CI, Kraft R, Restifo LL. Defective neuronal development in the mushroom bodies of Drosophila fragile X mental retardation 1 mutants. J Neurosci. 2004;24:5798–5809. doi: 10.1523/JNEUROSCI.1102-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, Jackson FR, Nelson DL, Hassan BA. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–972. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sharma Y, Cheung U, Larsen EW, Eberl DF. PPTGAL, a convenient Gal4 P-element vector for testing expression of enhancer fragments in Drosophila. Genesis. 2002;34:115–118. doi: 10.1002/gene.10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Sweeney NT, Li W, Gao FB. Genetic manipulation of single neurons in vivo reveals specific roles of Flamingo in neuronal morphogenesis. Dev Biol. 2002;247:76–88. doi: 10.1006/dbio.2002.0702. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, Eussen BE, van Ommen G-JB, Blonden LAJ, Riggins GJ, Chastain JL, Kunst CB, Galjaard H, Caskey CT, Nelson DL, Oostraa BA, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Yang L, Duan R, Chen D, Wang J, Chen D, Jin P. Fragile X mental retardation protein modulates the fate of germline stem cells in Drosophila. Hum Mol Genet. 2007;16:1814–1820. doi: 10.1093/hmg/ddm129. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]