Abstract
The morphology of insect antennae varies widely among species, but our understanding of antennal development comes almost solely from studies of one species - the fruit fly, Drosophila melanogaster. Moreover, this knowledge applies mostly to adult structures, since Drosophila lacks external larval appendages. In contrast to Drosophila, the red flour beetle, Tribolium castaneum, has both larval and adult antennae, which are very different from one another in morphology. Thus, Tribolium provides an ideal system to compare modes of antennal development both within and between species. Here we report that the Tribolium ortholog of spineless (Tc-ss) is required in both the larval and adult antennae. Knockdown of Tc-ss by RNAi during either larval or imaginal development causes transformation of the distal portion of the antennae to legs. Thus, the function of ss is conserved between Drosophila and Tribolium with respect to adult antennal specification, and also between Tribolium larval and adult antennal development. The similarity of the Tc-ss RNAi phenotype to that of a classically described Tribolium mutation, antennapedia (ap) (of no relationship to the Drosophila Hox gene of the same name), led us to characterize the original ap mutation and two newly identified ap alleles. Our mapping and phenotypic data suggest Tc-ss is the best candidate for the ap locus. These results represent a first step in characterizing larval and adult antennal patterning in Tribolium, which should provide important insights into the evolution of insect antennal development.
Keywords: Tribolium, antenna, spineless, appendage, insect
Introduction
Virtually all our knowledge about the specification of insect antennal identity comes from analysis of Drosophila imaginal development. These studies suggest that the key factor in determining antenna versus leg identity is the extent to which homothorax (hth) and Distal-less (Dll) expression overlap within the developing imaginal disc (Dong et al., 2000). Co-expression of these genes triggers a cascade of gene expression that ultimately leads to antennal identity (Dong et al. 2002; Emerald et al. 2003). An important target of these genes is spineless (ss) (Dong et al. 2002; Emmons et al. 2007), a member of the bHLH-PAS transcription factor family (Duncan et al. 1998). Loss of ss function causes transformation of the distal antennal segments (A3 and arista) to leg, indicating that ss plays a key role in the specification of antennal identity (Duncan et al. 1998; Struhl 1982). Like many genes involved in antennal development, ss is also expressed in the legs (Duncan et al. 1998) where it is required for normal development of the tarsal segments (Struhl 1982). However, expression in the antennae is more extensive and persistent than that in the legs, with a high level of expression apparently being important for antennal identity (Duncan et al. 1998). In addition to these appendage roles, ss has an additional function in bristle development as shown by the reduced bristle size of ss mutants.
Although ss is strongly expressed in the Drosophila embryonic antennal segment (Duncan et al. 1998; Emmons et al. 2007), it is not clear whether it actually plays a role in establishing embryonic antennal identity. The antennae of Drosophila larvae are represented only by sense organs, which in ss mutants are somewhat abnormal and/or sclerotized, but not obviously transformed (Duncan et al. 1998).
In contrast to Drosophila, the red flour beetle, Tribolium castaneum, has both larval and adult antennae. Antennal morphology is drastically different at these two life stages (compare Fig. 1d to 2a), suggesting that the antennal tissue undergoes substantial re-patterning during metamorphosis. This raises the question of whether the same genes specify and pattern antennae during embryonic and imaginal development. To begin to address this issue, we have examined the function of the Tribolium ss ortholog. We show here that RNAi-induced knockdown of Tc-ss during either embryonic or adult development causes transformation of antennae to legs, suggesting that at least some aspects of patterning are conserved between larval and adult antennae. We also revisit the classical mutation antennapedia (ap) - of no relationship to the Hox gene Antennapedia - which causes transformation of the antennae to legs (Englert and Bell 1963). We describe two new alleles of ap and provide phenotypic and mapping evidence that suggests ap corresponds to the Tc-ss locus.
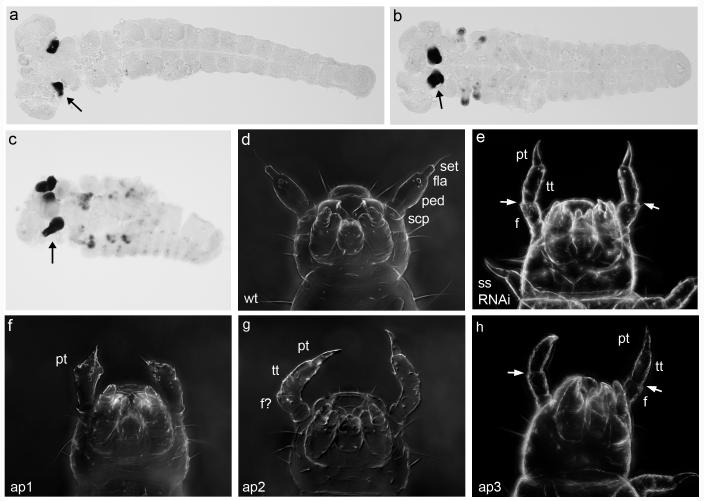
Fig. 1.
Embryonic expression and larval phenotypes. a-c Tc-ss expression in Tribolium embryos. An arrow points to the antennal expression in each panel. a Tc-ss expression in the antennae of an extended germ band stage embryo. b As the germband retracts, Tc-ss is expressed in the maxillary and labial appendages, as well as the antennae. c A dorsally closing embryo continues to show strong Tc-ss expression the antennae as well as patches of expression in the other appendages. Additional punctate staining probably corresponds to peripheral nervous system cells. d-h Cuticle preparations of first instar larvae. d Wild-type larval antennae have a basal scape (scp), a pedicel (ped), a short cylindrical flagellar segment (fla) and a terminal seta (set). The antennae of Tc-ss RNAi (e), ap1 (f), ap2 (g) and ap3 (h) larvae are transformed toward leg. Leg segments are identified where relevant: pretarsus (pt), tibiotarsus (tt) and femur (f). Arrows in e and h point to the clear boundary between femur and tibiotarsus.
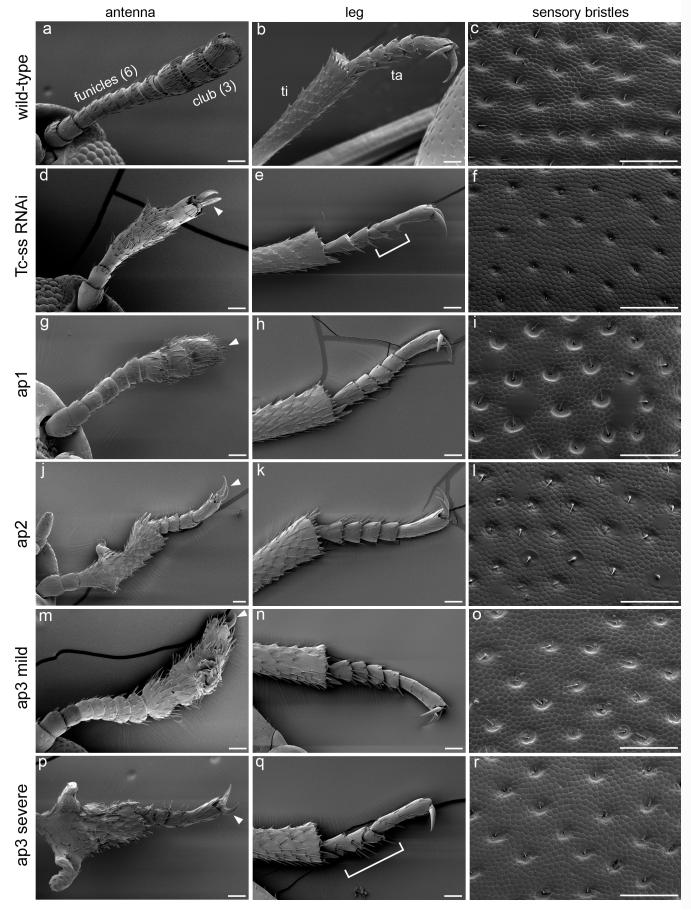
Fig. 2.
Scanning electron micrographs of adult structures. Scale bars represent 50 um. a The wild-type antenna has two basal segments, six funicle segments and three club segments. b The distal portion of a wild-type T2 leg consists of the tibia (ti), five tarsal segments (ta) and a terminal claw. c Sensory bristles on a wild-type pronotum. These structures are also shown for Tc-ss RNAi (d-f), ap1 (g-i), ap2 (j-l), mildly affected ap3 (m-o), and severely affected ap3 (p-r) adults. Brackets in e and q denote regions of tarsal fusion. Arrowheads point to terminal claws, which are indicative of leg identity.
Materials and Methods
Sequence Analysis
BLAST analyses against the Tribolium genome and predicted genes (Tribolium Genome Sequencing Consortium 2008) were performed at the NCBI (http://www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid=7070) and Baylor College of Medicine (http://www.hgsc.bcm.tmc.edu/blast.hgsc?organism=13) web sites. BLAST analyses against Drosophila melanogaster sequences were performed at FlyBase (http://flybase.bio.indiana.edu/blast/).
Gene Cloning
Predicted coding sequences (NCBI RefSeq XP_967876, GLEAN_11104 and GLEAN_11105) identified by BLAST searches with Drosophila Spineless were used to design primers (ss3 (5′-ggaattcaccgtctccaccg-3′) and ss4 (5′-acatactcccgctcctccattt-3′)) to amplify a 1.875 kb fragment of the predicted Tc-ss cDNA. The resulting cDNA fragment was cloned and sequenced (GenBank accession EU912437).
In situ hybridization
The 1.875 kb fragment of Tc-ss described above was used as template for riboprobe synthesis by standard methods. In situ hybridization was performed essentially as described by Nagaso et al. (2001)
RNAi
A 755 bp fragment of Tc-ss (bases 785-1539 of Genbank EU912437) was used as template for dsRNA synthesis. Injections of dsRNA into embryos (2 ug/ul), larvae (0.5-3.25 ug/ul) and pupae (1.225 ug/ul) were performed as previously described (Brown et al. 1999; Bucher et al. 2002; Tomoyasu and Denell 2004).
Documentation
Cuticle preparations were mounted either in Hoyer’s medium or as described by Shippy et al. (2008). Adult heads were mounted in ethanol on a depression slide and covered with a cover slip. Images of several focal planes were captured using a Nikon DXM1200F digital camera and 3-D reconstructions were made using Auto-Montage software (Syncrosopy). Scanning electron micrographs were obtained using a Hitachi S-3500N scanning electron microscope (Hitachi Science Systems) at the Kansas Agricultural Experiment Station Scanning Electron Microscope Laboratory, Kansas State University Entomology Department.
Beetle Genetics
Beetles were reared by standard methods on whole wheat flour supplemented with 5% brewers yeast. ap2 and ap3 were identified as spontaneous, recessive mutations in laboratory stocks of Df(HOMC)/mxpStbd and Ga-2, respectively. Complementation tests using either ap1 (to test ap2) or ap2 (to test ap3) indicated that these mutations were all alleles of the same locus.
Mapping
To identify a piggyBac insertion linked to ap, transgenic lines carrying GFP-marked transposons were crossed to the multiple marker strain (mms), which is homozygous for ap (Beeman and Brown 1999). GFP-positive progeny were then backcrossed to mms. The progeny were scored for GFP and ap to assess potential linkage. The insertion site junction of the line showing linkage (PU-105) was amplified by vectorette PCR as described by Lorenzen et al. (2007), cloned and sequenced (GenBank Accession EU912436). This sequence was then BLASTed against the Tribolium genome to determine the location of the PU-105 insertion. To estimate the cM positions of Tc-ss and PU-105, marker sequences from LG10 of the genetic linkage map (Lorenzen et al. 2005) were BLASTed against the Tribolium genome to identify those markers closest to the sequences of interest. Relevant marker sequences and their GenBank accession numbers are 17.K01s (CZ012512), 23.A01t (CZ012536) and 2D10 (CB334987 and CB334988).
Results and Discussion
Tribolium has a single spineless ortholog
We used Drosophila Spineless as the query sequence in a tBLASTn search of the Tribolium genome (Tribolium Genome Sequencing Consortium 2008) and identified a locus on LG10 predicted to encode a protein with extensive identity to Spineless (data not shown). A reciprocal BLAST against Drosophila recognized Spineless as the top hit for this sequence, suggesting one-to-one orthology. Therefore, we have named the Tribolium gene Tc-spineless (Tc-ss).
Tc-ss is expressed in the embryonic antennae
Tc-ss expression is first observed in the antennae at the extended germ band stage (Fig. 1a). During germband retraction, this expression domain becomes more extensive, eventually encompassing the entire distal portion of the antenna (Fig. 1b). Expression also appears in the maxillary and labial appendages (Fig. 1b), and later in small patches in the legs (Fig. 1c). Late in embryogenesis, Tc-ss is also expressed in a punctate pattern that probably represents cells of the peripheral nervous system (Fig. 1c).
Tc-ss is required for larval and adult antennal identity
To assess the function of Tc-ss, we first injected Tc-ss dsRNA into Tribolium embryos. The resulting larvae display transformations of the distal antennal segments to legs (Fig. 1e), consistent with the expression domain of Tc-ss. The basal antennal segment (scape) is unaffected (compare to wild-type in 1d), but the pedicel, flagellum and seta are replaced by a femur, tibiotarsus and pretarsal claw. Parental RNAi with Tc-ss produced progeny with the same phenotype (data not shown).
To determine the function of Tc-ss during adult development, we injected Tc-ss dsRNA into last instar larvae. The resulting adults show abnormal development of several structures. First, the antennae are transformed toward leg identity (Fig. 2d). The most basal antennal segments (the scape and pedicel) are unaffected. The funicle and club segments (which together make up the flagellum) are replaced by a tibia-like segment, a partial tarsus, and a terminal claw. Second, the thoracic legs show fusions and/or deletions of the tarsal segments (Fig. 2e). Third, the sensory bristles of the body wall are greatly reduced in length (Fig. 2f) compared to wild type (Fig. 2c).
These RNAi results indicate that the role of Tc-ss in adult development is very similar to the role of ss in Drosophila. One difference, however, is that the antennal transformations in Tribolium include more proximal leg segments. In Drosophila ss null mutants, the arista is transformed into tarsal segments, while A3 seems to lose any identifying characteristics (Duncan et al. 1998). In contrast, Tc-ss RNAi produces a fused segment with definite leg identity (probably tibia) in addition to the terminal tarsal region. As in Drosophila ss nulls, the tarsal regions of the transformed antennae as well as the thoracic legs are reduced.
The transformation of larval antenna to leg produced by Tc-ss RNAi is even more extensive than that of the adult antenna, in that it includes a femur in addition to a tibiotarsus and claw. (Consistent with this difference, the larval pedicel is affected while the adult pedicel remains intact.) Despite the apparent difference between the two developmental stages with respect to the proximal extent of Tc-ss function, our results suggest that at least some aspects of patterning are shared between embryonic and imaginal antennal development. It will be interesting to determine whether genes acting upstream and downstream of ss in Drosophila adult antennal specification also play similar roles in Tribolium adult and/or embryonic antennal development.
The antennapedia locus maps near Tc-spineless
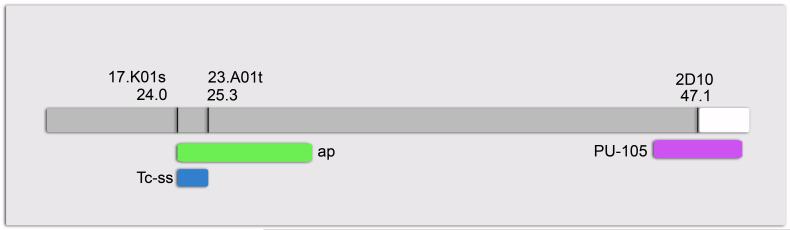
The transformation of antenna to leg produced by Tc-ss RNAi is reminiscent of the Tribolium antennapedia (ap) mutant phenotype described by Englert and Bell (1963). This mutation, which has no connection to the Hox gene Antennapedia, produces transformation of both larval and adult antennae to legs. Like Tc-ss, ap maps to LG10 (formerly LG8). To determine a more precise map position for ap, we identified a piggyBac insertion (PU-105) that showed linkage to ap. Out of 96 mapping progeny, we identified 19 recombinants between PU-105 and ap, indicating that these loci are separated by roughly 20 cM. The insertion site of PU-105 maps approximately 165 kb away from the distal-most marker (47.1 cM) on an SSCP-based genetic linkage map (Lorenzen et al. 2005). The recombination rate in this region of the genome is unknown, but we estimate that PU-105 maps somewhere between 45-49 cM on LG10 (Fig. 3). The proximity of PU-105 to the end of the linkage group means that ap almost certainly maps proximal to PU-105. Thus, allowing some room for error, we estimate that ap maps in the range of 24-30 cM on LG10 (Fig. 3).
Fig. 3.
Schematic diagram of a portion of LG10. Genetic markers shown are from the molecular map produced by Lorenzen et al. (2005). Approximate positions of ap (green), Tc-ss (blue), and PU-105 (purple) are shown below the chromosome (gray). The undefined region of the chromosome beyond the distal-most genetic marker is shown in white.
To evaluate candidate genes for the ap locus, we determined map positions for the Tribolium orthologs of genes important for antennal identity in Drosophila. In addition to spineless, these include tango (Emmons et al. 1999), hth (Casares and Mann 1998; Pai et al. 1998), extradenticle (Gonzalez-Crespo and Morata 1995; Rauskolb et al. 1995), Dll (Cohen and Jurgens 1989; Dong et al. 2000; Sato 1984; Sunkel and Whittle 1987), distal antenna (dan) and distal antenna-related (danr) (Emerald et al. 2003; Suzanne et al. 2003). Tc-Dll has previously been mapped to LG7 (Beeman and Brown 1999) and thus can be removed from consideration. We found that Tc-homothorax and the single Tribolium ortholog of dan/danr also map to LG7, while Tc-tango maps to the X chromosome, eliminating these genes as candidates for ap. Tc-extradenticle, like ap, maps to LG10. However, its position near a marker at 15.8 cM is quite far from the estimated position of ap. Interestingly, Tc-ss maps to LG10 between markers at 24.0 cM and 25.3 cM (Fig. 3), putting it within range of the ap locus. Although finer mapping is needed, the similar map positions of Tc-ss and ap make Tc-ss the most likely candidate gene for the ap locus.
antennapedia mutant phenotypes are similar to the effects of Tc-ss RNAi
In addition to the original ap allele (Englert and Bell 1963), which we will hereafter call ap1, we have isolated two new recessive alleles of this locus (see Materials and Methods). To further investigate whether ap mutations might affect the Tc-ss locus, we carefully examined the phenotype produced by each ap allele. In the descriptions below, we have specifically noted whether structures affected by Tc-ss RNAi are altered in the ap mutants.
ap1 causes partial transformation of antenna toward leg at both the larval (Fig. 1f) and adult (Fig. 2g) stages. The most obvious leg-like feature in ap1 larval antennae is a distal pretarsal claw. In adults, the expressivity of the phenotype is somewhat variable, but typically a distal tarsal claw and a fused region with some degree of leg identity are present. The funicle segments are either partially fused or altogether absent. The two basal segments always retain antennal identity. The legs (Fig. 2h) appear normal, as do the bristles of the body wall (Fig. 2i).
ap2 causes a more complete transformation to leg than ap1 in both larvae (Fig. 1g) and adults (Fig. 2j). The larval pedicel, flagellum and seta are replaced by a fused segment (apparently tibiotarsus and femur) and pretarsus. In adults, the funicle and club segments are transformed into tibia and tarsal segments. Similar to ap1, the legs (Fig. 2k) and bristles show no defects (Fig. 2l).
ap3 homozygous larvae display a severe antenna to leg transformation (Fig. 1h), similar to that seen for ap2. One difference in ap3 is the presence of an obvious boundary between the tibiotarsus and femur of the transformed antenna, a feature that is also seen in ss RNAi larvae. The antennal transformations of ap3 adults are quite variable. Mildly affected individuals (Fig. 2m) display normal funicles but fused club segments. Interestingly, one or more tarsal segments and a terminal claw are sometimes present distal to the three club segments. Other ap3 antennae (Fig. 2p) resemble those of ap2 homozygotes. In contrast to the other alleles of ap, ap3 homozygotes sometimes show leg defects, namely fusion or deletion of the adult tarsal segments (Fig. 2q), as well as a reduction in bristle length (Fig. 2r). These additional phenotypes are very similar to those produced by Tc-ss RNAi.
To try to confirm the relationship between ap and ss, we sequenced the coding region (CDS) of ss from ap1, ap2 and ap3 homozyogtes. However, we found no changes predicted to affect the encoded protein. This result is not particularly surprising, given that none of the ap alleles appear to be nulls (see below).
Despite the lack of a definitive mutant lesion, several lines of evidence suggest that Tc-ss is the gene affected by ap mutations. First, both genes map to a similar position on LG10. Second, the phenotypes of ap mutations, particularly the pleiotropic effects of ap3, are very similar to those produced by Tc-ss RNAi. Third, the existence of different classes of ap alleles, some affecting only the antennae and others affecting the legs and bristles as well, is reminiscent of Drosophila ss. While null ss alleles affect the antennae, legs and bristles (similar to the most severe effect of ap3), many alleles affect only the antennae (as seen for ap1 and ap2). In Drosophila, these different classes of alleles seem to be the result of the complex regulatory structure of the ss locus. Separate enhancer regions driving expression in antennae, legs and bristles have been identified (Emmons et al. 2007). The antennal enhancers are located some distance upstream of the ss transcription unit, while the leg and bristle enhancers are within or just upstream of the transcription unit. Therefore, antenna-specific ss alleles can be explained by deletions or rearrangements that affect only the antennal enhancers (Emmons et al. 2007).
If, indeed, ap mutations are Tc-ss alleles, we would predict that the ap1 and ap2 mutations affect only antennal expression. It is interesting that both of these alleles affect antennal identity in larvae as well as adults. In Drosophila, there are at least two embryonic antennal segment enhancers for ss. Both of these map near imaginal antenna enhancers, but at least one of them is clearly a separate element. Unfortunately the evolutionary distance between Drosophila and Tribolium (∼300 million years) is too great to permit conservation-based recognition of cis-regulatory elements, but if the regulatory structure of Tc-ss is similar to Drosophila ss, we might expect ap1 and ap2 to be deletions or rearrangements with breakpoints upstream of Tc-ss. Such lesions might affect expression of Tc-ss in both embryonic and adult antennae, but not adult legs or sensory bristles.
In contrast to ap1 and ap2, the ap3 lesion probably affects all aspects of Tc-ss expression. However, this allele is unlikely to be a null, since not all ap3 homozygotes show the severe phenotype. It is possible that a second site mutation is responsible for the variation observed in ap3 homozygotes. However, the Ga-2 background of ap3 is highly inbred and believed to be homozygous at most loci (Lorenzen et al. 2005). It is more likely that the variable phenotype of ap3 is caused by differences in gene expression levels among individuals.
The results described here highlight the advantages of using Tribolium as a model organism to study the patterning of larval and adult antennae. The two-pronged approach of using both forward genetics and candidate genes from the Drosophila adult antennal paradigm, promises to provide important insights into Tribolium development, as well as the evolution of insect antennae.
Acknowledgements
We thank Kathy Leonard, Canda Harvey and Michelle Coleman for technical assistance and Yoshinori Tomoyasu, Renata Bolognesi, Sherry Miller and Sue Brown for helpful discussions and comments on the manuscript. We thank Gregor Bucher, Markus Weber and Martin Klingler for the PU-105 line. We are also grateful to Kent Hampton for expert assistance with the scanning electron microscope. S.Y. was a Howard Hughes Medical Institute Undergraduate Research Scholar. This work was also supported by grants from the Terry C. Johnson Center for Basic Cancer Research at Kansas State University, the National Science Foundation (IOB0321882) and the National Institutes of Health (R01HD029594).
References
- Beeman RW, Brown SJ. RAPD-based genetic linkage maps of Tribolium castaneum. Genetics. 1999;153:333–8. doi: 10.1093/genetics/153.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SJ, Mahaffey JP, Lorenzen MD, Denell RE, Mahaffey JW. Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evol Dev. 1999;1:11–5. doi: 10.1046/j.1525-142x.1999.99013.x. [DOI] [PubMed] [Google Scholar]
- Bucher G, Scholten J, Klingler M. Parental RNAi in Tribolium (Coleoptera) Curr Biol. 2002;12:R85–6. doi: 10.1016/s0960-9822(02)00666-8. [DOI] [PubMed] [Google Scholar]
- Casares F, Mann RS. Control of antennal versus leg development in Drosophila. Nature. 1998;392:723–6. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- Cohen SM, Jurgens G. Proximal-distal pattern formation in Drosophila: cell autonomous requirement for Distal-less gene activity in limb development. Embo J. 1989;8:2045–2055. doi: 10.1002/j.1460-2075.1989.tb03613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong PD, Chu J, Panganiban G. Coexpression of the homeobox genes Distal-less and homothorax determines Drosophila antennal identity. Development. 2000;127:209–16. doi: 10.1242/dev.127.2.209. [DOI] [PubMed] [Google Scholar]
- Dong PD, Dicks JS, Panganiban G. Distal-less and homothorax regulate multiple targets to pattern the Drosophila antenna. Development. 2002;129:1967–74. doi: 10.1242/dev.129.8.1967. [DOI] [PubMed] [Google Scholar]
- Duncan DM, Burgess EA, Duncan I. Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes Dev. 1998;12:1290–303. doi: 10.1101/gad.12.9.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerald BS, Curtiss J, Mlodzik M, Cohen SM. Distal antenna and distal antenna related encode nuclear proteins containing pipsqueak motifs involved in antenna development in Drosophila. Development. 2003;130:1171–80. doi: 10.1242/dev.00323. [DOI] [PubMed] [Google Scholar]
- Emmons RB, Duncan D, Duncan I. Regulation of the Drosophila distal antennal determinant spineless. Dev Biol. 2007;302:412–26. doi: 10.1016/j.ydbio.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons RB, Duncan D, Estes PA, Kiefel P, Mosher JT, Sonnenfeld M, Ward MP, Duncan I, Crews ST. The spineless-aristapedia and tango bHLH-PAS proteins interact to control antennal and tarsal development in Drosophila. Development. 1999;126:3937–45. doi: 10.1242/dev.126.17.3937. [DOI] [PubMed] [Google Scholar]
- Englert DC, Bell AE. “antennapedia”: An unusual antennal mutation in Tribolium castaneum. Ann Entomol Soc Am. 1963;56:123–24. [Google Scholar]
- Gonzalez-Crespo S, Morata G. Control of Drosophila adult pattern by extradenticle. Development. 1995;121:2117–25. doi: 10.1242/dev.121.7.2117. [DOI] [PubMed] [Google Scholar]
- Lorenzen MD, Doyungan Z, Savard J, Snow K, Crumly LR, Shippy TD, Stuart JJ, Brown SJ, Beeman RW. Genetic linkage maps of the red flour beetle, Tribolium castaneum, based on bacterial artificial chromosomes and expressed sequence tags. Genetics. 2005;170:741–7. doi: 10.1534/genetics.104.032227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen MD, Kimzey T, Shippy TD, Brown SJ, Denell RE, Beeman RW. piggyBac-based insertional mutagenesis in Tribolium castaneum using donor/helper hybrids. Insect Mol Biol. 2007;16:265–75. doi: 10.1111/j.1365-2583.2007.00727.x. [DOI] [PubMed] [Google Scholar]
- Nagaso H, Murata T, Day N, Yokoyama KK. Simultaneous detection of RNA and protein by in situ hybridization and immunological staining. J Histochem Cytochem. 2001;49:1177–82. doi: 10.1177/002215540104900911. [DOI] [PubMed] [Google Scholar]
- Pai CY, Kuo TS, Jaw TJ, Kurant E, Chen CT, Bessarab DA, Salzberg A, Sun YH. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev. 1998;12:435–46. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Smith KM, Peifer M, Wieschaus E. extradenticle determines segmental identities throughout Drosophila development. Development. 1995;121:3663–73. doi: 10.1242/dev.121.11.3663. [DOI] [PubMed] [Google Scholar]
- Sato T. A new homoeotic mutation affecting antennae and legs. Drosophila Information Service. 1984;60:180–182. [Google Scholar]
- Shippy TD, Ronshaugen M, Cande J, He J, Beeman RW, Levine M, Brown SJ, Denell RE. Analysis of the Tribolium homeotic complex: insights into mechanisms constraining insect Hox clusters. Dev Genes Evol. 2008;218:127–39. doi: 10.1007/s00427-008-0213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. Spineless-aristapedia: a homeotic gene that does not control the development of specific compartments in Drosophila. Genetics. 1982;102:737–49. doi: 10.1093/genetics/102.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel CE, Whittle JR. Brista: a gene involved in the specification and differentiation of distal cephalic and thoracic structures in Drosophila melanogaster. Roux’s Arch Dev Biol. 1987;196:124–132. doi: 10.1007/BF00402034. [DOI] [PubMed] [Google Scholar]
- Suzanne M, Estella C, Calleja M, Sanchez-Herrero E. The hernandez and fernandez genes of Drosophila specify eye and antenna. Dev Biol. 2003;260:465–83. doi: 10.1016/s0012-1606(03)00249-5. [DOI] [PubMed] [Google Scholar]
- Tomoyasu Y, Denell RE. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev Genes Evol. 2004;214:575–8. doi: 10.1007/s00427-004-0434-0. [DOI] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–55. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]