Abstract
The skeletal muscle (RyR1) and cardiac muscle (RyR2) ryanodine receptor calcium release channels contain a single, conserved calmodulin (CaM) binding domain, yet are differentially regulated by CaM. Here, we report that high-affinity [35S]CaM binding to RyR1 is driven by favorable enthalpic and entropic contributions at Ca2+ concentrations from <0.01 to 100 μM. At 0.15 μM Ca2+, [35S]CaM bound to RyR2 with decreased affinity and binding enthalpy compared with RyR1. The rates of [35S]CaM dissociation from RyR1 increased as the temperature was raised, whereas at 0.15 μM Ca2+ the rate from RyR2 was little affected. The results suggest major differences in the energetics of CaM binding to and dissociation from RyR1 and RyR2.
Keywords: Ca2+ release channel, sarcoplasmic reticulum, ryanodine receptor, binding enthalpy, binding entropy
Introduction
In skeletal and cardiac muscle, the release of Ca2+ ions through ryanodine receptor calcium release channels (RyRs) into the cytoplasm leads to muscle contraction. RyR1 is present at high levels in skeletal muscle, and RyR2 is the dominant isoform in cardiac muscle. A muscle action potential initiates L-type Ca2+ channel protein conformational changes that either permit an influx of extracellular Ca2+ (in cardiac muscle)[1] or alter the conformation of the RyRs by a direct physical interaction (in skeletal muscle)[2] with both mechanisms leading to the release of Ca2+ from the sarcoplasmic reticulum (SR) via the RyRs and subsequent muscle contraction. The skeletal muscle and cardiac muscle RyRs are massive ion channels that are composed of four RyR 560 kDa peptide subunits, four small 12 kDa FK506 binding proteins (FKBP), and various associated proteins that include calmodulin.[3-5]
Calmodulin (CaM) is a small cytoplasmic Ca2+ binding protein that is comprised of two globular domains separated by a central alpha helical domain. Each of the N- and C-terminal domains contains two EF-hand Ca2+ binding sites.[6-8] The two Ca2+ binding sites in the C-lobe have a higher Ca2+ affinity than the two sites in the N-lobe, resulting in nearly full occupancy at 100 μM Ca2+, binding of only one or two Ca2+ to the C-lobe at <1 μM Ca2+, and little binding at <0.01 μM free Ca2+. A notable property of the RyRs is that they interact with both the Ca2+-free (apoCaM) and Ca2+-bound (CaCaM) forms of CaM. CaM inhibits RyR1 and RyR2 at micromolar Ca2+ concentrations, whereas at submicromolar Ca2+ concentrations, RyR1 is activated but RyR2 is inhibited by CaM.[9-13] CaM binding to the intact receptors indicates that the RyRs have a single, high-affinity binding domain for apoCaM and CaCaM.[9][10][13][14] Use of CaM mutants and fragments of CaM binding domain, and structural studies have suggested that the C-lobe of CaM tightly binds to the highly conserved CaM binding site, affording the N-lobe of CaM to interact with other RyR regions.[15-17] To better understand the mechanism of CaM regulation of RyRs, the present study compares the thermodynamic parameters of CaM binding to RyR1 and RyR2.
Materials and Methods
Materials
[3H]Ryanodine was obtained from Perkin Elmer (Boston, MA), Tran35S-Label from MP Biomedicals (Irvine, CA), unlabeled ryanodine from EMD Biosciences (La Jolla, CA), and complete protease inhibitors from Roche Diagnostics (Indianapolis, IN). Unlabeled CaM and [35S]CaM were prepared as described.[13]
SR vesicle preparations
SR vesicles enriched in RyRs were isolated in presence of protease inhibitors from rabbit hind limb and back muscle[18] and canine cardiac muscle.[19]
[35S]Calmodulin binding
Effects of temperature on calmodulin binding to RyR1 and RyR2 were determined using CaM metabolically labeled with 35S.[13] Unless otherwise indicated, SR membranes (100-150 μg/mL) were incubated for 2-5 h with [35S]CaM in 150 mM KCl, 20 mM imidazole (pH 7.0), 5 mM reduced glutathione, 0.1 mg/mL bovine serum albumin (Sigma A-0281), 0.2 mM Pefabloc, 20 μM leupeptin with either 100 μM (200 μM CaCl2, 100 μM EGTA), 0.15 μM (0.55 mM CaCl2, 1 mM EGTA) or <0.01 μM (5 mM EGTA, no added CaCl2) free Ca2+. Equilibrium was assumed to have been reached when no additional binding was measured at more extended times. Nonspecific binding was determined using a 100-1000 fold excess of unlabeled calmodulin. SR membranes were sedimented by centrifugation in a Beckman Airfuge at the same temperature as used for incubation for 30 min at 90,000g, which should have minimally affected binding because it was performed in presence of constant concentration of free CaM in the solution. Bound [35S]CaM was determined by liquid scintillation counting following solubilization of pellets in 50 mM Tris HCl buffer (pH 8.5) containing 2% sodium dodecylsulfate.
The time course of [35S]CaM dissociation was determined with the use of a filter assay. To minimize nonspecific binding of [35S]CaM, Whatman GF/B filters were blocked for 1 h in 0.15M KCl, 10 mM KPipes, pH 7.0 buffer containing 10 mg/mL bovine serum albumin. Vesicles on the filters were washed with 3 × 5 mL of 0.15 KCl, 10 mM KPipes, pH 7.0 buffer containing 0.1 mg/mL bovine serum albumin.
[3H]Ryanodine binding
Bmax values of [3H]ryanodine binding were determined using a saturating [3H]ryanodine concentration (30 nM) in 0.6M KCl buffer.[13] Bmax values of [3H]ryanodine binding to RyR1 and RyR2 were 7-15 and 3-6 pmol/mg or 28-60 and 12-24 pmol/mg of [35S]CaM binding sites, respectively, as there are four CaM binding sites per high affinity [3H]ryanodine binding site.[13]
Biochemical assays and data analyses
Free Ca2+ concentrations of ≥1 μM were determined with the use of a Ca2+ selective electrode and concentrations of <1 μM with the use of Ca2+ indicator dye Fluo-3. Results are given as mean ± SE.
Results
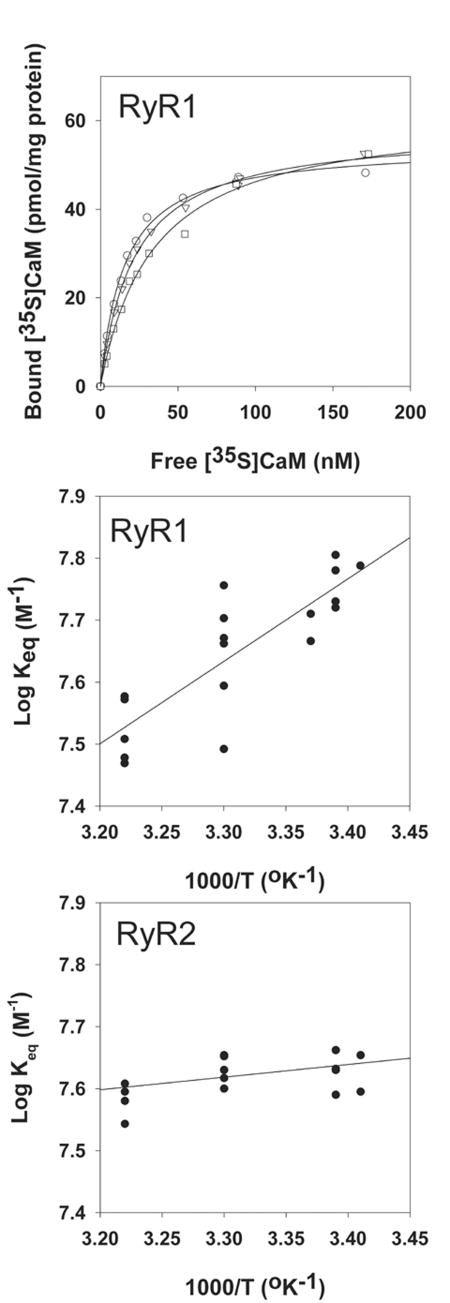
Skeletal muscle and cardiac muscle high molecular weight proteins (now known to be the RyRs) are the major proteins labeled by azido-[125I]CaM in cardiac and skeletal SR membranes.[20] [35S]CaM binding studies confirmed that RyR1 is the principle CaM binding protein of skeletal muscle SR membranes.[21] Figure 1 (top panel) shows three [35S]CaM binding curves to skeletal muscle SR membranes in 0.15 μM free Ca2+ media at temperatures ranging from 22 to 37°C. Bound amounts corresponded to 4.5 ± 0.5 mols of [35S]CaM per mol bound [3H]ryanodine or ∼ 1 bound CaM per RyR1 subunit, as there is only one high affinity [3H]ryanodine binding site per RyR tetramer.[13] Inspection of the binding curves indicated that an increase in temperature decreased CaM binding affinity to RyR1, that is, complex formation was driven by a favorable enthalpic contribution. van't Hoff analysis of CaM binding data yielded averaged enthalpy of -26 ± 4 kJ/mol (Fig. 1, middle panel). Table I compares the averaged thermodynamic parameters of CaM binding to RyR1 at 26°C and 0.15 μM free Ca2+ with those at <0.01 μM free Ca2+ (apoCaM binding) and 100 μM free Ca2+ (CaCaM binding). Increase in free Ca2+ concentration from <0.01 μM to 0.15 μM increased the affinity and entropy of CaM binding to RyR1, whereas binding enthalpy was decreased. Further increase in Ca2+ concentration to 100 μM resulted in only small changes in the three parameters. In contrast to RyR1, CaM binding to RyR2 proceeded with an enthalpy near zero and increased entropy at 0.15 μM Ca2+, which suggests that the reaction was entropically driven (Fig. 1, bottom panel; Table I). Thermodynamic parameters of CaM binding to RyR2 were not determined at <0.01 μM Ca2+ because of a low binding affinity, and not at 100 μM Ca2+ because CaM bound to additional sites most likely present in other proteins of cardiac SR membrane fractions.[13]
Figure 1.
Effect of temperature on [35S]CaM equilibrium binding to RyR1. (Top panel) Examples of [35S]CaM binding to skeletal muscle SR membranes. Membranes were incubated as described in “Materials and Methods” section at 22°C (○), 30°C (▽) or 37°C (□) in 0.15 μM free Ca2+ media. Data were fitted by regression analysis (solid lines) with the following Bmax (pmol/mg protein) and Keq (106 M-1) values, respectively: 54.4 ± 1.0 and 65.0 ± 4.0 at 22°C, 58 ± 0.8 and 46.0 ± 2.0 at 30°C, and 61.9 ± 2.1 and 29.3 ± 2.7 at 37°C. (Middle and bottom panels) Van't Hoff plots of [35S]CaM binding to RyR1 and RyR2, respectively. Averaged thermodynamic parameters obtained by linear regression analysis (solid lines) are shown in Table I. R2 and P values were 0.70 and <0.0001 (middle panel) and 0.25 and 0.06 (lower panel), respectively.
Table I. Thermodynamic Parameters of CaM Binding to RyR1 and RyR2.
| [Ca2+] (μM) | Keq (106M-1) | ΔH (KJ/mol) | ΔS (J/mol degree) | |
|---|---|---|---|---|
| RyR1 | <0.01 | 38 ± 4 | -38 ± 7 | 23 ± 22 |
| 0.15 | 50 ± 4 | -26 ± 4 | 62 ± 14 | |
| 100 | 60 ± 5 | -22 ± 7 | 77 ± 24 | |
| RyR2 | 0.15 | 24 ± 2 | -4 ± 2 | 98 ± 7 |
Keq values and thermodynamic parameters of [35S]CaM binding to RyRs at 26°C. Keq and ΔH values were determined by linear regression analysis, as shown in Figure 1.
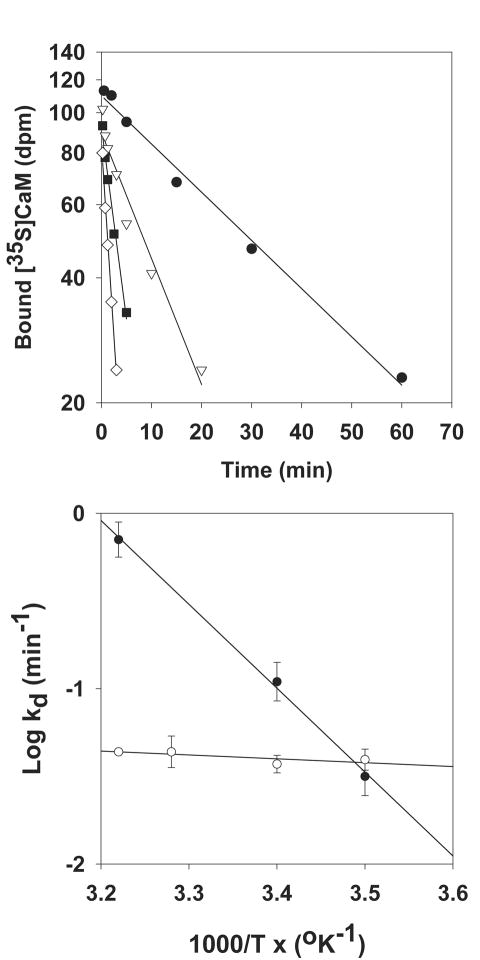
[35S]CaM dissociation experiments were performed to determine the effects of temperature on the stability of the CaM-RyR complexes. As shown in Figure 2(top panel), the rate of [35S]CaM displacement from RyR1 by 60-fold excess of unlabeled CaM increased at 0.15 μM free Ca2+ as the temperature was raised from 12 to 37°C. An Arrhenius plot of the rate constants yielded averaged activation energy Ed = 84 ± 5 kJ/mol (Fig. 2, bottom panel). Table II shows that the dissociation rate constant for RyR1 was decreased twofold at 26°C in presence of 100 μM Ca2+ compared with <0.01 μM Ca2+, whereas the activation energy of dissociation was independent of Ca2+ concentration. In contrast to RyR1, the dissociation rate constant of CaM from RyR2 was nearly independent of temperature at 0.15 μM Ca2+, yielding an activation energy of dissociation 20-fold lower for RyR2 than RyR1. Taken together, data of Tables I and II suggest major differences in the thermodynamics of interaction between CaM and RyR1 or RyR2.
Figure 2.
Effect of temperature on [35S]CaM dissociation from skeletal SR membranes. (Top panel) Skeletal muscle SR membranes were labeled with [35S]CaM by incubation for 2 h at 24°C in media containing 0.15 μM free Ca2+ and 100 nM [35S]CaM. [35S]CaM dissociation was initiated at 12°C (■), 23°C (▽), 32°C (■), or 37°C (○) by diluting membranes 60-fold into 0.15 μM free Ca2+ media containing 100 nM unlabeled CaM. Dissociation constants (kd) were obtained by linear regression analysis (solid lines). (Bottom panel) Arrhenius plots of CaM dissociation from RyR1 (■) and RyR2 (○), respectively. Averaged activation energies (Ed) obtained by linear regression analysis (solid lines) are shown in Table II. R2 and p values were 0.97 and <0.0001 (■) and 0.01 and 0.73 (○), respectively.
Table II. Rate Constants and Activation Energies of CaM Dissociation fromRyR1 and RyR2.
| [Ca2+] (μM) | Kd (min-1) | Ed (kJ/mol) | |
|---|---|---|---|
| RyR1 | <0.01 | 0.20 ± 0.05 | 91 ± 13 |
| 0.15 | 0.15 ± 0.03 | 84 ± 5 | |
| 100 | 0.10 ± 0.02 | 98 ± 8 | |
| RyR2 | 0.15 | 0.41 ± 0.05 | 4 ± 12 |
Dissociation rate constants (kd) and activation energies (Ed) of [35S]CaM dissociation from RyR1 and RyR2 at 26°C. Ed values were determined by linear regression analysis, as shown in Figure 2.
Discussion
Cryo-electron microscopy has indicated that apoCaM and CaCaM exert their functional effects by binding to an overlapping region that is a considerable distance away from the effector site (ion pore) of RyR1,[22] which suggests that CaM exerts its function via long-range interactions involving appreciable parts of the massive RyRs. CaM binding to the intact receptors confirmed that the RyRs have a single conserved binding domain for apoCaM and CaCaM.[9][10][13][14] The CaM binding domain in RyR1 is highly conserved among the mammalian RyRs; however, differences in the interaction of the CaM with the RyRs have been observed. At submicromolar Ca2+ concentrations, RyR1 is activated whereas RyR2 is inhibited by CaM.[13] Site directed mutagenesis in CaM binding domain affected CaM binding and regulation to a different extent in the two isoforms.[23] Consistent with this observation, comparison of thermodynamics shows that CaM interacts with the two RyRs in different ways. At 0.15 μM Ca2+, RyR2 CaM complex formation was driven by entropy with only a small enthalpic contribution, in contrast to RyR1, which showed similar enthalpic and entropic gains on CaM binding. Furthermore, at 0.15 μM Ca2+ the activation energy of CaM dissociation was much lower for RyR2 than RyR1. A more detailed understanding of the conformational changes and associated energetics of CaM binding will require that the solution structures of the RyRs and RyR-CaM complexes become available.
Acknowledgments
Funded by:
National Institutes of Health; Grant Number: AR 018687, HL 073051
American Heart Association; Grant Number: 0665361U
Macromolecular Interactions Facility of University of North Carolina at Chapel Hill
Abbreviations
- apoCaM
Ca2+-free CaM
- CaM
calmodulin
- CaCaM
Ca2+-bound CaM
- RyR
ryanodine receptor
- RyR1
skeletal muscle RyR
- RyR2
cardiac muscle RyR
- SR
sarcoplasmic reticulum
References
- 1.Fabiato A. Calcium-induced release of calcium from the cardiac sarcoplasmic reticulum. Am J Physiol. 1983;245:C1–C14. doi: 10.1152/ajpcell.1983.245.1.C1. [DOI] [PubMed] [Google Scholar]
- 2.Rios E, Pizarro G. Voltage sensor of excitation-contraction coupling in skeletal muscle. Physiol Rev. 1991;71:849–908. doi: 10.1152/physrev.1991.71.3.849. [DOI] [PubMed] [Google Scholar]
- 3.Fill M, Copello JA. Ryanodine receptor calcium release channels. Physiol Rev. 2002;82:893–922. doi: 10.1152/physrev.00013.2002. [DOI] [PubMed] [Google Scholar]
- 4.Franzini-Armstrong C, Protasi F. Ryanodine receptors of striated muscles: a complex channel capable of multiple interactions. Physiol Rev. 1997;77:699–729. doi: 10.1152/physrev.1997.77.3.699. [DOI] [PubMed] [Google Scholar]
- 5.Meissner G. Regulation of mammalian ryanodine receptors. Frontiers Biosci. 2002;7:d2072–d2080. doi: 10.2741/A899. [DOI] [PubMed] [Google Scholar]
- 6.Babu YS, Sack JS, Greenhough TJ, Bugg CE, Means AR, Cook WJ. Three-dimensional structure of calmodulin. Nature. 1985;315:37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- 7.Klee CB, Vanaman TC. Calmodulin Adv Protein Chem. 1982;35:213–321. doi: 10.1016/s0065-3233(08)60470-2. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, Tanaka T, Ikura M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nature Struct Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 9.Fruen BR, Bardy JM, Byrem TM, Strasburg GM, Louis CF. Differential Ca2+ sensitivity of skeletal and cardiac muscle ryanodine receptors in the presence of calmodulin. Am J Physiol. 2000;279:C724–C733. doi: 10.1152/ajpcell.2000.279.3.C724. [DOI] [PubMed] [Google Scholar]
- 10.Rodney GG, Williams BY, Strasburg GM, Beckingham K, Hamilton SL. Regulation of RYR1 activity by Ca2+ and calmodulin. Biochemistry. 2000;39:7807–7812. doi: 10.1021/bi0005660. [DOI] [PubMed] [Google Scholar]
- 11.Buratti R, Prestipino G, Menegazzi P, Treves S, Zorzato F. Calcium dependent activation of skeletal muscle Ca2+ release channel (ryanodine receptor) by calmodulin. Biochem Biophys Res Com. 1995;213:1082–1090. doi: 10.1006/bbrc.1995.2238. [DOI] [PubMed] [Google Scholar]
- 12.Tripathy A, Xu L, Mann G, Meissner G. Calmodulin activation and inhibition of skeletal muscle Ca2+ release channel (ryanodine receptor) Biophys J. 1995;69:106–119. doi: 10.1016/S0006-3495(95)79880-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balshaw DM, Xu L, Yamaguchi N, Pasek DA, Meissner G. Calmodulin binding and inhibition of cardiac muscle calcium release channel (ryanodine receptor) J Biol Chem. 2001;276:20144–20153. doi: 10.1074/jbc.M010771200. [DOI] [PubMed] [Google Scholar]
- 14.Moore CP, Rodney G, Zhang JZ, Santacruz-Toloza L, Strasburg G, Hamilton SL. Apocalmodulin and Ca2+ calmodulin bind to the same region on the skeletal muscle Ca2+ release channel. Biochemistry. 1999;38:8532–8537. doi: 10.1021/bi9907431. [DOI] [PubMed] [Google Scholar]
- 15.Xiong LW, Newman RA, Rodney GG, Thomas O, Zhang JZ, Persechini A, Shea MA, Hamilton SL. Lobe-dependent regulation of ryanodine receptor type 1 by calmodulin. J Biol Chem. 2002;277:40862–40870. doi: 10.1074/jbc.M206763200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Zhang JZ, Danila CI, Hamilton SL. A noncontiguous, intersubunit binding site for calmodulin on the skeletal muscle Ca2+ release channel. J Biol Chem. 2003;278:8348–8355. doi: 10.1074/jbc.M209565200. [DOI] [PubMed] [Google Scholar]
- 17.Maximciuc AA, Putkey JA, Shamoo Y, MacKenzie KR. Complex of calmodulin with a ryanodine receptor target reveals a novel, flexible binding mode. Structure. 2006;14:1547–1556. doi: 10.1016/j.str.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Meissner G. Adenine nucleotide stimulation of Ca2+-induced Ca2+ release in sarcoplasmic reticulum. J Biol Chem. 1984;259:2365–2374. [PubMed] [Google Scholar]
- 19.Meissner G, Henderson JS. Rapid calcium release from cardiac sarcoplasmic reticulum vesicles is dependent on Ca2+ and is modulated by Mg2+, adenine nucleotide, and calmodulin. J Biol Chem. 1987;262:3065–3073. [PubMed] [Google Scholar]
- 20.Seiler S, Wegener AD, Whang DD, Hathaway DR, Jones LR. High molecular weight proteins in cardiac and skeletal muscle junctional sarcoplasmic reticulum vesicles bind calmodulin, are phosphorylated, and are degraded by Ca2+-activated protease. J Biol Chem. 1984;259:8550–8557. [PubMed] [Google Scholar]
- 21.Balshaw DM, Yamaguchi N, Meissner G. Modulation of intracellular calcium-release channels by calmodulin. J Membr Biol. 2002;185:1–8. doi: 10.1007/s00232-001-0111-4. [DOI] [PubMed] [Google Scholar]
- 22.Samso M, Wagenknecht T. Apocalmodulin and Ca2+-calmodulin bind to neighboring locations on the ryanodine receptor. J Biol Chem. 2002;277:1349–1353. doi: 10.1074/jbc.M109196200. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi N, Xu L, Pasek DA, Evans KE, Chen SR, Meissner G. Calmodulin regulation and identification of calmodulin binding region of type-3 ryanodine receptor calcium release channel. Biochemistry. 2005;44:15074–15081. doi: 10.1021/bi051251t. [DOI] [PubMed] [Google Scholar]