Abstract
The zebrafish is emerging as a prominent model system for studying the genetics of human development and disease. Genetic alterations that underlie each mutant model can exist in the form of single base changes, balanced chromosomal rearrangements, or genetic imbalances. To detect genetic imbalances in an unbiased genome-wide fashion, array comparative genomic hybridization (CGH) can be used. We have developed a 5 Mb resolution array CGH platform specifically for the zebrafish. This platform contains 286 BAC clones, enriched for orthologous sequences of human oncogenes and tumor suppressor genes. Each BAC clone has been end-sequenced and cytogenetically assigned to a specific location within the zebrafish genome, allowing for ease of integration of array CGH data with the current version of the genome assembly. This platform has been applied to three zebrafish cancer models. Significant genomic imbalances were detected in each model, identifying different regions which may potentially play a role in tumorigenesis. Hence, this platform should be a useful resource for genetic dissection of additional zebrafish developmental and disease models as well as a benchmark for future array CGH platform development.
INTRODUCTION
Comparative genomic hybridization (CGH) allows for the scanning of entire genomes for variations in DNA copy number (Kallioniemi et al., 1992; Pinkel and Albertson, 2005). In a typical CGH experiment, total genomic DNA is isolated from a test and a reference sample, differentially labeled with fluorescent molecules (e.g., Cy3 and Cy5), and then competitively hybridized upon a representative genome. Originally, metaphase chromosomes were used to represent the genome (Kallioniemi et al., 1992). However, chromosomal CGH had limited resolution (approximately 5-10 Mb for single copy gains and losses) and relied upon routine preparations of optimal chromosome metaphase spreads. To circumvent some of these technical difficulties, array-based CGH technology was developed (Solinas-Toldo et al., 1997; Pinkel et al., 1998). Array CGH provides a higher resolution platform by replacing metaphase chromosomes with cloned DNA fragments, PCR products, or oligonucleotides that are mapped to the genome sequence (Cai et al., 2002; Fiegler et al., 2007). These DNA fragments are then spotted on a glass slide, and the resolution of the array is ultimately dependent upon the insert size and number of non-overlapping genomic elements on the array. Detection of copy number gains and losses is based upon changes in the signal fluorescence ratios (Cy3:Cy5) for a given genomic element with gains or losses being identified when ratios diverge significantly from the expected 1:1 ratio.
Genomic amplifications and deletions are hallmarks of cancers and array CGH has been a useful tool in the detection of genomic imbalances associated with human cancers (Pinkel et al., 1998; Nowak et al., 2005; Davies et al., 2005). Array CGH has also been used to characterize cancer models in other species, such as rodents (Hodgson et al., 2001; Cai et al., 2002; Sander et al., 2005; Lakshmi et al., 2006), canines (Thomas et al., 2007), and even nematode mutants (Mayden et al., 2007). We describe here the first array CGH platform developed for the zebrafish, which is becoming an increasingly popular model system in biomedical research (reviewed in Lieschke and Currie, 2007). External fertilization, transparent embryos, the ability of females to produce hundreds of eggs every week, small size, and low cost associated with maintenance make the zebrafish an attractive model system. In addition, the zebrafish acquiescence to transgenic and forward genetic strategies, provides the capacity for extensive genetic screens of developmental pathways and more recently has been used to identify novel pathways in the molecular genetics of cancer. To enhance cancer gene discovery in zebrafish models, we have developed an array CGH platform enriched with sequences orthologous to human oncogenes and tumor suppressor genes to assess for genomic imbalances in tumor specimens for this model organism.
MATERIALS AND METHODS
Development of Zebrafish Cancer Array
Protein sequences of human oncogenes and tumor suppressor genes were retrieved from Entrez Gene at the NCBI website (http://www.ncbi.nlm.nih.gov/). Sequences were then BLAST searched against finished and unfinished zebrafish BAC clone sequences. Individual BLAST results were then examined manually for orthology. BAC clones containing sequence domains that encoded orthologs of human proteins were identified. BAC clones were from the CHORI-211 (zC), Danio Key (zK), and CHORI-73 (zH) zebrafish BAC DNA libraries. Plasmid DNA from the clones was prepared for further analysis. Briefly, BAC clones were streaked on LBagar plates containing the appropriate antibiotic and grown overnight at 37°C. A single colony was inoculated in TB-media containing the appropriate antibiotic and placed in a shaking incubator at 37°C for 16 hours. From this culture, DNA was isolated following the Qiagen Plasmid Midi Kit protocol (Valencia, CA) and then amplified using the Repli-G Midi kit as outlined in the manufacturer’s protocol (Qiagen, Valencia, CA). DNA was quantified on a Nanodrop 1000 spectrophotometer. Amplified BAC clones were end-sequenced for clone verification and mapped on zebrafish metaphase chromosome spreads by fluorescence in situ hybridization (FISH) as described previously (Freeman et al., 2007). BAC clones that gave a unique signal with minimal non-specific hybridization were chosen for inclusion on the array.
Aliquots of the amplified DNA isolated from the BAC clones chosen for inclusion on the array were then sent to Spectral Genomics (Perkin Elmer, Houston, TX) for array spotting. For printing, BAC clone DNA was denatured by dissolving the DNA in 25 μl 50 mM NaOH and then neutralized in 25 μl 0.1M Tris-HCl immediately before spotting. BAC clone DNA was spotted at a concentration of ∼150 ng/μl. Arrays were printed by spotting the neutralized BAC DNA solution onto acid cleaned plain glass slides using a GMS 417 arrayer. Each BAC clone was printed in triplicate in sixteen 7 × 9 blocks. In addition to the BAC clone DNA, blocks contained blank spots with no DNA as an internal background control. Two separate sections of sixteen blocks were spotted on each glass slide allowing for two separate hybridizations to be conducted on each slide. Once printing was completed, slides were treated at 80°C for 2 hours and then cured in 80% ethanol at 65°C for 5 hours, rinsed with 80% ethanol, and air-dried. BAC arrays were then stored at room temperature until use.
Three Transgenic Zebrafish Cancer Models
This zebrafish BAC array CGH platform was applied to three transgenic zebrafish cancer models: (1) rhabdomyosarcoma (RMS; Langenau et al., 2007), (2) T-cell acute lymphoblastic leukemia (T-ALL; Langenau et al., 2003; Langenau et al., 2005; Feng et al., 2007), and (3) melanoma (Patton et al., 2005). RMS transient transgenic fish were obtained by injecting rag2-kRASG12D along with rag2-GFP similar to how previously described (Langenau et al., 2007). Fish with tumors were scored under a fluorescent microscope. GFP+ tumor tissue was isolated from normal tissue as indicated by Rag2-GFP labeling. Rag2-GFP demarks the boundaries of the tumor population and cells not labeled were used as the reference. DNA was extracted using the Gentra Puregene DNA isolation kit.
The T-ALL model is a rag2-loxp-dsRED2-loxp-EFFP-mMyc; hsp70-Cre double transgenic fish. In this model, T-ALL is induced by heat activation at 37°C for 45 minutes at 6 dpf (Feng et al., 2007). The normal tissue (i.e., non-tumor tissue) was collected from the tail-fin of each tumor fish when the fish developed localized thymic lymphoma. Tumor cells were collected once fish were leukemic. Tumor cells were enriched (over 93% purity) by GFP-based FACS sorting. DNA was extracted from the tumor cells using the Gentra Puregene DNA isolation kit, while DNA was extracted from the tail-fin tissues using embryonic lysis buffer with subsequent purification by phenol-chloroform extraction.
The fish for the melanoma model express the human BRAF(V600E) oncogenic variant under the control of the melanocyte-specific mitfa promoter (Patton et al., 2005). This transgene is stable in that it is integrated into a chromosome and transmitted through the germline. These fish also contain a TP53 mutation of the M214K variant described in Berghmans et al. (2005). DNA from tumors and matched normal (i.e., non-tumor) tissue were prepared by standard phenol-chloroform extractions.
All fish were cared for under Institutional Animal Care and Use Committee oversight following approved protocols for humane treatment.
DNA Preparation, Probe Labeling, and Hybridization
Total genomic DNA was obtained from the three transgenic zebrafish cancer models (i.e., RMS, T-ALL, and melanoma) as described above. DNA was also isolated from normal (i.e., non-cancerous) tissues of the same fish and used as reference DNA. Genomic DNA (1 μg) from the test and genomic DNA (1 μg) from the reference sample was labeled with Cy3-dCTP or Cy5-dCTP by random priming using the BioPrime Labeling Kit, in which fluorescent molecules are incorporated while the genomic DNA is replicated (Invitrogen, Carlsbad, CA). Unincorporated nucleotides were removed using a Microcon YM-30 filter (Millipore). The concentration of DNA and the incorporation of dyes were then measured. For array hybridization, 10 μg of labeled tumor DNA was co-precipitated with 10 μg of reference DNA, and either 20 μg of zebrafish Cot1 DNA (prepared as described in Lee and Smith, 2004) or 20 μg of salmon sperm DNA. The DNA pellet was dissolved in 40 μl hybridization buffer (50% formamide, 10% dextran sulfate in 2X SSC) and denatured at 75°C for 10 minutes. The probe mixture was placed at 42°C for 45 minutes to allow the Cot1 DNA to preanneal and block repetitive sequences. The slide was placed on a 42°C heat block and 30 μl of the hybridization solution was injected under a 22 × 22 mm Lifterslip in 10 μl increments. Slides were then placed in a humidified Corning hybridization chamber and prehybridized at 42°C for 30 minutes. Subsequently, the slides were hybridized for 24-48 hours at 37°C.
After hybridization, the coverslips were removed by placing the slide in 2X SSC, 0.05% Tween at 43°C for 5 minutes. Slides were transferred into a preheated solution of 50% formamide, 2X SSC, 0.1% Tween at 43°C for 15 minutes in a Petri dish, washed in a prewarmed solution of 2X SSC, 0.05% Tween at 43°C for 10 minutes in a rocking Petri dish, and followed by a wash at room temperature in 1X PBS, 0.05% Tween for 10 minutes. Slides were finally rinsed in ddH2O and dried under nitrogen gas.
A total of 21 zebrafish samples (i.e., 7 fish with RMS, 8 fish with T-ALL, and 6 fish with melanoma) were analyzed. Each array was run in duplicate as a dye-swap experimental design to minimize false positives.
Array Imaging and Analysis
A two-color scan of the arrays was conducted using the GenePix 4000B Scanner (Molecular Devices, Union City, CA) at a 5 μm resolution. Using GenePix Pro image analysis software, Cy3 and Cy5 fluorescence intensities of each DNA spot was quantified. Background intensity was then subtracted and the data imported into an excel spreadsheet for further data processing. The Cy3/Cy5 fluorescence ratios were calculated and transformed into log2 values. The ratios were normalized using block median normalization for each of the sixteen blocks to account for any block to block variation. Following normalization, the mean of the three replicate values for each specific BAC clone was calculated. Individual values not within 10% of the mean of the three replicates for that clone were removed from the analysis and deemed as invalid. Two of three replicate values were needed to pass this criterion for each BAC clone to be included in further analysis. In addition, the overall number of invalid clones were calculated and had to be less than 10% to analyze the hybridization. The average log2 ratios were then plotted graphically on chromosome-specific profiles based upon the clone location identified from FISH mapping. Gains or losses were identified in those regions deviating from the expected 1:1 ratio value (or a log2 value of 0). Each hybridization was treated as a separate experiment (i.e., the initial hybridization was treated as experiment 1 and the dye-swap hybridization was treated as experiment 2) and analysis was conducted separately for each hybridization. This allowed for two replicate hybridizations of the same test.
In a dye-swap experimental design, the two ratio curves should be reciprocal, allowing for the quick identification of false positives. A gain in DNA copy number is depicted by a positive deviation from log2 = 0 for the Cy3:Cy5 ratio, whereas a loss in DNA copy number will result in a negative deviation from log2 = 0 for the Cy3:Cy5 ratio (plotted in blue). For true changes in DNA copy number, the dye-swap hybridization (plotted in red) will represent a mirror image of the initial hybridization. Ratios greater than ±0.30 in both of the experiments were considered as a significant genomic imbalance. This value was determined based on routine variation exhibited in self-to-self experiments on this array CGH platform.
FISH Validation
A subset of the BAC clones, which were observed to have significant genomic imbalances in 2 or more cancer samples, were validated by FISH. Two-color FISH was performed on paraffin sections of specific cancer types for confirmation of the array CGH calls. In addition, non-tumor material was also placed on each slide for comparison of signal frequency and intensity. Labeling of the BAC clone DNA probes was done as described in Freeman et al. (2007) with the clone exhibiting a copy number change labeled with spectrum orange and another BAC clone, used as a reference probe, labeled with spectrum green.
For the FISH hybridization, slides were placed in three solutions of xylene for 15 minutes at room temperature. Slides were then dehydrated twice in 100% ethanol for 2 minutes and then incubated in a preheated solution of 1X TE at 100°C for 15 minutes. Slides were washed in 1X PBS at room temperature for 5 minutes and digested for 20 minutes by pepsin treatment at 37°C. Fresh pepsin was added to the slides and digestion was allowed to proceed for an additional 20 minutes at 37°C. Slides were washed in 1X PBS for 5 minutes at room temperature and placed in formalin for 10 minutes. A 1X PBS wash for 5 minutes was repeated at room temperature and the slides dehydrated in an ethanol series of 70%, 90%, and 100% for 2 minutes each at room temperature. The BAC clone DNA probes were added to the slides under a coverslip and sealed with rubber cement, denatured at 94°C for 3 minutes, and then incubated at 37°C in a humidified chamber in the dark for 48 hours.
Following hybridization, slides were washed in 0.5X SSC for 5 minutes at 74°C and washed three times in 1X PBS, 0.025% Tween for 2 minutes at room temperature. Slides were counterstained with DAPI (Vector Laboratories, Burlingame, CA) and analyzed by fluorescence microscopy on an Olympus BX-51 fluorescence microscope equipped with narrow band pass filters for spectrum orange and spectrum green dyes. Images were captured using a Photometrics KAF1400 CCD camera and Applied Imaging Genus Software system. At least 100 cells were analyzed per sample.
RESULTS
Development of Zebrafish Cancer Array CGH Platform
A panel of 357 BAC clones, each containing sequences orthologous to human oncogenes and tumors suppressor genes, was cytogenetically localized using fluorescence in situ hybridization (FISH). Seven BAC clones (2.0%) hybridized to multiple locations within the genome, including one pan-centromeric clone (i.e., hybridized near the centromeres of all chromosomes). Seventy-three BAC clones (20.4%) only had non-specific signals, 30 BAC clones (8.4%) gave weak hybridization signals, and 40 BAC clones (11.2%) had no detectable FISH signals.
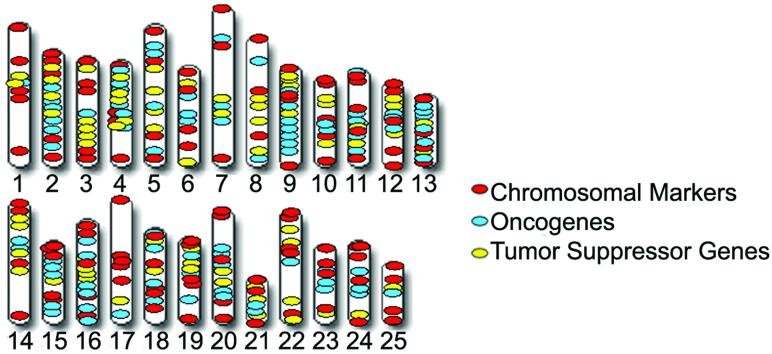
The remaining 207 clones (58.0%) resulted in robust, unique FISH signals, with 4 of these clones localizing to the heterochromatic region of the long arm of LG chromosome 4 and subsequently excluded from the array. From the remaining 203 clones, 98 clones containing orthologous sequences to human oncogenes and 79 clones containing orthologous sequences to human tumor suppressor genes were cytogenetically mapped and chosen for inclusion on the array platform. The remaining 26 clones were discarded from the analyses due to mapping inconsistencies. An additional 109 BAC clones (previously mapped cytogenetically, Lee and Smith, 2004), which served as chromosomal markers to achieve increased coverage along the length of each chromosome, were also included in this array design (Fig. 1; Table 1; Supplementary Table S1). This resulted in a total of 286 BAC clones on the platform for an average spacing of approximately 5 Mb.
Figure 1. Chromosome coverage of BAC clones included on the zebrafish BAC-based array CGH platform.
There is a total of 286 BAC clones for an average spacing of 5 Mb. The array platform is enriched with BAC clones which contain orthologous sequences to human oncogenes (i.e., 98 clones) and tumor suppressor genes (i.e., 79 clones). In addition, 109 BAC clones were also included to improve overall chromosome coverage. All BAC clones were confirmed by end-sequencing and cytogenetically mapped to their LG chromosome assignment upon zebrafish metaphase chromosome spreads.
Table 1.
LG chromosome assignment of the three different categories of BAC clones chosen for inclusion on the zebrafish cancer array
| LG Chromosome | Chromosomal Marker | Oncogene | Tumor Suppressor Gene | Total No. of BAC Clones on LG Chromosome |
|---|---|---|---|---|
| 1 | 5 | 2 | 3 | 10 |
| 2 | 5 | 8 | 4 | 17 |
| 3 | 5 | 1 | 5 | 11 |
| 4 | 4 | 6 | 5 | 15 |
| 5 | 4 | 4 | 4 | 12 |
| 6 | 4 | 4 | 3 | 11 |
| 7 | 3 | 3 | 2 | 8 |
| 8 | 3 | 2 | 3 | 8 |
| 9 | 4 | 7 | 5 | 16 |
| 10 | 5 | 4 | 3 | 12 |
| 11 | 4 | 9 | 3 | 16 |
| 12 | 5 | 3 | 4 | 12 |
| 13 | 4 | 6 | 4 | 14 |
| 14 | 3 | 2 | 5 | 10 |
| 15 | 5 | 5 | 1 | 11 |
| 16 | 5 | 7 | 4 | 16 |
| 17 | 5 | 1 | 2 | 8 |
| 18 | 5 | 4 | 2 | 11 |
| 19 | 5 | 3 | 4 | 12 |
| 20 | 5 | 7 | 2 | 14 |
| 21 | 3 | 2 | 3 | 8 |
| 22 | 5 | 1 | 4 | 10 |
| 23 | 4 | 3 | 1 | 8 |
| 24 | 5 | 3 | 1 | 9 |
| 25 | 4 | 1 | 2 | 7 |
| Total | 109 | 98 | 79 | 286 |
Chromosomal assignments from the FISH mapping data was compared to the chromosomal assignment of the BAC clones by the Zebrafish Genome Project using the Zebrafish Genome Fingerprinting Project Clone database (WebFPC; www.sanger.ac.uk/cgibin/Projects/D_rerio/WebFPCreport.cgi). Chromosomal assignments for the BAC clones agreed for 211 of the clones (73.8%), while only 8 BAC clones (2.8%) had discrepant chromosomal assignments (Supplementary Table S2). The remaining 67 BAC clones (23.4%) have not yet been assigned to a specific chromosomal location in the WebFPC database (Supplementary Table S2).
Array CGH Analysis
A series of 6 self-to-self hybridizations were initially performed to assess array performance and hybridization quality (Supplementary Fig. S1A). These 6 experiments included hybridizations of both AB and Tu wild-type strain zebrafish. Self-to-self hybridizations resulted in an average standard deviation of 0.27. Based on this variation observed from the self-to-self hybridizations, a deviation value of ±0.30 was established for calling significant losses or gains, when observed in both experiments of the dye-swap set. Using this calling threshold there were no significant genomic imbalances called in the 6 self-to-self hybridizations. In addition, within strain (i.e., AB fish versus AB fish and Tu fish versus Tu fish) and among strain (i.e., AB fish versus Tu fish) comparisons were completed and were observed to be normal (Supplementary Fig. S1B-S1D). To demonstrate the application of this array, a total of 21 zebrafish, representing 3 transgenic zebrafish cancer models, were then analyzed on the array CGH platform (e.g., Supplementary Fig. S1E).
Zebrafish RMS cancer model
The first transgenic model, which develops RAS-induced rhabdomyosarcoma (RMS) by 10 days of life (Langenau et al., 2007), was analyzed on the array. Seven rhabdomyosarcoma specimens were analyzed on our custom zebrafish BAC-based array and observed to have 6 to 14 significant genomic imbalances per sample (Supplementary Table S3). In the RMS samples, genomic imbalances were present in contiguous BAC clones (as defined by FISH mapping for those BAC clones included on the array platform) indicating the potential for the presence of chromosomal alterations larger than the size of an individual BAC clone. For example, a loss was observed in BAC clones zC085P24 and zC165B19, which were FISH mapped near the middle of the long arm of LG chromosome 9, in RMS sample #3. In addition, a gain in zK006P15 and zK014B13, which were FISH mapped near the centromere of LG chromosome 17, was observed in RMS samples #2, #4, #5, #6, and #7. Lastly, a gain in zK192M14 and zC009D01, which were FISH mapped near the long arm telomere of LG chromosome 22, was present in RMS samples #5 and #7.
Although no BAC clones were seen as losses in multiple RMS samples, 11 BAC clones were observed as gains in two or more tumors (Table 2). The BAC clones observed to have significant gains among multiple samples were cytogenetically mapped to LG chromosomes 1 (zK242H09), 11 (zK013A21), 13 (zK006L12), 17 (zK014B13 and zK006P15), 18 (zK014D24 and zK003B08), 22 (zK002J07, zC009D01, and zK192M14), and 23 (zC214H13). LG chromosomes 17, 18, and 22 all had multiple BAC clones with a significant gain in genomic material. To confirm array CGH results, seven of the BAC clones (i.e., zK242H09, zK006L12, zK014B13, zK006P15, zK014D24, zK002J07, and zC009D01) were chosen for FISH validation. FISH was performed on both tumor and non-tumor tissue from 10 additional fish (e.g., Fig. 2) to equal a total number of 17 zebrafish interrogated for genomic imbalances in these seven regions of DNA sequence. Amplification events were observed for all seven of the BAC clones on the additional tumor tissue samples in comparison to matched normal tissue with FISH. The number of samples exhibiting amplification events was dependent on the specific BAC clone (Supplementary Table S4).
Table 2.
Genomic imbalances in the zebrafish RMS model observed in multiple fish
| BAC Clonea | LG Chromosome Assignment by FISH | Relative Position Along Chromosome | Initial Designation of BAC Clone (Marker/Oncogene/TSG)b | Current Gene Annotation (Nov. 2007) | No. of Samples with a Significant Genomic Imbalance (N = 7) | Percent of Samples with a Significant Genomic Imbalance | Gain/Loss | FISH Confirmed |
|---|---|---|---|---|---|---|---|---|
| zK242H09 | 1 | 7.5 | Oncogene | DENND4A | 6 | 86% | Gain | Yes |
| zK013A21 | 11 | 7 | TSG | COMT | 3 | 43% | Gain | NAc |
| CXXC1 | ||||||||
| GNL3L | ||||||||
| GRM7 | ||||||||
| HCFC1 | ||||||||
| MBD1 | ||||||||
| mGluR8b | ||||||||
| MITF | ||||||||
| OPN1MW | ||||||||
| RAB7 | ||||||||
| RHO | ||||||||
| zK006L12 | 13 | 1.5 | Marker | ABCC2 | 4 | 57% | Gain | Yes |
| CNTNAP3 | ||||||||
| COX15 | ||||||||
| CUTC | ||||||||
| KCNIP2 | ||||||||
| KIAA1305 | ||||||||
| NRXN1 | ||||||||
| TP53BP2 | ||||||||
| zK006P15 | 17 | 6 | Marker | PAPSS2 | 5 | 71% | Gain | Yes |
| zK014B13 | 17 | 6.5 | Marker | ARHGEF10 | 7 | 100% | Gain | Yes |
| CLN8 | ||||||||
| DLGAP2 | ||||||||
| DLGAP4 | ||||||||
| PPIE | ||||||||
| zK003B08 | 18 | 2 | Oncogene | MAGI2 | 3 | 43% | Gain | NAc |
| SIX4 | ||||||||
| TBCB | ||||||||
| YAP1 | ||||||||
| zK014D24 | 18 | 8 | Marker | ARHGEF17 | 6 | 86% | Gain | Yes |
| RELL1 | ||||||||
| TNFRSF19L | ||||||||
| zK002J07 | 22 | 0.5 | Marker | CENTB2 | 7 | 100% | Gain | Yes |
| HTR2B | ||||||||
| PIM3 | ||||||||
| PP1R2 | ||||||||
| PSMD1 | ||||||||
| ZBED1 | ||||||||
| zK192M14 | 22 | 8.5 | TSG | EDG1 | 2 | 29% | Gain | NAc |
| EPS15L1 | ||||||||
| KLF2 | ||||||||
| RC3H1 | ||||||||
| SERPINC1 | ||||||||
| STIL | ||||||||
| ZBTB37 | ||||||||
| zC009D01 | 22 | 9.5 | Marker | No hits | 7 | 100% | Gain | Yes |
| zC214H13 | 23 | 8.5 | TSG | ATXN1 | 2 | 29% | Gain | NAc |
| GTPBP5 | ||||||||
| IRF4 | ||||||||
| KCNK9 | ||||||||
| LSM14B | ||||||||
| L1CAM | ||||||||
| MITF | ||||||||
| OGFR | ||||||||
| SAMHD1 |
zC denotes the CHORI-211 zebrafish BAC clone library and zK denotes the Danio Key zebrafish BAC clone library
Please see Table S1 for original gene annotation
NA = FISH confirmation was not completed for these clones
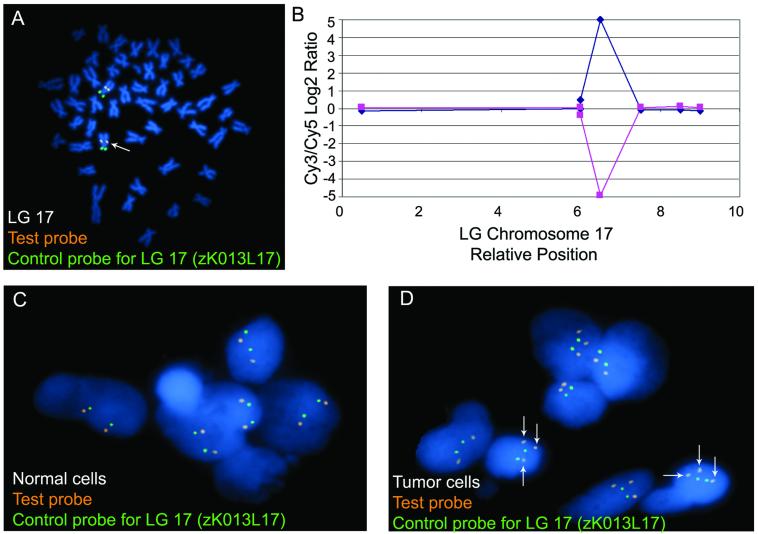
Figure 2. Application and FISH confirmation of a genomic imbalance in a zebrafish cancer model.
(A) Cytogenetic localization of the test clone (zK014B13, labeled in orange) near the middle of the long arm of LG chromosome 17 (denoted by white arrow). A BAC clone (zK013L17), which is the near-telomeric marker for the short arm of LG chromosome 17, is labeled in green. (B) A gain on LG chromosome 17 (in the test probe, zK014B13, labeled in A.) as detected by array CGH in the transgenic RMS zebrafish model. (C) Test probe (zK014B13, labeled in orange) and control probe (zK013L17, labeled in green) co-hybridized upon normal cells. Two signals for each probe were observed at equal intensity. (D) Test probe (zK014B13, labeled in orange) and control probe (zK013L17, labeled in green) co-hybridized upon tumor cells. Two signals of equal intensity were observed for the control probe, while there were three signals for the test probe in approximately half of the cells with all signals having a slight increase in signal intensity in comparison to the signal intensity of the test probe in the normal cells.
Zebrafish T-ALL cancer model
The second zebrafish cancer model analyzed on the array CGH platform was a zebrafish transgenic line for Myc-induced T-cell acute lymphoblastic leukemia (T-ALL; Langenau et al., 2003; Langenau et al., 2005; Feng et al., 2007). A total of 8 zebrafish were analyzed and found to have anywhere from 1 to 17 significant genomic imbalances per sample (Supplementary Table S5). As with the RMS samples, genomic imbalances were observed in contiguous BAC clones (as defined by FISH mapping for those BAC clones included on the array platform) indicating the potential for the presence of chromosomal alterations larger than the size of an individual BAC clone. T-ALL sample #2 had a gain in BAC clones zK006P15 and zK014B13, which were FISH mapped near the centromere of LG chromosome 17. A gain was also observed in zK097O05 and zK221H15, which were FISH mapped near the middle of the long arm of LG chromosome 20, in T-ALL samples #1 and #2. Genomic imbalances were also called for two chromosomal segments involving three BAC clones. A gain was observed in BAC clones zC004P07, zK025L23, and zK255G14, which were FISH mapped to LG chromosome 9, in T-ALL sample #8. Integrating these three contiguous BAC clones with the current build of the zebrafish genome (UCSC, Zv7) indicates these BAC clones span a 3.8 Mb segment of the genome. T-ALL sample #1 had a gain in zC226M16, zK151G10, and zK166N08, cytogenetically mapped to LG chromosome 23. Integrating the genomic location of these three contiguous BAC clones indicates these BAC clones encompass an 8.5 Mb segment of the genome. In addition, T-ALL sample #6 had a gain called for 8 of the 13 BAC clones cytogenetically mapped to LG chromosome 18.
Common gains and losses were observed among multiple T-ALL samples (Table 3). Multiple samples were observed to have a gain in a BAC clone(s) on LG chromosomes 1 (zK242H09), 17 (zK014B13), 18 (zK014D24), 20 (zK221H15 and zK097O05), and 22 (zK002J07 and zC009D01). In addition, a loss was seen in two samples for two different BAC clones on LG chromosome 14 (zC159I08 and zC117N19). FISH validation was completed for 7 of these BAC clones (i.e., zK242H09, zC159I08, zC117N19, zK014B13, zK014D24, zK002J07, and zC009D01). FISH was performed on tumor and matched non-tumor tissue from the same zebrafish also analyzed on the array CGH platform. Genomic imbalance calls by array CGH were confirmed by FISH for 6 of the 7 clones (Table 3). In addition, a correlation between the array CGH log2 values and the FISH results was observed in the 5 BAC clones with gains. The BAC clones with the largest log2 ratio change (zK014B13 and zK002J07) had the greatest increase in signal intensity and also had multiple signals present in the cells. BAC clone zC009D01 was observed to have a large increase in signal intensity, but no multiple signals present. The signal intensity of the BAC clone zK242H09 was less intense in comparison to zC009D01, but slightly more intense than zK014D24. In addition, only two signals were present in the cells. BAC clone zK014D24 had only a slight increase in signal intensity, but multiple signals in the cells were observed. Overall the level of signal intensity in combination with the presence or absence of multiple signals observed in the FISH experiments correlated with the rankings of the log2 values from the array CGH experiments for the T-ALL samples.
Table 3.
Genomic imbalances in the zebrafish T-ALL model observed in multiple fish.
| BAC Clonea | LG Chromosome Assignment by FISH | Relative Position Along Chromosome | Initial Designation of BAC Clone (Marker/Oncogene/TSG)b | Current Gene Annotation (Nov. 2007) | No. of Samples with a Significant Genomic Imbalance (N = 8) | Percent of Samples with a Significant Genomic Imbalance | Gain/Loss | FISH Confirmed |
|---|---|---|---|---|---|---|---|---|
| zK242H09 | 1 | 7.5 | Oncogene | DENND4A | 6 | 75% | Gain | Yes |
| zC117N19 | 14 | 0.5 | Marker | PCDHA3 | 2 | 25% | Loss | No |
| PCDHAC2 | ||||||||
| PCDHGA5 | ||||||||
| PCDHGA11 | ||||||||
| zC159I08 | 14 | 4.5 | Oncogene | AMIGO2 | 2 | 25% | Loss | Yes |
| MXRA5 | ||||||||
| PEG10 | ||||||||
| PNMA1 | ||||||||
| RAB33A | ||||||||
| RNF4 | ||||||||
| SELT | ||||||||
| zK014B13 | 17 | 6.5 | Marker | ARHGEF10 | 7 | 88% | Gain | Yes |
| CLN8 | ||||||||
| DLGAP2 | ||||||||
| DLGAP4 | ||||||||
| PPIE | ||||||||
| zK014D24 | 18 | 8 | Marker | ARHGEF17 | 6 | 75% | Gain | Yes |
| RELL1 | ||||||||
| TNFRSF19L | ||||||||
| zK097O05 | 20 | 7 | Oncogene | MLL2 | 2 | 25% | Gain | NAc |
| PDC | ||||||||
| PLA2G4A | ||||||||
| PRG4 | ||||||||
| PTGS2 | ||||||||
| SCYL3 | ||||||||
| TPR | ||||||||
| zK221H15 | 20 | 7 | Marker | CAD | 2 | 25% | Gain | NAc |
| CPS1 | ||||||||
| DNAJC5 | ||||||||
| IFT172 | ||||||||
| RDHE2 | ||||||||
| TRIM54 | ||||||||
| zK002J07 | 22 | 0.5 | Marker | CENTB2 | 7 | 88% | Gain | Yes |
| HTR2B | ||||||||
| PIM3 | ||||||||
| PP1R2 | ||||||||
| PSMD1 | ||||||||
| ZBED1 | ||||||||
| zC009D01 | 22 | 9.5 | Marker | No hits | 7 | 88% | Gain | Yes |
zC denotes the CHORI-211 zebrafish BAC clone library and zK denotes the Danio Key zebrafish BAC clone library
Please see Table S1 for original gene annotation
NA = FISH confirmation was not completed for these clones
Zebrafish melanoma cancer model
The third set of samples interrogated on the array CGH platform was from a model for melanoma. This transgenic zebrafish model expresses a mutant form of the BRAF gene, under control of the melanocyte-specific mitfa promoter (Patton et al., 2005). In TP53-deficient zebrafish, activated BRAF induces the formation of melanocyte lesions that develop into melanomas. Six melanoma tumor specimens were analyzed with 6 to 28 significant genomic imbalances observed per sample (Supplementary Table S6). Genomic imbalances were observed encompassing contiguous BAC clones (as defined by FISH mapping of the BAC clones on the array platform) as seen in the other two zebrafish cancer models. Ten genomic imbalance events included two contiguous BAC clones, four genomic imbalances included three contiguous BAC clones, and two genomic imbalances included four contiguous BAC clones (Supplementary Table S7). In addition, 14 of the 15 BAC clones were called as a loss on LG chromosome 4 in melanoma sample #3 and half of the clones on LG chromosome 16 in melanoma sample #5 were also called as a loss (Supplementary Table S6).
Genomic imbalances observed in more than one sample included gains in five BAC clones in half of the tumor specimen (zK143E20, zC194E15, zC171D11, zC197G15, and zC195I16), and losses in 2 BAC clones in half of the tumor specimen (zC239E06 and zK255N06) (Table 4). In addition, one-third of the samples exhibited a gain in 8 BAC clones (zK031B10, zK219L12, zK201G07, zK023K06, zC206A19, zK192M14, zC226M16, and zK151G10) and a loss in 3 BAC clones (zK021H14, zK222F08, and zC079A18) (Table 4). Common gains were observed predominantly in LG chromosomes 19 (zC143E20, zK219L12, zC194E15, and zK201G07), 22 (zC171D11, zC197G15, zC206A19, and zK192M14), and 23 (zC195I16, zC226M16, and zK151G10), while losses were primarily in LG chromosome 4 (zC239E06, zK021H14, zK222F08, and zC079A18). Sufficient material was not available to conduct FISH confirmations of the array CGH results in this set of tumors.
Table 4.
Genomic imbalances in the zebrafish melanoma model observed in multiple fish.
| BAC Clonea | LG Chromosome Assignment by FISH | Relative Position Along Chromosome | Initial Designation of BAC Clone (Marker/Oncogene/TSG)b | Current Gene Annotation (Nov. 2007) | No. of Samples with a Significant Genomic Imbalance (N = 6) | Percent of Samples with a Significant Genomic Imbalance | Gain/Loss |
|---|---|---|---|---|---|---|---|
| zK031B10 | 2 | 6.5 | Oncogene | PRKCI | 2 | 33% | Gain |
| SKIL | |||||||
| SLC24A6 | |||||||
| zK021H14 | 4 | 2 | Oncogene | DMBT1 | 2 | 33% | Loss |
| zK222F08 | 4 | 2.5 | Oncogene | BRAF | 2 | 33% | Loss |
| BTBD11 | |||||||
| CRY1 | |||||||
| MRPS33 | |||||||
| TNNT2 | |||||||
| zC239E06 | 4 | 2.5 | TSG | CDKN1B | 3 | 50% | Loss |
| PPARA | |||||||
| WNT7B | |||||||
| zC079A18 | 4 | 9 | Marker | ZDB-GENE- | 2 | 33% | Loss |
| 060503-161 | |||||||
| ZDB-GENE- | |||||||
| 060503-255 | |||||||
| ZDB-GENE- | |||||||
| 060503-433 | |||||||
| zK255N06 | 16 | 8 | Oncogene | ALKBH8 | 3 | 50% | Loss |
| DYRK1A | |||||||
| FBL | |||||||
| KCNJ5 | |||||||
| zC143E20 | 19 | 4.5 | Oncogene | MYCL1 | 3 | 50% | Gain |
| TRIT1 | |||||||
| zK219L12 | 19 | 4.5 | Marker | IRX1 | 2 | 33% | Gain |
| PEG10 | |||||||
| zC194E15 | 19 | 8 | Oncogene | KIAA1522 | 3 | 50% | Gain |
| MFSD2 | |||||||
| MYCL1 | |||||||
| TRIT1 | |||||||
| YARS | |||||||
| zK201G07 | 19 | 9 | TSG | ANP32E | 2 | 33% | Gain |
| PLEKHO1 | |||||||
| POGZ | |||||||
| PSMB4 | |||||||
| RDH13 | |||||||
| SUHW4 | |||||||
| VPS45 | |||||||
| zK023K06 | 20 | 4 | Oncogene | EPHA7 | 2 | 33% | Gain |
| ZNF452 | |||||||
| zC171D11 | 22 | 4.5 | TSG | AK097143 | 3 | 50% | Gain |
| EPS15L1 | |||||||
| KIAA1509 | |||||||
| KLF2 | |||||||
| RC3H1 | |||||||
| STIL | |||||||
| zC206A19 | 22 | 4.5 | Marker | MIER2 | 2 | 33% | Gain |
| PPAP2C | |||||||
| TLE1 | |||||||
| ZBED5 | |||||||
| zC197G15 | 22 | 5 | Oncogene | GPR52 | 3 | 50% | Gain |
| HNRPM | |||||||
| IFI44L | |||||||
| MARCH2 | |||||||
| PIP5K1C | |||||||
| RAB11B | |||||||
| RABGAP1L | |||||||
| zK192M14 | 22 | 8.5 | TSG | EDG1 | 2 | 33% | Gain |
| EPS15L1 | |||||||
| KLF2 | |||||||
| RC3H1 | |||||||
| SERPINC1 | |||||||
| STIL | |||||||
| ZBTB37 | |||||||
| zC195I16 | 23 | 3.5 | Oncogene | C20orf11 | 3 | 50% | Gain |
| DNAJC5 | |||||||
| FYN | |||||||
| HRIHFB2255 | |||||||
| NOL4 | |||||||
| TPD52L2 | |||||||
| TRIM54 | |||||||
| TRIM55 | |||||||
| zC226M16 | 23 | 4 | Marker | FLJ20489 | 2 | 33% | Gain |
| MUC3 | |||||||
| zK151G10 | 23 | 5 | Oncogene | ERBB3 | 2 | 33% | Gain |
| FAM62A | |||||||
| PA2G4 | |||||||
| WNK3 |
zC denotes the CHORI-211 zebrafish BAC clone library and zK denotes the Danio Key zebrafish BAC clone library
Please see Table S1 for original gene annotation
Comparison of the imbalanced genomic regions in the three zebrafish cancer models
Common genomic regions were observed to be imbalanced in individuals between the three cancer models. When evaluating the imbalanced genomic regions between the three cancer models, the RMS and T-ALL samples had seven common BAC clones imbalanced (zK242H09, zK006L12, zK006P15, zK014B13, zK014D24, zK002J07, and zC009D01). The RMS and melanoma samples had five common BAC clones with genomic imbalances (zC165B19, zK030A08, zK013A21, zC194E15, and zK192M14). The T-ALL and melanoma samples had six BAC clones with genomic imbalances in individuals from both cancer models (zC117N19, zC241L24, zC255L12, zC226M16, zK151G10, and zK166N08). In addition, two BAC clones were imbalanced in individuals in all three cancer models (zK003B08 and zC014G13). Similarities were also present between the three cancer models when looking at genomic imbalances involving contiguous BAC clones. A gain on chromosome 17 in BAC clones zK006P15 and zK014B13 (relative positions 6 and 6.5, respectively) was observed in five of the RMS samples and also in one of the T-ALL samples. This is also the case for a gained genomic region on chromosome 23 in one of the T-ALL samples and in two of the melanoma samples. In the T-ALL sample (sample #1) BAC clones zC226M16, zK151G10, and zK166N08 were gained. In melanoma sample #3, these three BAC clones along with zC195I16 were gained, whereas in melanoma sample #4 only two of the BAC clones were observed as a gain (zC226M16 and zK151G10). Overall these commonalities demonstrate potential for these genomic regions to possibly play a role in a common pathway for tumorigenesis in multiple cancer models.
DISCUSSION
The zebrafish has recently been established as an important model system for studying the molecular pathways of cancer (reviewed in Berghmans et al., 2005). To interrogate the genomes of zebrafish cancer models, we developed a genome-wide array CGH platform enriched with sequences containing genes orthologous to human oncogenes and tumor suppressor genes. The initial oncogene or tumor suppressor gene of interest for the specific BAC clones is listed in Supplementary Table S1 (annotation from Zv5). With the sequencing of the zebrafish genome about 80% complete, annotation and assembly of the genome is still preliminary. The latest build of the zebrafish genome (Zv7) was recently released by the Wellcome Trust Sanger Institute (www.sanger.ac.uk/Projects/D_rerio/). The BAC clones chosen for inclusion on this array CGH platform containing orthologous sequences to human oncogenes and tumor suppressor genes and in addition, those BAC clones significantly imbalanced in at least one of the zebrafish cancer models (which were designated as markers), were reannotated using Zv7 (Supplementary Table S1).
One important feature of this array is that each BAC clone chosen for inclusion on the array has been assigned to a precise cytogenetic location by FISH, allowing for direct integration with the zebrafish reference genome. When comparing the chromosomal assignment by cytogenetic mapping in comparison to the WebFPC database, 211 BAC clones (73.8%) were assigned to the same chromosome and only 8 BAC clones (2.8%) had different chromosomal assignments (Supplementary Table S2). The remaining 67 BAC clones (23.4%) have not yet been annotated and assigned to a specific chromosomal location in the WebFPC database (Supplementary Table S2), but using contig assignments these BAC clones can be integrated with the reference sequence. In addition, cytogenetic localization of the BAC clones by FISH allows for confirmable studies of the array CGH results using FISH on cells from the tumor samples of interest. BAC clones exhibiting gains or losses by array CGH analysis in this study, which were subsequently tested by FISH, were all confirmed, except for one of the losses in the T-ALL samples on LG chromosome 14.
Overall the number of genomic imbalances per individual in the RMS and T-ALL samples was fairly comparable (6 to 14 and 1 to 17, respectively), with the melanoma samples having more genomic imbalances per individual (6 to 28) in comparison to the other two cancer models. In addition, there were similarities in the specific BAC clones observed to be imbalanced within and between the three cancer models (Tables 2, 3 and 4). These analogous regions harboring genomic imbalances in multiple samples within and between the cancer models indicate potential for these regions to possibly be involved in common pathways of tumorigenesis.
Conservation of molecular processes between human and zebrafish tumors has been previously reported (Lam et al., 2006). Using the updated gene annotation list for the significantly imbalanced BAC clones for the three zebrafish cancer models, speculations about genes involved in the respective human cancer and future investigations into identifying genes and/or regulatory elements that are gained or lost during tumorigenesis can start to be hypothesized. For example in the zebrafish T-ALL model, genomic imbalances were observed in BAC clones containing genes which have been reported to play a role in hematological malignancies. EP300 (zK271F03), a transcriptional co-activator for the TAL1/SCL oncoprotein involved in T-ALL (Huang et al., 1999), was gained in one of the zebrafish T-ALL samples. HOXB3 (zK097O05), which is over expressed in myeloid leukemia (Lawrence et al., 1996; Chiba, 1998) was also gained in one of the zebrafish T-ALL samples. MLL2, which is associated with amplifications in leukemia (Prasad et al., 1997; Fitzgerald and Diaz, 1999), was gained in 2 samples. P2RY8 (zK255G14), an oncogene associated with biphenotypic acute leukemia (Fujiwara et al., 2007), was gained in one of the samples. In addition, imbalances were also observed in BAC clones containing genes reported to play a role in a variety of other cancers including hepatocellular carcinoma (PIM3; Fujii et al., 2005), pancreatic cancer (PIM3 and RAB20; Amillet et al., 2006; Li et al., 2006;), gastric adenoma-adenocarcinoma (PIM3; Zheng et al., 2008), glioblastoma (COL4A2; van den Boom et al., 2003; Ruano et al., 2006), mantle cell lymphoma (GLI2; Hedge et al., 2008), renal cell carcinoma (DIRC2 and HSPABP1; Bodmer et al., 2002; 2003), and thyroid cancers (APP; Krause et al., 2008). In the zebrafish melanoma model, KIT (zK245P14) was lost in one melanoma sample and KIT expression is reported to be lost in a majority of human melanomas during the transition from melanoma in situ to metastatic disease (Ryu et al., 2007). MITF (zK013A21) is gained in one melanoma sample. The MITF genomic locus is amplified and protein is over expressed in melanomas (Garraway et al., 2005). MYC (zC143E20 and zC194E15), which was gained in three melanoma samples, has also been reported to be amplified in many melanomas (Lin et al., 2008). In addition, the BRAF oncogene, which is amplified in human melanomas (Curtin et al., 2005; Lin et al., 2008) may show genomic imbalances in the zebrafish melanoma model. The mitfa:BRAFv600E transgene that drives formation of the zebrafish melanomas is integrated on chromosome 19 (C. Ceol and L. Zon, data not shown). The chromosomal region corresponding to zK219L12, the BAC clone most proximal to the transgene integration site, is gained in two of the melanoma tumors. These data suggest there may be conserved chromosomal abnormalities in human and zebrafish melanoma. It will be interesting in future investigations to compare this data to higher resolution platforms in the two species as a means to focus in on these imbalanced genomic regions and to identify genes that may play a role in tumorigenesis.
When applying this array CGH platform, normal DNA from the same fish should be used whenever possible as the reference DNA to ensure that the genomic imbalances observed are tumor-specific and not simply copy number variants among the different fish tested. Copy number variation has recently been implicated as a widespread phenomenon among humans and other vertebrates (Iafrate et al., 2004; Li et al., 2004; Sebat et al., 2004; Adams et al., 2005; Freeman et al., 2006; Perry et al., 2006; Redon et al., 2006; Graubert et al., 2007). Hence, the widespread presence of copy number variants complicates the analysis of tumor genomes, since comparisons are usually made between the DNA from a tumor specimen and genomic DNA from another individual presumed to be normal. When interpreting the data, genomic imbalances related to the tumors (i.e., somatic copy number changes associated with the tumor) must then be differentiated from germline copy number variants. With the zebrafish cancer models studied here, this is not an issue, since normal DNA can be obtained from the same fish allowing for the detection of tumor-specific genomic alterations. While copy number variation within the zebrafish genome has not yet been well established, it is likely that the genome of the zebrafish will also contain a substantial amount of copy variable regions between individual fish. In this study, normal DNA from the same fish was used as the reference DNA to avoid mistakenly identifying germline copy number variants as tumor-related genomic imbalances.
The zebrafish BAC-based array CGH platform is proving to be an excellent approach for the initial genome-wide identification of imbalances in zebrafish cancer models. This platform provides a valuable resource for interrogating the zebrafish genome and could be used in future applications to detect genomic imbalances in additional zebrafish developmental and disease mutants. In addition, this array platform may be helpful in beginning to assess copy number variation in the zebrafish genome. While this platform has lower resolution in comparison to array CGH platforms of other species, this BAC-based array is not dependent on fully sequenced DNA regions. Using the current zebrafish reference genome sequence to produce an array CGH platform (e.g., an oligonucleotide array) may limit coverage in non-annotated sequences. This BAC-based array CGH platform will complement higher resolution platforms that are produced in the future as the sequencing and annotation of the zebrafish nears completion.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Adeola Adeniyi, Cheryn Couter, and Stephanie Dallaire (Brigham and Women’s Hospital, Boston, MA, USA) for their technical assistance.
Supported by: This work was supported in part by National Institutes of Health grant R01-CA111560 (CL) and by a Ruth L. Kirschstein National Research Service Award (JLF).
REFERENCES
- Adams DJ, Dermitzakis ET, Cox T, Smith J, Davies R, Banerjee R, Bonfield J, Mullikin JC, Chung YJ, Rogers J, Bradley A. Complex haplotypes, copy number polymorphisms and coding variation in two recently divergent mouse strains. Nat Genet. 2005;37:532–536. doi: 10.1038/ng1551. [DOI] [PubMed] [Google Scholar]
- Amillet JM, Ferbus D, Real FX, Antony C, Muleris M, Gress TM, Goubin G. Characterization of human Rab20 overexpressed in exocrine pancreatic carcinoma. Hum Pathol. 2006;37:256–263. doi: 10.1016/j.humpath.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Berghmans S, Jette C, Langenau D, Hsu K, Stewart R, Look T, Kanki JP. Making waves in cancer research: new models in the zebrafish. Biotechniques. 2005;39:227–237. doi: 10.2144/05392RV02. [DOI] [PubMed] [Google Scholar]
- Bodmer D, Schepens M, Eleveld MJ, Schoenmakers EF, Geurts van Kessel A. Disruption of a novel gene, DIRC3-HSPBAP1 fusion transcipts in a case of familial renal cell cancer and t(2;3)(q35;q21) 2003 doi: 10.1002/gcc.10243. [DOI] [PubMed] [Google Scholar]
- Bodmer D, Eleveld M, Kater-Baats E, Janssen I, Janssen B, Weterman M, Schoenmakers E, Nickerson M, Linehan M, Zbar B, van Kessel AG. Disruption of a novel MFS transporter gene, DIRC2, by a familial renal cell carcinoma-associated t(2;3)(q35;q21) Hum Mol Genet. 2002;11:641–649. doi: 10.1093/hmg/11.6.641. [DOI] [PubMed] [Google Scholar]
- Cai W, Mao J, Chow C, Damani S, Balmain A, Bradley A. Genome-wide detection of chromosomal imbalances in tumors using BAC microarrays. Nat Biotech. 2002;20:393–396. doi: 10.1038/nbt0402-393. [DOI] [PubMed] [Google Scholar]
- Chiba S. Homeobox genes in normal hematopoiesis and leukemogenesis. Int J Hematol. 1998;68:343–353. doi: 10.1016/s0925-5710(98)00093-0. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, LeBoit PE, Pinkel D, Bastian BC. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Davies JJ, Wilson IM, Lam WL. Array CGH technologies and their applications to cancer genomes. Chromosome Res. 2005;13:237–248. doi: 10.1007/s10577-005-2168-x. [DOI] [PubMed] [Google Scholar]
- Feng H, Langenau DM, Madge JA, Quinkertz A, Gutierrez A, Neuberg DS, Kanki JP, Look AT. Heat-shock induction of T-cell lymphoma/leukemia in conditional Cre/lox-regulated transgenic zebrafish. Brit J Haematol. 2007;138:169–175. doi: 10.1111/j.1365-2141.2007.06625.x. [DOI] [PubMed] [Google Scholar]
- Fiegler H, Redon R, Carter NP. Construction and use of spotted large-insert clone DNA microarrays for the detection of genomic copy number changes. Nat Protocols. 2007;2:577–587. doi: 10.1038/nprot.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald KT, Diaz MO. MLL2: A new mammalian member of the trx/MLL family of genes. Genomics. 1999;59:187–192. doi: 10.1006/geno.1999.5860. [DOI] [PubMed] [Google Scholar]
- Freeman JL, Adeniyi A, Banerjee R, Dallaire S, Maguire SF, Chi J, Ng BL, Zepeda C, Humphray S, Rogers J, Zhou Y, Zon LI, Carter NP, Yang F, Lee C. Definition of the zebrafish genome using flow cytometry and cytogenetic mapping. BMC Genomics. 2007;8:195. doi: 10.1186/1471-2164-8-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME, Carter NP, Scherer SW, Lee C. Copy number variation: New insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- Fujii C, Nakamoto Y, Lu P, Tsuneyama K, Popivanova BK, Kaneko S, Mukaida N. Int J Cancer. 2005;114:209–218. doi: 10.1002/ijc.20719. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Yamashita Y, Choi YL, Watanabe H, Kurashina K, Soda M, Enomoto M, Hatanaka H, Takada S, Ozawa K, Mano H. Transforming activity of purinergic receptor P2Y, G protein coupled, 8 revealed by retroviral expression screening. Leuk Lymphoma. 2007;48:978–986. doi: 10.1080/10428190701225882. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, Lee C, Wagner SN, Li C, Golub TR, Rimm DL, Meyerson ML, Fisher DE, Sellers WR. Integrative genomic analyses identify MITF as a lineage survival oncogenes amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, Eis PS, Shannon WD, Li X, McLeod HL, Cheverud J, Ley JL. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genetics. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde GV, Munger CM, Emanuel K, Joshi AD, Greiner TC, Weisenburger DD, Vose JM, Joshi SS. Targeting of sonic hedgehog-GLI signaling: a potential strategy to improve therapy for mantel cell lymphoma. Mol Cancer Ther. 2008;7:1450–1460. doi: 10.1158/1535-7163.MCT-07-2118. [DOI] [PubMed] [Google Scholar]
- Hodgson G, Hager JH, Volik S, Hariono S, Wernick M, Moore D, Albertson DG, Pinkel D, Collins C, Hanahan D, Gray JW. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet. 2001;29:459–464. doi: 10.1038/ng771. [DOI] [PubMed] [Google Scholar]
- Huang S, Qiu Y, Stein RW, Brandt SJ. p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene. 1999;18:4958–4967. doi: 10.1038/sj.onc.1202889. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Krause K, Karger S, Sheu SY, Aigner T, Kursawe R, Gimm O, Schmid KW, Dralle H, Fuhrer D. Evidence for a role of the amyloid precursor protein in thyroid carcinogenesis. J Endocrinol. 2008 doi: 10.1677/JOE-08-0005. 0:JOE-08-0005. [DOI] [PubMed] [Google Scholar]
- Lakshmi B, Hall IM, Egan C, Alexander J, Leotta A, Healy J, Zender L, Spector MS, Xue W, Lowe SW, Wigler M, Lucito R. Mouse genomic representational oligonucleotide microarray analysis: Detection of copy number variations in normal and tumor specimens. Proc Natl Acad Sci USA. 2006;103:11234–11239. doi: 10.1073/pnas.0602984103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SH, Wu YL, Vega VB, Miller LD, Spitsbergen J, Tong Y, Zhan H, Govindarajan KR, Lee S, Mathavan S, Murthy KR, Buhler DR, Liu ET, Gong Z. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat Biotech. 2006;24:73–75. doi: 10.1038/nbt1169. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, Goessling W, Neuberg DS, Kunkel LM, Zon LI. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21:1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2005;102:6068–6073. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, Lin S, Prochownik E, Trede NS, Zon LI, Look TA. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- Lawrence HJ, Sauvageau G, Humphries RK, Largman C. The role of HOX homeobox genes in normal and leukemic hematopoiesis. Stem Cells. 1996;14:281–291. doi: 10.1002/stem.140281. [DOI] [PubMed] [Google Scholar]
- Lee C, Smith A. Molecular cytogenetic methodologies and bacterial artificial chromosome (BAC) probe panel resource for genomic analyses in zebrafish. Method Cell Biol. 2004;77:241–254. doi: 10.1016/s0091-679x(04)77013-2. [DOI] [PubMed] [Google Scholar]
- Li J, Jiang T, Mao JH, Balmain A, Peterson L, Harris C, Rao PH, Havlak P, Gibbs R, Cai WW. Genomic segmental polymorphisms in inbred mouse strains. Nat Genet. 2004;36:952–954. doi: 10.1038/ng1417. [DOI] [PubMed] [Google Scholar]
- Li YY, Popivanova BK, Nagai Y, Ishikura H, Fujii C, Mukaida N. Pim-3, a proto-oncogene with serine/threonine kinase activity, is aberrantly expressed in human pancreatic cancer and phosphorylates bad to block bad-mediated apoptosis in human pancreatic cancer cell lines. Cancer Res. 2006;66:6741–6747. doi: 10.1158/0008-5472.CAN-05-4272. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, Hodi FS, Dranoff G, Sellers WR, Thomas RK, Meyerson M, Golub TR, Dummer R, Herlyn M, Getz G, Garraway LA. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maydan JS, Filbotte S, Edgley ML, Lau J, Selzer RR, Richmond TA, Pofahl NJ, Thomas JH, Moerman DG. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Res. 2007;17:337–347. doi: 10.1101/gr.5690307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak NJ, Gaile D, Conroy JM, McQuaid D, Cowell J, Carter R, Goggins MG, Hruban RH, Maitra A. Genome-wide aberrations in pancreatic adenocarcinoma. Cancer Genet Cytogen. 2005;161:36–50. doi: 10.1016/j.cancergencyto.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, Berghmans S, Mayhall EA, Traver D, Fletcher CDM, Aster JC, Granter SR, Look AT, Lee C, Fisher DE, Zon LI. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Perry GH, Tchinda J, McGrath SD, Zhang J, Picker SR, Caceres AM, Iafrate AJ, Tyler-Smith C, Scherer SW, Eichler EE, Stone AC, Lee C. Hotspots for copy number variation in chimpanzees and humans. Proc Natl Acad Sci USA. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Albertson DG. Comparative genomic hybridization. Annu Rev Genomics Hum Genet. 2005;6:331–354. doi: 10.1146/annurev.genom.6.080604.162140. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo W-L, Chen C, Dairkee SH, Ljung B, Gray JW. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Prasad R, Zhadanov AB, Sedkov Y, Bullrich F, Druck T, Rallapalli R, Yano T, Alder H, Croce CM, Huebner K, Mazo A, Canaani E. Structure and expression pattern of human ALR, a novel gene with strong homology to ALL-1 involved in acute leukemia and to Drosophila trithorax. Oncogene. 1997;15:549–560. doi: 10.1038/sj.onc.1201211. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles M. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano Y, Mollejo M, Ribalta T, Fiano C, Camacho FI, Gomez E, de Lope AR, Hernandez-Moneo JL, Martinez P, Melendez B. Identification of novel candidate target genes in amplicons of Glioblastoma multiforme tumors detected by expression and CGH microarray profiling. Mol Cancer. 2006;5:39. doi: 10.1186/1476-4598-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu B, Kim DS, Deluca AM, Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS ONE. 2007;2:e594. doi: 10.1371/journal.pone.0000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander S, Bullinger L, Karlsson A, Giuriato S, Hernandez-Boussard T, Felsher DW, Pollack JR. Comparative genomic hybridization on mouse cDNA microarrays and its application to a murine lymphoma model. Oncogene. 2005;24:6101–6107. doi: 10.1038/sj.onc.1208751. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- Thomas R, Duke SE, Bloom SK, Breen TE, Young AC, Feiste E, Seiser EL, Tsai PC, Langford CF, Ellis P, Karlsson EK, Lindblad-Toh K, Breen M. A cytogenetically characterized, genome-anchored 10-Mb BAC set and CGH array for the domestic dog. J Hered. 2007;98:474–484. doi: 10.1093/jhered/esm053. [DOI] [PubMed] [Google Scholar]
- van den Boom J, Wolter M, Kuick R, Misek DE, Youkilis AS, Wechsler DS, Sommer C, Reifenberger G, Hanash SM. Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am J Pathol. 2003;163:1033–1043. doi: 10.1016/S0002-9440(10)63463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng HC, Tsuneyama K, Takahashi H, Miwa S, Sugiyama T, Popivanova BK, Fujii C, Nomoto K, Mukaida N, Takano Y. Aberrant Pim-3 expression is involved in gastric adenoma-adenocarcinoma sequence and cancer progression. J Cancer Res Clin Oncol. 2008;134:481–488. doi: 10.1007/s00432-007-0310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.