Abstract
The Rad9-Rad1-Hus1 (9-1-1) cell cycle checkpoint complex plays a key role in the DNA damage response. Cells with a defective 9-1-1 complex have been shown to be sensitive to apoptosis induced by certain types of genotoxic stress. However, the mechanism linking the loss of a functional 9-1-1 complex to the cell death machinery has yet to be determined. Here, we report that etoposide treatment dramatically upregulates the BH3-only proteins, Bim and Puma, in Hus1-deficient cells. Inhibition of either Bim or Puma expression in Hus1-knockout cells confers significant resistance to etoposide-induced apoptosis, while knockdown of both proteins results in further resistance, suggesting that Bim and Puma cooperate in sensitizing Hus1-deficient cells to etoposide treatment. Moreover, we found that Rad9 collaborates with Bim and Puma to sensitize Hus1-deficient cells to etoposide-induced apoptosis. In response to DNA damage, Rad9 localizes to chromatin in Hus1-wild-type cells, whereas in Hus1-deficient cells it is predominantly located in the cytoplasm where it binds to Bcl-2. Taken together, these results suggest that loss of Hus1 sensitizes cells to etoposide-induced apoptosis, not only by inducing Bim and Puma expression, but also by releasing Rad9 into the cytosol to augment mitochondrial apoptosis.
Keywords: Hus1, Rad9, Bim, Puma, DNA damage, apoptosis
Introduction
The genomes of eukaryotic cells are constantly being subjected to exogenous and endogenous genotoxic stresses. Damage resulting from exposure to these stresses threatens cell survival and can lead to cancer, as well as other genetic diseases. In order to preserve genomic integrity and ensure that an accurate copy of the genome is passed on to subsequent generations, cells have evolved a core surveillance machinery known as cell cycle checkpoints. These checkpoints sense damaged or abnormally structured DNA and coordinate cell cycle progression with DNA repair. In cases when damage is excessive or repair is unfavorable, the cell death machinery is activated in order to eliminate damaged cells (Melo and Toczyski, 2002; Niida and Nakanishi, 2006; Zhou and Elledge, 2000).
In mammalian cells, the DNA damage response is regulated by two primary signaling pathways. The ATM-mediated pathway is activated in response to DNA damaging agents that induce double-strand breaks (Shiloh, 2003), whereas the ATR-dependent pathway responds to a broad spectrum of genotoxic stresses including those that inhibit replication and induce single-strand DNA breaks or bulky DNA lesions (Abraham, 2001; Bakkenist and Kastan, 2004). Certain DNA damaging agents, such as the topoisomerase II poison, etoposide, activate both the ATM- and ATR-dependent pathways by inducing both double- and single-strand breaks in DNA (Montecucco and Biamonti, 2007). Upon activation, ATM and ATR act in collaboration with members of the Rad family in order to relay the damage signal to downstream transducer and effector proteins (Chen et al., 2001; Niida and Nakanishi, 2006; Weiss et al., 2002; Zou et al., 2002). Three members of the Rad family, Rad9, Rad1 and Hus1, form a heterotrimeric clamp that acts as a putative sensor for DNA damage (Burtelow et al., 2001; Volkmer and Karnitz, 1999). Another member of the Rad family, Rad17, along with the four small subunits of the replication factor C complex, is responsible for loading the Rad9-Rad1-Hus1 (9-1-1) complex onto DNA in response to various types of DNA damage (Bermudez et al., 2003; Lindsey-Boltz et al., 2001; Rauen et al., 2000).
As the 9-1-1 complex plays an apical role in the DNA damage response, loss of a functional complex affects many downstream checkpoint pathways. Impaired function of the 9-1-1 complex results in defects in cell cycle arrest, an increase in chromosomal abnormalities and increased sensitivity to genotoxic stresses (Bao et al., 2004; Dang et al., 2005; Hopkins et al., 2004; Kinzel et al., 2002; Wang et al., 2003; Weiss et al., 2000; Weiss et al., 2003). Moreover, loss of Rad9 or Hus1 results in embryonic lethality, which is, at least in part, attributable to widespread apoptosis during embryogenesis (Hopkins et al., 2004; Weiss et al., 2000). Although Hus1-/- mouse embryonic fibroblast (MEF) cells have a cellular proliferative defect, crossing of Hus1+/- mice to a p21-/- background results in Hus1-/-p21-/- MEFs that are viable and can be grown in culture (Weiss et al., 2000). In addition to playing a key role in the regulation of DNA damage checkpoints, Rad9 also has been shown to induce apoptosis through its interaction with the pro-survival Bcl-2 and Bcl-xL proteins (Ishii et al., 2005; Komatsu et al., 2000b; Yoshida et al., 2002; Yoshida et al., 2003). Furthermore, Rad9 can be cleaved by caspase-3, resulting in disruption of the 9-1-1 complex and release of the BH3-containing fragment of Rad9 into the cytosol where it binds to Bcl-xL to potentiate the apoptotic response (Lee et al., 2003). These studies demonstrate that the members of the 9-1-1 complex are not only important for initiating cell cycle checkpoints in response to DNA damage, but that these proteins also play a role in the regulation of apoptosis.
Apoptosis is mediated through two major pathways: the extrinsic pathway, which is activated by ligation of death receptors, and the intrinsic or stress-induced, mitochondrial pathway. The members of the Bcl-2 family play a central role in the regulation of apoptosis induced through the mitochondrial pathway (Cory et al., 2003). The Bcl-2 family consists of both anti-apoptotic and pro-apoptotic members. Proteins, such as Bcl-2 and Bcl-xL, are anti-apoptotic and thus prevent activation of apoptosis through the mitochondrial pathway. The pro-apoptotic Bcl-2 family members can be subdivided into the multi-domain proteins, including Bax and Bak, and the BH3-only proteins, which include Bim and Puma, among others. The BH3-only proteins act as sensors for damage signals and induce apoptosis by neutralizing the anti-apoptotic proteins or by directly activating the multi-domain pro-apoptotic proteins of the Bcl-2 family to release apoptogenic factors from the mitochondria (Strasser, 2005). Therefore, the members of the Bcl-2 family ultimately control the decision of whether a cell is to live or die, based on the relative ratio of anti- to pro-apoptotic proteins (Adams and Cory, 2007; Oltvai and Korsmeyer, 1994).
While loss of Hus1 results in increased sensitivity to DNA damage-induced apoptosis, the molecular mechanism by which this occurs has yet to be elucidated. In this study, we demonstrate that loss of Hus1 sensitizes cells to etoposide treatment through the upregulation of the BH3-only proteins, Bim and Puma. Furthermore, loss of Hus1 results in a defect in the binding of Rad9 to chromatin and release of Rad9 into the cytosol, which in turn enhances the interaction of Rad9 with Bcl-2 to amplify the apoptotic response.
Results
Loss of Hus1 sensitizes cells to etoposide-induced apoptosis
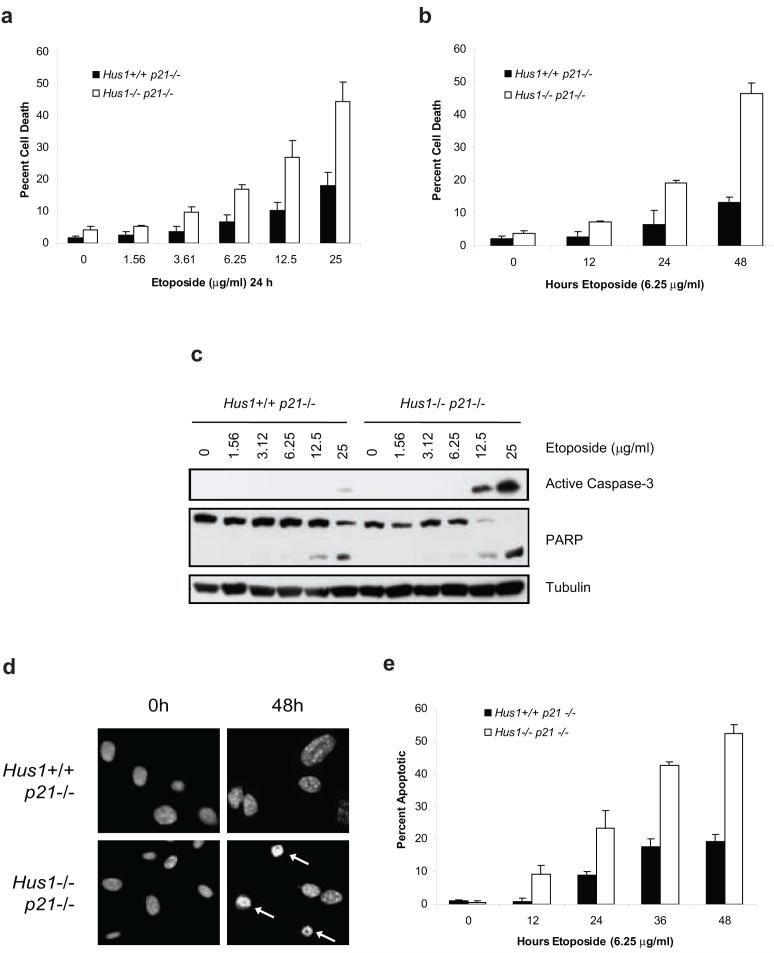
Knockout of Hus1 results in cell cycle checkpoint defects and enhanced cell death in response to DNA damage induced by hydroxyurea and UV radiation (Weiss et al., 2000; Weiss et al., 2003; Weiss et al., 2002). In this study, we examine the sensitivity of Hus1-deficient cells to etoposide, one of the most potent drugs used for cancer therapy (Montecucco and Biamonti, 2007). In order to determine whether loss of Hus1 would sensitize cells to etoposide-induced cell death, Hus1+/+p21-/- and Hus1-/-p21-/- MEFs were treated with increasing doses of etoposide for 24 h. Measurement of cell death by trypan blue exclusion assay revealed that knockout of Hus1 greatly enhanced the dose-dependent susceptibility of MEFs to etoposide (Figure 1a). Consistently, the hypersensitivity of Hus1-deficient cells to etoposide-induced cell death also occurred in a time-dependent manner (Figure 1b).
Figure 1.
Loss of Hus1 sensitizes cells to apoptosis induced by etoposide treatment. (a) Hus1+/+p21-/- and Hus1-/- p21-/- MEFs were treated with increasing doses of etoposide for 24 h and viability was determined by trypan blue exclusion assay (mean ± s.d.; n=3). (b) Hus1+/+p21-/- and Hus1-/-p21-/- MEFs were treated with 6.25 μg/ml etoposide for varying time points and subjected to trypan blue exclusion assay (mean ± s.d.; n=3). (c) Cells were treated as in (a). Total cell lysate was normalized for protein content and subjected to SDS-PAGE/immunoblot analysis. (d) Hus1+/+p21-/- and Hus1-/-p21-/- MEFs were treated with 6.25 μg/ml etoposide for 0 or 48 h. Apoptosis was determined by examination of nuclear morphology. Arrows indicate apoptotic nuclei. (e) Hus1+/+p21-/- and Hus1-/-p21-/- MEFs were treated with 6.25 μg/ml etoposide for the times indicated. The percent of apoptotic cells was quantified based on nuclear morphology (mean ± s.d.; n=3).
To determine whether the increase of cell death in Hus1-deficient cells is due to enhanced induction of apoptosis, activation of caspase-3, as well as cleavage of its downstream substrate, poly(ADP-ribosyl) transferase (PARP), were examined by immunoblot analysis. Hus1-/-p21-/- cells exhibited a robust induction of caspase-3 processing, which correlated with PARP cleavage, upon 24 h treatment with 12.5 μg/ml etoposide that was further enhanced at a higher dose (Figure 1c). In contrast, Hus1+/+p21-/- cells showed only slight activation of caspase-3 and minimal PARP cleavage even upon treatment with the highest dose of etoposide (Figure 1c). Induction of apoptosis in response to etoposide treatment was further analyzed by examination of nuclear morphology for chromatin condensation and nuclear fragmentation. As shown in Figures 1d and e, Hus1-deficient cells were almost three times more sensitive to etoposide-induced apoptosis. Taken together, these results suggest that MEFs that lack Hus1 are not only sensitive to hydroxyurea and UV radiation as previously described, but that these cells are also sensitive to DNA damage induced by topoisomerase II poisons, such as the chemotherapeutic drug, etoposide.
Loss of Hus1 enhances Bim and Puma expression at both the protein and mRNA level in response to DNA damage
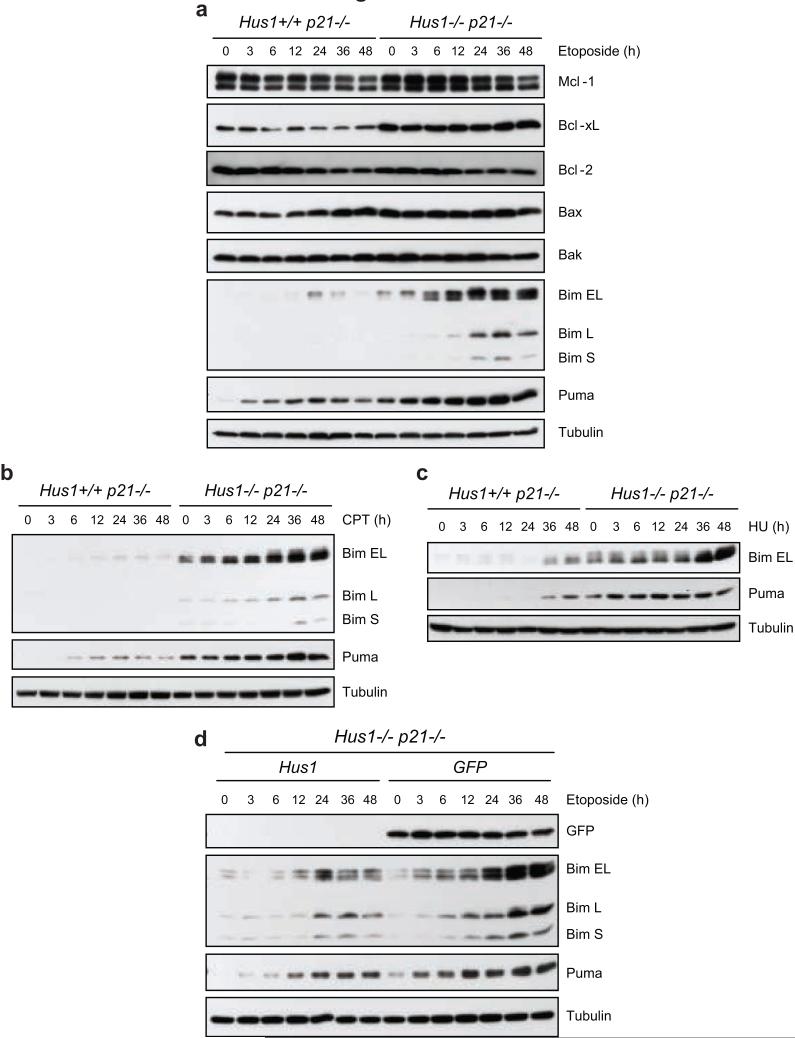
Since our results indicate that loss of Hus1 sensitizes cells to etoposide-induced apoptosis, the expression levels of members of the Bcl-2 family were examined (Figure 2a). In response to etoposide treatment, the expression levels of anti-apoptotic Bcl-2-like proteins and pro-apoptotic multi-domain proteins remained relatively stable in both Hus1+/+p21-/- and Hus1-/-p21-/- MEFs. Notably, the basal level of Bcl-xL was higher in Hus1-/-p21-/- cells compared to Hus1+/+p21-/- cells, presumably to neutralize the elevated Bax expression in cells lacking Hus1 (Weiss et al., 2000). Interestingly, the expression of the BH3-only proteins, Bim and Puma, was induced following etoposide treatment. Whereas the expression of these proteins was only slightly induced and peaked at 24 h in Hus1+/+p21-/- cells, the upregulation of all three isoforms of Bim, as well as Puma, was much more dramatic and persisted to later time points in Hus1-/-p21-/- cells. Similar results were seen after treatment with other DNA damaging agents including camptothecin, an inhibitor of topoisomerase I, and hydroxyurea, an inhibitor of DNA replication (Figures 2b and c, respectively).
Figure 2.
Loss of Hus1 results in upregulation of Bim and Puma expression in response to DNA damage. (a, b, c) Hus1+/+p21-/- and Hus1-/-p21-/- MEFs were treated with (a) 6.25 μg/ml etoposide, (b) 500 nM camptothecin (CPT) or (c) 50 μM hydroxyurea (HU) for the indicated time points. Total cell lysate was prepared and analyzed by SDS-PAGE/immunoblot using the indicated antibodies. (d) Hus1-/-p21-/- MEFs stably expressing Hus1 or GFP were treated with 6.25 μg/ml etoposide for varying time points. The expression of Bim and Puma was examined by SDS-PAGE/immunoblot analysis.
To confirm that the etoposide-induced upregulation of Bim and Puma expression is a direct result of loss of Hus1, we examined whether restoration of Hus1 expression would suppress the induction of these BH3-only proteins in response to etoposide treatment. To this end, Hus1-/-p21-/- MEFs that were infected with retrovirus to express either Hus1 (Hus1-/-p21-/- Hus1) or control GFP (Hus1-/-p21-/- GFP) (Weiss et al., 2002) were treated with etoposide for varying time points and the expression of Bim and Puma was examined. Expression of Hus1, but not control GFP, significantly reduced etoposide-induced expression of Bim and Puma in Hus1-/-p21-/- MEFs. These results indicate that loss of Hus1 results in upregulation of Bim and Puma in response to etoposide-induced DNA damage.
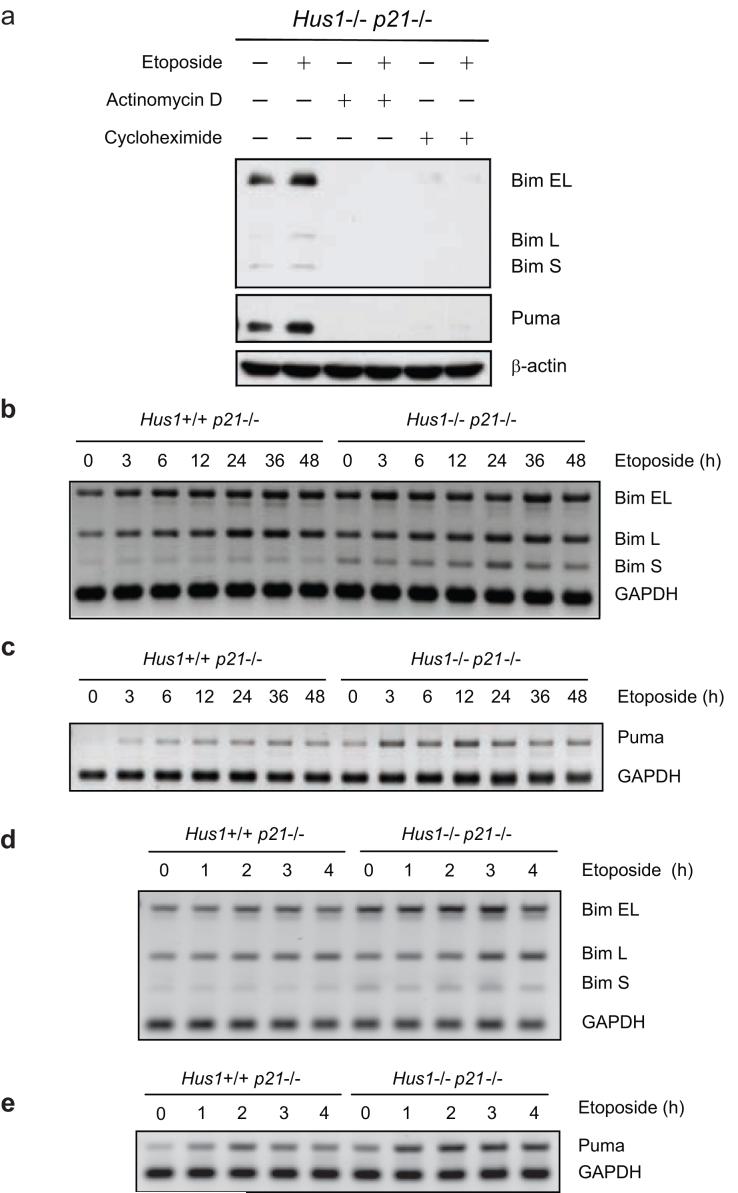
The activity of BH3-only proteins can be regulated by various means including transcriptional upregulation, post-translational modification, proteasomal degradation and sequestration to cytoskeletal components (Puthalakath and Strasser, 2002; Willis and Adams, 2005). To examine whether the induction of Bim and Puma expression observed in Hus1-deficient cells is regulated at the transcriptional level after exposure to etoposide, Hus1-/-p21-/- cells were treated with etoposide in the presence of a transcriptional inhibitor, actinomycin D, a translational inhibitor, cycloheximide, or control DMSO. As shown in Figure 3a, treatment with either actinomycin D or cycloheximide abrogated the expression of Bim, as well as Puma, even in the presence of etoposide. In contrast, the expression of these BH3-only proteins was significantly induced in response to etoposide treatment in the control DMSO treated cells (Figure 3a). This indicates that upregulation of both Bim and Puma in response to etoposide-induced DNA damage occurs at the transcriptional level. Indeed, semi-quantitive RT-PCR analyses revealed that the expression of Bim and Puma mRNAs are increased in response to etoposide treatment (Figures 3b-e and Supplementary Figure 1). The levels of Bim and Puma mRNAs continued to increase until approximately 24 to 36 h and then decreased at later time points, presumably due to induction of cell death. Notably, a greater upregulation of Bim, especially BimS, the most potent isoform, and Puma mRNAs was observed in Hus1-deficient cells, as compared to Hus1-wild-type cells. Consistently, when cells were treated with a higher dose of etoposide for a shorter time course, a clear induction of Bim and Puma mRNAs occurred in a time-dependent manner, with more dramatic increases seen in Hus1-/-p21-/- cells (Figures 3d and e). Notably, the protein expression levels (Figure 2a) of Bim and Puma in Hus1-/-p21-/- cells after etoposide treatment are higher than their mRNA levels (Figures 3b, c), suggesting that etoposide-mediated upregulation of Bim and Puma is regulated through both transcriptional and post-transcriptional mechanisms.
Figure 3.
Induction of Bim and Puma expression in response to etoposide treatment is regulated at the transcriptional level. (a) Hus1-/-p21-/- MEFs were treated with control DMSO (-), 1 μg/ml actinomycin D or 5 μg/ml cycloheximide alone or in combination with 3.125 μg/ml etoposide for 24 h. Total cell lysate was prepared and the expression of Bim and Puma was analyzed by SDS-PAGE/immunoblot. (b, c) Hus1+/+p21-/- and Hus1-/-p21-/- MEFs were treated with 6.25 μg/ml etoposide for 0, 3, 6, 12, 24, 36 or 48 h. Semi-quantitative RT-PCR was used to examine the mRNA levels of (b) Bim and (c) Puma. (d, e) Hus1+/+p21-/- and Hus1-/-p21-/- MEFs were treated with 25 μg/ml etoposide for 0, 1, 2, 3 or 4 h. Semi-quantitative RT-PCR was used to examine the mRNA levels of (d) Bim and (e) Puma.
Knockdown of Bim and Puma confers resistance to etoposide-induced apoptosis in Hus1-deficient cells
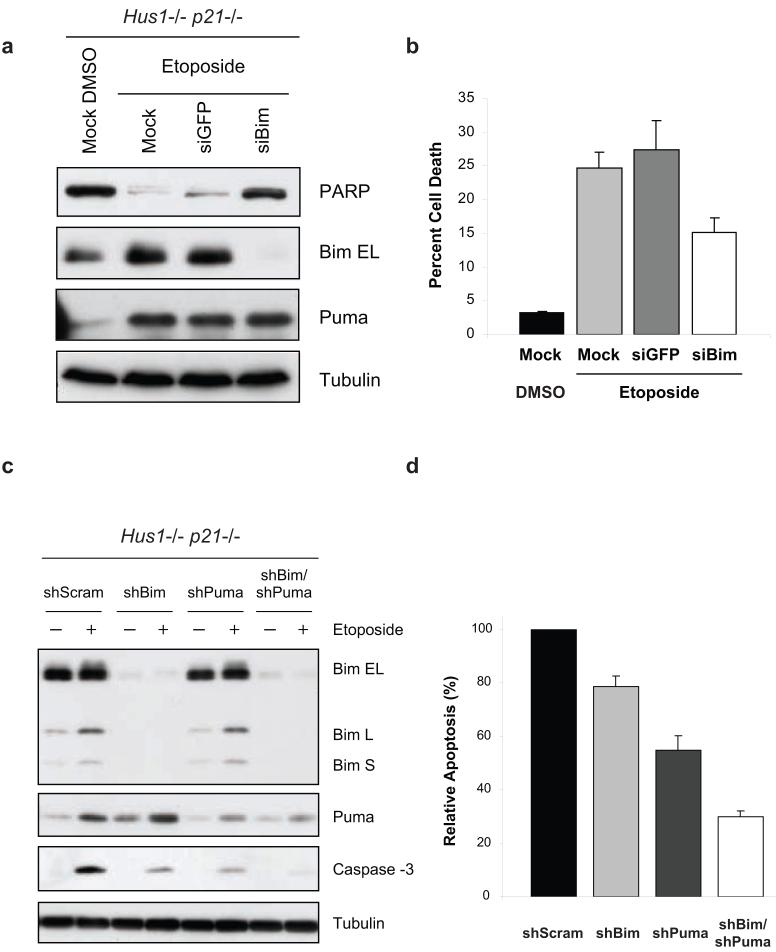
Our results clearly show that loss of Hus1 not only results in the upregulation of Bim and Puma expression, but also promotes caspase-3 activation and cell death induced by etoposide treatment. Since BH3-only proteins play a key role in the initiation of apoptosis (Huang and Strasser, 2000; Puthalakath and Strasser, 2002; Willis and Adams, 2005), we examined whether the upregulation of Bim and Puma is responsible for sensitizing Hus1-deficient cells to etoposide treatment. As a dramatic induction of all three isoforms of Bim was observed in response to etoposide treatment, we first examined whether inhibition of Bim expression would suppress DNA damage-induced cell death in Hus1-/-p21-/- MEFs. Transfection of siRNA specific for Bim abrogated its expression, even upon treatment with etoposide (Figure 4a). Knockdown of Bim expression resulted in a decrease in PARP cleavage (Figure 4a) and partial resistance to DNA damage-induced cell death (Figure 4b), as compared to siGFP or mock transfected cells. These results suggest that upregulation of Bim expression contributes to the sensitivity of Hus1-deficient cells to etoposide-induced apoptosis. We next examined whether the upregulation of Puma expression is also involved in sensitizing Hus1-deficient cells to etoposide treatment. To this end, a lentiviral delivery system was used to transduce Hus1-/-p21-/- MEFs with shRNA targeting Puma or Bim, or a control scrambled shRNA. Whereas etoposide treatment resulted in the induction of Bim and Puma expression in control shScrambled expressing cells, the upregulation of these proteins was suppressed by their respective shRNA (Figure 4c). Consistent with the siBim results shown in Figure 4a, expression of shBim suppressed etoposide-induced apoptosis (Figure 4c and d). Moreover, knockdown of Puma expression significantly suppressed etoposide-induced caspase-3 activation and apoptosis, as compared to control cells (Figures 4c and d). Since knockdown of either Bim or Puma alone only partially suppressed etoposide-induced cell death, we next examined whether Bim and Puma are acting redundantly or synergistically to induce apoptosis in response to etoposide treatment. To this end, shBim expressing Hus1-/-p21-/- MEFs were infected with lentivirus expressing shRNA targeting Puma, which resulted in efficient knockdown of Puma expression (Figure 4c). Importantly, knockdown of both Bim and Puma resulted in further inhibition of caspase-3 processing and apoptosis, when compared to Hus1-/-p21-/- cells expressing shBim or shPuma alone (Figures 4c and d). Taken together, these results indicate that Bim and Puma cooperate in sensitizing Hus1-deficient cells to etoposide-induced apoptosis.
Figure 4.
Suppression of Bim and Puma expression confers resistance to etoposide-induced apoptosis in Hus1-deficient cells. (a, b) Hus1-/-p21-/- MEFs were mock transfected or transiently transfected with siRNA targeting GFP or Bim. Thirty-six hours after transfection, the cells were treated with control DMSO or 6.25 μg/ml etoposide for 30 h. (a) Whole cell lysate was subjected to SDS-PAGE/immunoblot analysis with antibodies to PARP (full length PARP is shown), Bim, Puma and Tubulin. (b) Viability was determined by trypan blue exclusion assay (mean ± s.d.; n=2). The DMSO-treated mock transfected cells were used to show the basal levels of Bim expression and cell death. (c, d) Hus1-/-p21-/- MEFs were infected with lentivirus expressing shRNA targeting Bim, Puma, Bim and Puma, or a control scrambled shRNA (shScram). After selection on Puromycin, the cells were treated with 12.5 μg/ml etoposide or control DMSO for 16 h. (c) Knockdown of Bim and Puma was confirmed by SDS-PAGE/immunoblot analysis. (d) Induction of apoptosis was measured by caspase-3 activity assay. The caspase-3 activity of control DMSO treated cells was subtracted from the amount of caspase-3 activity observed in the etoposide treated cells. The data are represented as percent relative apoptosis as normalized to the control infected cells (mean ± s.d.; n=3).
Loss of Hus1 enhances the binding of Rad9 to Bcl-2 to potentiate the apoptotic response
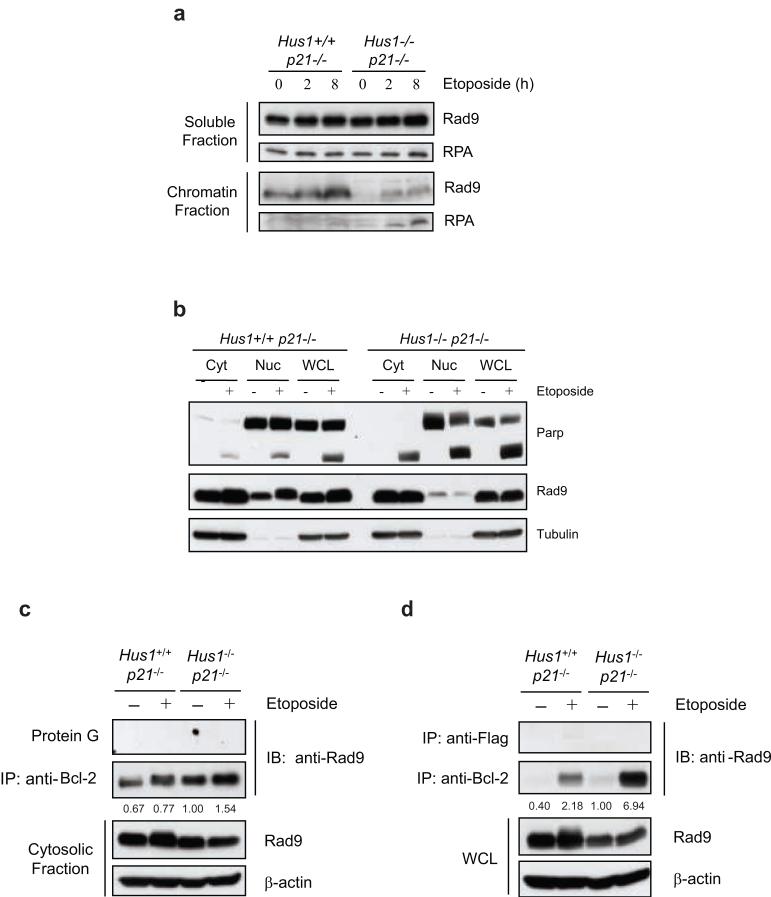
It has been shown that DNA damage promotes the binding of the 9-1-1 checkpoint complex to chromatin to initiate the DNA damage response and facilitate the activation of downstream proteins (Parrilla-Castellar et al., 2004; Zhou and Elledge, 2000). Consistently, Rad9 binding to chromatin was enhanced in Hus1-wild-type cells in a time-dependent manner after etoposide treatment (Figure 5a). In contrast, chromatin bound Rad9 was barely detectable in Hus1-deficient cells, although a slight increase was noticeable after etoposide treatment. These results are consistent with previous findings which show that HU- and UV-induced binding of Rad9 to the chromatin is decreased in Hus1-/- cells (Zou et al., 2002). Taken together, these results indicate that loss of Hus1 results in a defect in the binding of Rad9 to chromatin in response to DNA damage.
Figure 5.
Rad9 is predominantly detected in the cytoplasm of Hus1-deficient cells where it binds to Bcl-2 in response to DNA damage. (a) Hus1+/+p21-/- and Hus1-/-p21-/- MEFs were treated with 12.5 μg/ml etoposide for 0, 2 or 8 h and subjected to subcellular fractionation. The resulting chromatin bound and soluble fractions were analyzed by SDS-PAGE/immunoblot using antibodies specific for Rad9 and RPA as a control. (b) Hus1+/+p21-/- and Hus1-/-p21-/- MEFs were treated with 12.5 μg/ml etoposide or control DMSO for 12 h and subjected to subcellular fractionation. The resulting cytosolic (Cyt) and nuclear (Nuc) fractions, along with whole cell lysate (WCL), were analyzed by SDS-PAGE/immunoblot. (c, d) Hus1+/+p21-/- and Hus1-/-p21-/- MEFs were treated with 12.5 μg/ml etoposide or control DMSO for 12 h. (c) The cytosolic fractions of Hus1+/+p21-/- and Hus1-/-p21-/- MEFs were subjected to immunoprecipitation in the absence or presence of anti-Bcl-2 monoclonal antibody. The resulting immune complexes were analyzed by SDS-PAGE/immunoblot. (d) Whole cell lysate (WCL) was subjected to immunoprecipitation with anti-Bcl-2 or control anti-Flag monoclonal antibodies. The resulting immunocomplexes and WCL were analyzed by SDS-PAGE/immunoblot. The amount of Rad9 in the immunocomplexes was quantified and normalized to cytosolic (c) or total (d) Rad9. The levels of Bcl-2 bound Rad9 are listed relative to those of untreated Hus1-/-p21-/- cells, which were set as 1.0.
We and others have previously shown that Rad9 interacts with Bcl-2 or Bcl-xL through a BH3-like domain within its N-terminus to promote apoptosis following DNA damage (Ishii et al., 2005; Komatsu et al., 2000a; Komatsu et al., 2000b; Lee et al., 2003; Yoshida et al., 2002; Yoshida et al., 2003). These results indicate that Rad9 not only has functions in the nucleus as a member of a DNA damage checkpoint complex, but also in the cytosol as an inducer of apoptosis. Therefore, the effect of loss of Hus1 on the intracellular localization of Rad9 was examined. Consistent with previous studies (Burtelow et al., 2000), Rad9 was detected in both the nuclear and cytosolic fractions of Hus1+/+p21-/- cells when analyzed by subcellular fractionation (Figure 5b). Moreover, etoposide treatment resulted in Rad9 accumulation and hyperphosphorylation in the nucleus of Hus1+/+p21-/- cells (Figure 5b). In contrast, Rad9 was primarily detected in the cytosolic fraction of Hus1-deficient cells and remained hypophosphorylated even upon DNA damage (Figure 5b). These results suggest that Rad9 chromatin binding and hyperphosphorylation are Hus1-dependent.
In order to determine whether Rad9 binds to Bcl-2 family members during apoptosis, the interaction between Rad9 and Bcl-2 was examined in Hus1+/+p21-/- and Hus1-/-p21-/- cells in response to etoposide treatment. Since the majority of Rad9 was detected in the cytosolic fraction of Hus1-deficient cells, coimmunoprecipitation of cytosolic Rad9 with Bcl-2 was performed. While etoposide treatment enhanced Rad9 interaction with Bcl-2, a significant amount of cytosolic Rad9 was bound to Bcl-2 even in the absence of DNA damage (Figure 5c). Thus, it is possible that subcellular fractionation using a hypotonic buffer may alter the conformation or localization of Rad9 and Bcl-2, which affects their interaction. Indeed, it has been shown that Rad9 can leak from the nucleus during subcellular fractionation even in the absence of DNA damage (Burtelow et al., 2000). In order to confirm the interaction of Rad9 and Bcl-2, the coimmunoprecipitation was repeated using whole cell lysates. As Figure 5d shows, a minimal amount of Rad9 was bound to Bcl-2 in the absence of DNA damage, regardless of Hus1 status. Treatment with etoposide resulted in an induction of Rad9 binding to Bcl-2 that was much greater in Hus1-deficient cells as compared to Hus1-wild-type cells. These results suggest that, in response to DNA damage, Rad9 may also contribute to the enhanced sensitivity of Hus1-deficient cells through its interaction with anti-apoptotic Bcl-2 family members.
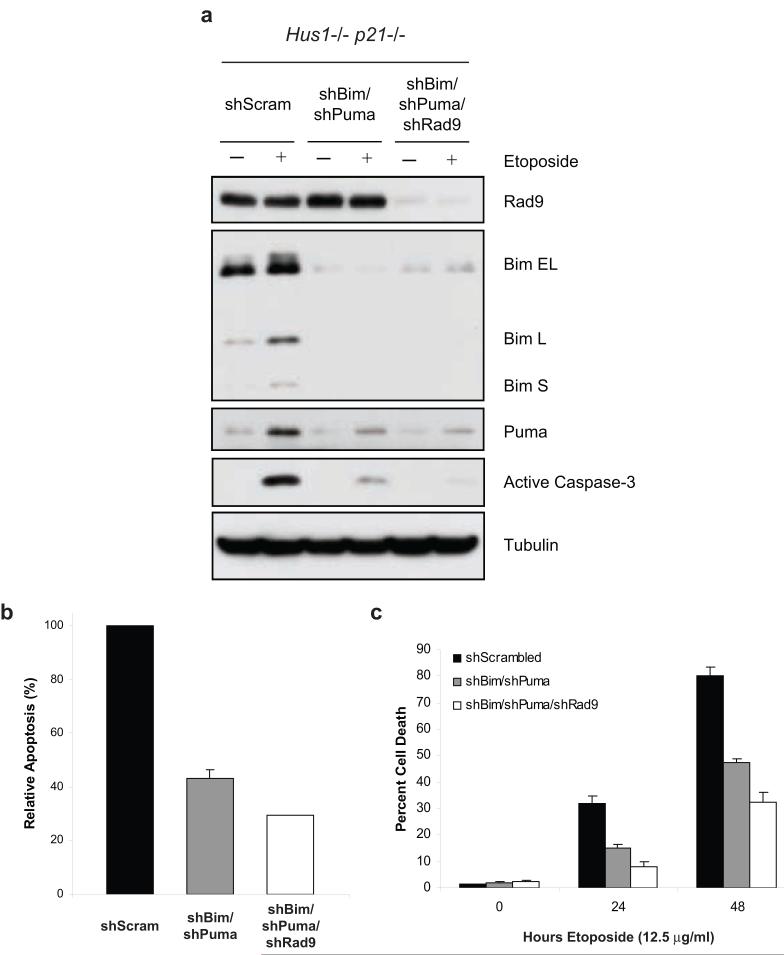
We therefore examined whether Rad9 cooperates with Bim and Puma to sensitize Hus1-deficient cells to etoposide-induced apoptosis. Hus1-/-p21-/- MEFs stably expressing shBim and shPuma were infected with lentivirus expressing shRNA targeting Rad9. Indeed, knockdown of Rad9 resulted in further inhibition of caspase-3 activation, as compared to the shBim and shPuma expressing cells (Figures 6a and b). Consistently, suppression of Rad9 expression in the shBim and shPuma expressing cells conferred further resistance to etoposide-induced cell death (Figure 6c). Taken together, these results indicate that Rad9 acts in collaboration with Bim and Puma to sensitize Hus1-deficient cells to etoposide-induced apoptosis.
Figure 6.
Rad9 collaborates with Bim and Puma to sensitize Hus1-deficient cells to etoposide-induced apoptosis. (a, b) Hus1-/-p21-/- MEFs stably expressing shBim and shPuma were infected with lentivirus expressing shRNA targeting Rad9. The cells were treated with 12.5 μg/ml etoposide or control DMSO for 16 h. (a) Knockdown of Rad9, as well as Bim and Puma, was confirmed by SDS-PAGE/immunoblot analysis. (b) Induction of apoptosis was measured using a caspase-3 activity assay. (c) Hus1-/-p21-/- MEFs stably expressing control shRNA, shBim and shPuma, or shBim and shPuma plus shRad9 were treated with 12.5 μg/ml etoposide for 0, 24 or 48 h. Cell death was determined by trypan blue exclusion assay (mean ± s.d.; n=3).
Discussion
In this study, we have demonstrated that loss of Hus1 results in the upregulation of the BH3-only proteins, Bim and Puma, which is partially responsible for sensitizing Hus1-deficient cells to etoposide-induced apoptosis. In addition, we found that in the absence of Hus1, Rad9 functions as a BH3-only protein and cooperates with Bim and Puma to promote apoptosis in response to etoposide treatment. There are currently two models for the activation of apoptosis by the Bcl-2 family members: the direct model and the hierarchy model (Galonek and Hardwick, 2006). The direct model proposes that BH3-only proteins have varying levels of potency due to their ability to bind various Bcl-2-like family members. Thus, Bim, Puma and tBid are the most potent as they can bind all of the anti-apoptotic Bcl-2 family members, whereas Noxa and Bad are less potent as they can only bind to a subset of the Bcl-2-like proteins. In the hierarchy model, on the other hand, Bim and Puma, as well as tBid, are more potent as they act downstream of the other BH3-only proteins and the Bcl-2-like proteins and can bind directly to the multi-domain pro-apoptotic proteins, resulting in their activation and the induction of apoptosis. This model suggests that loss of both Bim and Puma would result in complete inhibition of apoptosis mediated through the intrinsic pathway. Our results show that knockdown of both Bim and Puma diminishes the hypersensitivity of Hus1-deficient cells to etoposide-induced apoptosis, indicating that both Bim and Puma indeed play a central role in the activation of this programmed cell death pathway. Interestingly, suppression of Rad9 expression in the Bim and Puma double-knockdown cells resulted in further inhibition of DNA damage-induced apoptosis. Therefore, our data argue in favor of the direct model and suggest that it is the ratio of the BH3-only proteins to the Bcl-2 like proteins that regulates apoptosis.
Our results demonstrate that Bim and Puma mRNA expression is induced by etoposide treatment (Figure 3). Among known transcription factors, FoxO3a, p53 and E2F1 are the best candidates for regulators of Bim and Puma expression in response to etoposide treatment, as these transcription factors have been shown to upregulate Bim and Puma in response to DNA damage (Dijkers et al., 2000; Hershko and Ginsberg, 2004; Nakano and Vousden, 2001; Sunters et al., 2003; Yang et al., 2006). However, knockdown of either FoxO3a or E2F1 in Hus1-deficient cells did not inhibit etoposide-induced upregulation of Bim or Puma expression (data not shown). In addition, knockout of p53 resulted in only a slight suppression of Bim and moderate inhibition of Puma expression induced by etoposide treatment (data not shown). These results suggest that p53 is indeed involved in upregulating Puma expression and to a lesser extent Bim expression, but that other factors are also responsible for the induction of Bim and Puma in response to etoposide treatment. While we cannot rule out the possibility that loss of p53, FoxO3a or E2F1 results in compensation by other transcription factors, our results suggest that these transcription factors are not essential for the upregulation of Bim and Puma in response to etoposide treatment in Hus1-deficient cells. Therefore, further studies are needed to identify the transcription factors that are responsible for etoposide-induced Bim and Puma expression in cells lacking Hus1.
Rad9 has been shown to play a role in multiple cellular processes including: regulation of cell cycle checkpoints, transcriptional activation of p53 targets, initiation of DNA repair and when DNA repair is unfavorable, induction of apoptosis (Lieberman, 2006). It has been suggested that the primary function of Rad9 is to act as a sensor in the DNA damage response pathway to promote survival by initiating cell cycle arrest and facilitating DNA repair (Brandt et al., 2006; Loegering et al., 2004). Thus, it is not surprising that loss of Rad9 sensitizes cells to DNA damage as these cells lack the ability to activate appropriate cell cycle checkpoints and DNA repair. On the other hand, Rad9 may function as a pro-apoptotic factor in cells with unrepairable, excessively damaged DNA or a disrupted 9-1-1 complex, such as through loss of Hus1. Hus1-deficient cells have a defective cell cycle checkpoint, thus making them more sensitive to DNA damage. In the absence of Hus1, Rad9 was found to be mostly located in the cytosol, where it formed perinuclear foci and associated with Bcl-2 in response to DNA damage (Figures 5 and data not shown). These results suggest that loss of Hus1 results in an abrogation of the nuclear functions of Rad9 and an augmentation of its pro-apoptotic functions.
Taken together, the results presented in this report indicate that the 9-1-1 complex plays a critical role in the suppression of etoposide-induced apoptosis by regulating the induction of the BH3-only proteins, Bim and Puma. Loss of Hus1 results in enhanced upregulation of these BH3-only proteins that initiate mitochondrial apoptosis in response to DNA damage. Moreover, disruption of the 9-1-1 complex, by loss of Hus1, switches Rad9 from functioning as a mediator of cell cycle checkpoints to an inducer of apoptosis. Thus, the 9-1-1 complex may act as a checkpoint sensor to decide whether a cell should survive or undergo apoptosis in response to DNA damage.
Materials and methods
Cell culture and transfection
Hus1+/+p21 -/- and Hus1-/-p21-/- cells (Weiss et al., 2000), as well as Hus1-/-p21-/- + GFP and Hus1-/- p21-/- + Hus1 cells (Weiss et al., 2002), were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 1.0 mM L-glutamine, 0.1 mM MEM nonessential amino acids, 100 μg/ml streptomycin and 100 U/ml penicillin. The 21-nucleotide siRNA duplexes targeting Bim and GFP were synthesized and purified by Dharmacon (Lafayette, CO). The siRNA sequence targeting mouse Bim mRNA was 5′-AAUUACAACUGUUACGCUUUA-3′. The siRNA specific for GFP has previously been described (Hirai and Wang, 2002). Hus1-/-p21-/- MEFs were transfected with 10 μg of siRNA per 1 × 106 cells using the Nucleofector system. After transfection, the cells were allowed to recover for 36 h. The medium, containing any cells which may have died due to transfection, was removed and replaced with treatment medium containing control DMSO or etoposide. The pLKO.1-based lentiviral shRNAs targeting Bim (TRCN0000009692), Puma (TRCN0000009710) and Rad9 (TRCN0000012638) were purchased from Open Biosystems (Huntsville, AL). The pLKO.1-based scrambled control shRNA vector was purchased from Sigma (St. Louis, MO). Recombinant lentivirus was produced by co-transfecting the appropriate shRNA plasmid with the ViraPower Packaging Mix (Invitrogen, Carlsbad, CA) into 293FT cells. The resulting supernatant containing shRNA-expressing lentivirus was used to transduce Hus1-/- p21-/- MEFs according to the manufacturer’s protocol.
Analysis of cell death and apoptosis
Cell death was assessed by trypan blue exclusion assay. Apoptosis was scored by the presence of nuclear chromatin condensation and DNA fragmentation and evaluated by fluorescence microscopy. Briefly, cells were harvested, fixed in 4% paraformaldehyde for 10 min at room temperature and washed with PBS. Cell nuclei were stained with 0.5 μg/ml bis-benzimide trihydrochloride (Hoechst 33258, Molecular Probes, Eugene, OR). At least 200 cells were counted for each sample and percent apoptosis was calculated [(apoptotic nuclei) / (all nuclei) × 100]. The induction of apoptosis was analyzed using a caspase-3 activity assay (Sigma) and by examination of caspase-3 processing and PARP cleavage by SDS-PAGE/immunoblot analysis.
Semi-quantitative reverse transcription-PCR
Semi-quantitative reverse transcription-PCR was performed using the Qiagen (Valencia, CA) OneStep RT-PCR system according to the manufacturer’s recommendations. The primers for Bim and GAPDH have been previously described (Wong et al., 2005). The primers for Puma are 5′-GTGATCCGGACACGAAGACT-3′ and 5′-GACTCTAAGTGCTGCTGGGC-3′.
Subcellular fractionation and coimmunoprecipitation
Subcellular fractionation was performed as previously described (Wang et al., 1996). Chromatin fractionation was carried out as described previously (Mendez and Stillman, 2000). Coimmunoprecipitation of Rad9 with Bcl-2 was performed as previously described with minor modifications (Yoshida et al., 2002; Yoshida et al., 2003). Briefly, whole cell lysate was prepared in 1% NP-40 lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.1% SDS, 1 mM Na3VO4, 1 mM PMSF, 1 mM DTT, 10 mM NaF, 10 μg/ml aprotinin, 10 μg/ml leupeptin and 10μ g/ml pepstatin A). One milligram of the lysate was precleared by incubation with protein G agarose beads for 1 h at 4 °C. The precleared lysate was then incubated with anti-Bcl-2 (BD Pharmingen, Cat#554218) or anti-Flag monoclonal antibody as negative control overnight at 4 °C. The immunocomplexes were then incubated with protein G agarose beads for 1.5 h at 4 °C. After extensive washing in lysis buffer, the resulting immunocomplexes were subjected to SDS-PAGE/immunoblot analysis with anti-Rad9 polyclonal antibody. For coimmunoprecipitation analysis of the cytosolic fraction, NP-40, SDS and NaCl were added to a final concentration of 1%, 0.1% and 150 mM, respectively, after fractionation.
Supplementary Material
Acknowledgements
We thank the Analytical Microscopy Core facility at the H. Lee Moffitt Cancer Center and Research Institute for their technical assistance. This work was supported by grants from the National Institutes of Health (CA90315 to H-G.W. and CA108773 to R.S.W.) and the Department of Defense Breast Cancer Research Program (BC050563 to C.L.M).
References
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–96. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–37. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Bao S, Lu T, Wang X, Zheng H, Wang LE, Wei Q, et al. Disruption of the Rad9/Rad1/Hus1 (9-1-1) complex leads to checkpoint signaling and replication defects. Oncogene. 2004;23:5586–93. doi: 10.1038/sj.onc.1207753. [DOI] [PubMed] [Google Scholar]
- Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, et al. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci U S A. 2003;100:1633–8. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt PD, Helt CE, Keng PC, Bambara RA. The Rad9 protein enhances survival and promotes DNA repair following exposure to ionizing radiation. Biochem Biophys Res Commun. 2006;347:232–7. doi: 10.1016/j.bbrc.2006.06.064. [DOI] [PubMed] [Google Scholar]
- Burtelow MA, Kaufmann SH, Karnitz LM. Retention of the human Rad9 checkpoint complex in extraction-resistant nuclear complexes after DNA damage. J Biol Chem. 2000;275:26343–8. doi: 10.1074/jbc.M001244200. [DOI] [PubMed] [Google Scholar]
- Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage-responsive checkpoint complex. J Biol Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- Chen MJ, Lin YT, Lieberman HB, Chen G, Lee EY. ATM-dependent phosphorylation of human Rad9 is required for ionizing radiation-induced checkpoint activation. J Biol Chem. 2001;276:16580–6. doi: 10.1074/jbc.M008871200. [DOI] [PubMed] [Google Scholar]
- Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- Dang T, Bao S, Wang XF. Human Rad9 is required for the activation of S-phase checkpoint and the maintenance of chromosomal stability. Genes Cells. 2005;10:287–95. doi: 10.1111/j.1365-2443.2005.00840.x. [DOI] [PubMed] [Google Scholar]
- Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol. 2000;10:1201–4. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- Galonek HL, Hardwick JM. Upgrading the BCL-2 network. Nat Cell Biol. 2006;8:1317–9. doi: 10.1038/ncb1206-1317. [DOI] [PubMed] [Google Scholar]
- Hershko T, Ginsberg D. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem. 2004;279:8627–34. doi: 10.1074/jbc.M312866200. [DOI] [PubMed] [Google Scholar]
- Hirai I, Wang HG. A role of the C-terminal region of human Rad9 (hRad9) in nuclear transport of the hRad9 checkpoint complex. J Biol Chem. 2002;277:25722–7. doi: 10.1074/jbc.M203079200. [DOI] [PubMed] [Google Scholar]
- Hopkins KM, Auerbach W, Wang XY, Hande MP, Hang H, Wolgemuth DJ, et al. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol Cell Biol. 2004;24:7235–48. doi: 10.1128/MCB.24.16.7235-7248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DC, Strasser A. BH3-Only proteins-essential initiators of apoptotic cell death. Cell. 2000;103:839–42. doi: 10.1016/s0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]
- Ishii H, Inageta T, Mimori K, Saito T, Sasaki H, Isobe M, et al. Frag1, a homolog of alternative replication factor C subunits, links replication stress surveillance with apoptosis. Proc Natl Acad Sci U S A. 2005;102:9655–60. doi: 10.1073/pnas.0504222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzel B, Hall J, Natt F, Weiler J, Cohen D. Downregulation of Hus1 by antisense oligonucleotides enhances the sensitivity of human lung carcinoma cells to cisplatin. Cancer. 2002;94:1808–14. doi: 10.1002/cncr.10383. [DOI] [PubMed] [Google Scholar]
- Komatsu K, Hopkins KM, Lieberman HB, Wang H-G. Schizosaccharomyces pombe rad9 contains a BH3-like region and interacts with the anti-apoptotic protein bcl-2. FEBS Lett. 2000a;481:122–6. doi: 10.1016/s0014-5793(00)01975-x. [DOI] [PubMed] [Google Scholar]
- Komatsu K, Miyashita T, Hang H, Hopkins KM, Zheng W, Cuddeback S, et al. Human homologue of S. pombe Rad9 interacts with BCL-2/BCL-xL and promotes apoptosis. Nat Cell Biol. 2000b;2:1–6. doi: 10.1038/71316. [DOI] [PubMed] [Google Scholar]
- Lee MW, Hirai I, Wang H-G. Caspase-3-mediated cleavage of Rad9 during apoptosis. Oncogene. 2003;22:6340–6346. doi: 10.1038/sj.onc.1206729. [DOI] [PubMed] [Google Scholar]
- Lieberman HB. Rad9, an evolutionarily conserved gene with multiple functions for preserving genomic integrity. J Cell Biochem. 2006;97:690–7. doi: 10.1002/jcb.20759. [DOI] [PubMed] [Google Scholar]
- Lindsey-Boltz LA, Bermudez VP, Hurwitz J, Sancar A. Purification and characterization of human DNA damage checkpoint Rad complexes. Proc Natl Acad Sci U S A. 2001;98:11236–41. doi: 10.1073/pnas.201373498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loegering D, Arlander SJ, Hackbarth J, Vroman BT, Roos-Mattjus P, Hopkins KM, et al. Rad9 protects cells from topoisomerase poison-induced cell death. J Biol Chem. 2004;279:18641–7. doi: 10.1074/jbc.M313536200. [DOI] [PubMed] [Google Scholar]
- Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol. 2002;14:237–45. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–12. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A, Biamonti G. Cellular response to etoposide treatment. Cancer Lett. 2007;252:9–18. doi: 10.1016/j.canlet.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–94. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- Niida H, Nakanishi M. DNA damage checkpoints in mammals. Mutagenesis. 2006;21:3–9. doi: 10.1093/mutage/gei063. [DOI] [PubMed] [Google Scholar]
- Oltvai ZN, Korsmeyer SJ. Checkpoints of dueling dimers foil death wishes. Cell. 1994;79:189–192. doi: 10.1016/0092-8674(94)90188-0. [DOI] [PubMed] [Google Scholar]
- Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 2004;3:1009–14. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, Strasser A. Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ. 2002;9:505–12. doi: 10.1038/sj.cdd.4400998. [DOI] [PubMed] [Google Scholar]
- Rauen M, Burtelow MA, Dufault VM, Karnitz LM. The human checkpoint protein hRad17 interacts with the PCNA-like proteins hRad1, hHus1, and hRad9. J Biol Chem. 2000;275:29767–71. doi: 10.1074/jbc.M005782200. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Strasser A. The role of BH3-only proteins in the immune system. Nat Rev Immunol. 2005;5:189–200. doi: 10.1038/nri1568. [DOI] [PubMed] [Google Scholar]
- Sunters A, Fernandez de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003;278:49795–805. doi: 10.1074/jbc.M309523200. [DOI] [PubMed] [Google Scholar]
- Volkmer E, Karnitz LM. Human homologs of Schizosaccharomyces pombe rad1, hus1, and rad9 form a DNA damage-responsive protein complex. J Biol Chem. 1999;274:567–570. doi: 10.1074/jbc.274.2.567. [DOI] [PubMed] [Google Scholar]
- Wang H-G, Rapp UR, Reed JC. Bcl-2 targets the protein kinase Raf-1 to mitochondria. Cell. 1996;87:629–38. doi: 10.1016/s0092-8674(00)81383-5. [DOI] [PubMed] [Google Scholar]
- Wang X, Zou L, Zheng H, Wei Q, Elledge SJ, Li L. Genomic instability and endoreduplication triggered by RAD17 deletion. Genes Dev. 2003;17:965–70. doi: 10.1101/gad.1065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RS, Enoch T, Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 2000;14:1886–98. [PMC free article] [PubMed] [Google Scholar]
- Weiss RS, Leder P, Vaziri C. Critical role for mouse Hus1 in an S-phase DNA damage cell cycle checkpoint. Mol Cell Biol. 2003;23:791–803. doi: 10.1128/MCB.23.3.791-803.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RS, Matsuoka S, Elledge SJ, Leder P. Hus1 acts upstream of chk1 in a mammalian DNA damage response pathway. Curr Biol. 2002;12:73–7. doi: 10.1016/s0960-9822(01)00626-1. [DOI] [PubMed] [Google Scholar]
- Willis SN, Adams JM. Life in the balance: how BH3-only proteins induce apoptosis. Curr Opin Cell Biol. 2005;17:617–25. doi: 10.1016/j.ceb.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HK, Fricker M, Wyttenbach A, Villunger A, Michalak EM, Strasser A, et al. Mutually exclusive subsets of BH3-only proteins are activated by the p53 and c-Jun N-terminal kinase/c-Jun signaling pathways during cortical neuron apoptosis induced by arsenite. Mol Cell Biol. 2005;25:8732–47. doi: 10.1128/MCB.25.19.8732-8747.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Xia W, Hu MC. Ionizing radiation activates expression of FOXO3a, Fas ligand, and Bim, and induces cell apoptosis. Int J Oncol. 2006;29:643–8. [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Komatsu K, Wang HG, Kufe D. c-Abl tyrosine kinase regulates the human Rad9 checkpoint protein in response to DNA damage. Mol. Cell. Biol. 2002;22:3292–3300. doi: 10.1128/MCB.22.10.3292-3300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Wang HG, Miki Y, Kufe D. Protein kinase Cdelta is responsible for constitutive and DNA damage-induced phosphorylation of Rad9. Embo J. 2003;22:1431–41. doi: 10.1093/emboj/cdg134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–9. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.