Abstract
In the damaged heart, cardiac adaptation relies primarily on cardiomyocyte hypertrophy. The recent discovery of cardiac stem cells in the postnatal heart, however, suggests that these cells could participate in the response to stress via their capacity to regenerate cardiac tissues. Using models of cardiac hypertrophy and failure, we demonstrate that components of the Notch pathway are up-regulated in the hypertrophic heart. The Notch pathway is an evolutionarily conserved cell-to-cell communication system, which is crucial in many developmental processes. Notch also plays key roles in the regenerative capacity of self-renewing organs. In the heart, Notch1 signaling takes place in cardiomyocytes and in mesenchymal cardiac precursors and is activated secondary to stimulated Jagged1 expression on the surface of cardiomyocytes. Using mice lacking Notch1 expression specifically in the heart, we show that the Notch1 pathway controls pathophysiological cardiac remodeling. In the absence of Notch1, cardiac hypertrophy is exacerbated, fibrosis develops, function is altered, and the mortality rate increases. Therefore, in cardiomyocytes, Notch controls maturation, limits the extent of the hypertrophic response, and may thereby contribute to cell survival. In cardiac precursors, Notch prevents cardiogenic differentiation, favors proliferation, and may facilitate the expansion of a transient amplifying cell compartment.
In the Western world, cardiovascular diseases remain the leading cause of mortality and morbidity (1). Moreover, the increasing mean age of the population has altered the spectrum of cardiac diseases toward heart failure. Heart failure is a progressive disorder that is initiated by a loss of cardiomyocytes (2, 3). The primary event can be either acute, as in the case of myocardial infarction, or gradual, as in the case of hemodynamic overload, for instance in patients suffering from chronic hypertension. In the classical view, the heart contains at birth a predetermined number of cardiomyocytes, the loss of which cannot be compensated for, and the adaptive response of the heart to stress relies exclusively on cardiomyocyte hypertrophy. Several groups have, however, identified resident cells in the heart with properties of cardiac precursor cells (CPCs) (4–7). The existence of CPCs suggests that cardiac integrity also depends on a balance between cell death and cell production. Therefore, to understand the cellular mechanisms of cardiac adaptation, we need to identify pathways controlling cardiomyocyte growth as well as that regulating the level of the cardiac progenitor pool and its commitment toward the cardiogenic lineage.
The Notch signaling pathway is crucial in the development of metazoans (8, 9). It has also been implicated in the regeneration of adult self-renewing tissues (10, 11). In mammals, signaling occurs after the interaction of one of the four Notch receptors (Notch1–4) with membrane-bound ligands of the Jagged (Jagged1 and 2) and Delta-like (Delta-like1, 3, and 4) family. Therefore, the Notch pathway is essentially a communication system between two adjacent cells, a signal-sending cell expressing the ligand and a signal-receiving cell expressing the receptor. The Notch protein is synthesized as a single polypeptide that is cleaved by proteases during posttranslational processing. The two portions of the protein, however, remain associated and form a functional heterodimer on the cell surface. Upon activation, the Notch receptor is subjected to two additional cleavages by TACE (TNF-α converting enzyme) and by a multicomponent γ-secretase complex, thus releasing the Notch intracellular domain. This fragment enters the nucleus, binds to a transcription factor known as RBP-J in the mouse, recruits coactivators, and up-regulates target gene transcription. Prototypic target genes of Notch are bHLH (basic helix-loop-helix) repressors of the Hairy/enhancer of split (Hes) family. The Hes family includes Hes-1 and Herp (Hairy-related transcription factors) also known as Hey (12).
The biological effects of Notch activation are extremely context dependent. In certain situations, Notch restricts cell fate and maintains an undifferentiated state in uncommitted progenitors, whereas in others Notch produces inductive signaling that stimulates cells to adopt a particular fate (11). For instance, Notch preserves stem cell pools, regulates lateral inhibition, and controls asymmetrical divisions but has also been shown to induce terminal differentiation. Cell fate decisions and differentiation during hematopoiesis and lymphopoiesis are controlled by Notch. In hemangioblasts, Notch signaling could specify hematopoietic versus endothelial cell fate. In the adult, Notch regulates T versus B cell lineage decisions (13). Furthermore, Notch plays an important role in arteriovenous patterning, in regulating endothelial tip cell and vessel wall formation (14). The Notch pathway has also been implicated in cardiac morphogenesis (15, 16). Notch plays a crucial role in the regulation of endocardial epithelial-to-mesenchymal transformation in the primary heart tube during septation and formation of the valves (17, 18). Notch1 is the main receptor subtype found in the endocardial layer in the outflow tract of the developing heart (19). Interestingly, Notch1 haploinsufficiency in human results in heart disease characterized by cardiac valve calcification and ventricular septal defects (20). Furthermore, patients suffering from Alagille syndrome, an autosomal dominant disorder caused by mutations in the Jagged1 gene, demonstrate severe cardiac abnormalities including tetralogy of Fallot (21, 22). Therefore, the importance of the Notch pathway in cardiac development has been extensively studied but its role in the postnatal heart has not been investigated. In the present study, we demonstrate that cardiac remodeling, which develops secondary to hemodynamic overload or in response to chronic agonist stimulation in the adult heart, is in part regulated by the Notch1 receptor pathway. Importantly, Notch signaling is observed in both cardiomyocytes and mesenchymal CPCs. In cardiomyocytes, Notch represses cardiac gene expression and prevents an excessive and detrimental hypertrophic response. In CPCs, Notch appears to regulate proliferation and differentiation.
RESULTS
The Notch pathway is activated in cardiac myocyte and nonmyocyte cells
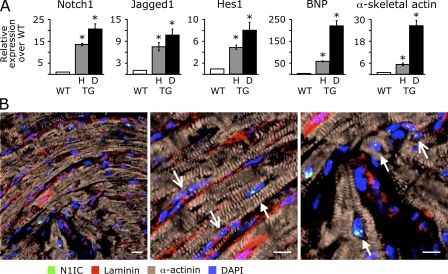
Agonist-induced cardiac hypertrophy in TG1306/1R transgenic mice with cardiac angiotensin II overproduction leads to a gradual transition from a compensatory state to heart failure (23, 24). To gain insight into the molecular mechanisms that play a role in maintaining cardiac integrity in the diseased heart, we performed a comparative study of gene expression between normal and stressed hearts using microarray analysis. TG1306/1R transgenics were separated into two phenotypic groups based on morphological parameters (cardiac weight index and histology) and either a moderate or a high up-regulation of markers of hypertrophy and failure (Table S1, available at http://www.jem.org/cgi/content/full/jem.20081427/DC1) and compared with age- and sex-matched WT. Microarray data revealed that several genes belonging to pathways important during cardiac morphogenesis were activated in hypertrophic and dilated adult hearts (Tables S2–S7). In particular, the activated genes were those encoding the Notch1 receptor, its ligand Jagged1, the transcription factor RBP-J, and the target gene of Notch Hes1. These results were confirmed using quantitative RT-PCR (Fig. 1 A). First, both dilated and hypertrophic hearts demonstrated significant up-regulation of brain natriuretic peptide (BNP) and α-skeletal actin expression, two markers of cardiac stress. BNP and α-skeletal actin were more expressed in the dilated than in the hypertrophic heart as expected. More importantly, there was a 15–20-fold increase in cardiac expression of the Notch1 receptor, whereas Jagged1 expression was up-regulated ∼10-fold. Expression of both the Notch1 receptor and its ligand was higher in the dilated hearts than in the hypertrophic hearts. Accordingly, Hes1 expression was also significantly up-regulated in the stressed adult hearts.
Figure 1.
Notch signaling is activated in cardiac myocytes and nonmyocyte cells in the adult heart. (A) Quantitative real-time RT-PCR analysis of gene expression showing the up-regulation of Notch1, Jagged1, and Hes1 in hypertrophic (H) and dilated (D) hearts of TG1306/1R transgenic mice relative to hearts of WT mice. The relative expression levels of BNP and α-skeletal actin are also shown. Asterisks denote significant difference between the groups (P < 0.05). n = 6 per group. Error bars show the mean ± SD. (B) Confocal immunofluorescence analysis of activated Notch1 in adult mouse myocardium. Tissue sections were stained with an antibody directed against the intracellular domain of proteolytically cleaved Notch1 (N1IC). Cardiomyocytes were revealed using anti–α-actinin antibody. Laminin immunostaining was used to delimit cell boundaries, and nuclei were stained with DAPI. Closed and open arrows indicate cardiomyocytes and CPCs, respectively. Bars, 10 μm.
To identify cardiac cells in which the Notch pathway was activated, we performed immunohistological analysis of adult heart tissue using antibodies directed against N1IC (the intracellular domain of Notch1), the cleaved portion of the Notch1 receptor. In initial experiments, we analyzed Notch activation in cardiac cells expressing the stem cell antigen (Sca) 1 because this marker has been previously used to identify resident CPCs (Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20081427/DC1). Clearly, N1IC staining was not limited to the Sca-1–positive population in the adult heart in WT mice or in TG1306/1R transgenics. Furthermore, not only nonmyocyte cells but also some cardiomyocytes were positive for N1IC staining. In different experiments, we isolated the mesenchymal fraction from neonatal mouse hearts, which was shown previously to contain Sca-1–positive precursor cells. Approximately 35% of cells stained positive for Sca-1 in this fraction. However, very few if any cells expressing Sca-1 demonstrated Notch1 activation (Fig. S1 B). Based on these results, we decided to use a different approach, which consisted of analyzing all cardiac N1IC-positive cells and not a preselected subpopulation. To distinguish between cardiac myocytes and nonmyocyte cells and to identify cellular boundaries, sarcomeric α-actinin and -laminin stainings were simultaneously performed. Fig. 1 B depicts a representative experiment, which demonstrates that N1IC was present in the nuclei of both cardiomyocytes (α-actinin positive, closed arrows) and nonmyocyte cells (α-actinin negative, open arrows). Not all cardiac myocytes demonstrated N1IC staining. Similarly, small N1IC-positive cardiac undifferentiated cells, which clustered in the interstitial mesenchyme between cardiac fibers, were readily detected.
The Notch pathway regulates the cardiac response to hypertrophic stress in vivo
To determine whether the Notch pathway plays a functional role during the development of cardiac hypertrophy, TG1306/1R transgenics were administrated with a γ-secretase inhibitor (LY411,575). This compound was previously shown to block both the proteolytic release of N1IC and the specific activation of Notch target gene expression (25). The drug was injected into 15-wk-old mice to investigate the role of Notch during the compensatory phase of the cardiac response to stress. To verify effective Notch inhibition, the percentage of marginal zone B (MZB) cells in the spleen (i.e., B220+ and CD21+, CD23− splenocytes), which strictly depend on active Notch signaling (26), was measured by FACS analysis (Fig. S2, A and B, available at http://www.jem.org/cgi/content/full/jem.20081427/DC1). Treatment with the γ-secretase inhibitor resulted in a dramatic decrease in MZB cells, demonstrating successful Notch inhibition. Importantly, the expression of Hes1, Hey1, and Hey2, as well as of Notch1 and Jagged1, was down-regulated in the hearts of transgenics receiving the inhibitor (Fig. 2 A). Treatment resulted in a concomitant stimulation of expression of cardiac makers of hypertrophy in treated hearts (Fig. 2 B). Furthermore, transgenic mice developed more hypertrophy upon Notch blockade than did vehicle-treated transgenics (Fig. 2, C and D; and Table I). Blood pressure and heart rate remained unaffected (Table I). Finally, echocardiography confirmed the thickening of the left ventricular wall and revealed a significantly increased fraction of shortening in the transgenic mice under γ-secretase inhibition. Administration of the γ-secretase inhibitor had no effects on cardiac structure and function in WT animals despite a demonstrated blockade of the Notch pathway (Fig. 2 and Table I).
Figure 2.
Inhibition of Notch signaling exacerbates cardiac hypertrophy. (A) Quantitative real time RT-PCR analysis of expression of Notch1, Jagged1, and Notch target genes Hes1, Hey1, and Hey2 in the hearts of WT and TG13061/R transgenic mice treated with the γ-secretase inhibitor Ly411,575 relative to their vehicle-treated counterparts. (B) Quantitative RT-PCR analysis of expression of markers of cardiac hypertrophy and failure in the hearts of WT and transgenic mice, treated as in A, relative to vehicle-treated mice. (C) Representative transverse histological sections showing exacerbated hypertrophy in hearts from TG1306/1R transgenic mice treated with the γ-secretase inhibitor Ly411,575 compared with control mice. (D) The heart weight/body weight ratio (cardiac weight index [CWI]) of vehicle- and γ-secretase inhibitor–treated transgenic and WT mice is represented. Asterisks denote significant difference between the groups (P < 0.05). n = 6–8 mice per group. Error bars show the mean ± SD. Bar, 2 mm.
Table I.
Physiological and echocardiographic parameters in mice treated with the γ-secretase inhibitor LY411,575
| WT
|
TG1306/1R
|
|||
|---|---|---|---|---|
| Vehicle (n = 5–6) | Inhibitor (n = 6) | Vehicle (n = 6) | Inhibitor (n = 6) | |
| Heart weight (mg) | 122.8 ± 7.1 | 125.5 ± 11.1 | 141.3 ± 7.8 | 183.7 ± 14.3** |
| Body weight (g) | 24.9 ± 1.0 | 25.0 ± 0.4 | 26.8 ± 0.9 | 26.1 ± 1.1 |
| Heart weight/body weight ratio (mg/g) | 4.9 ± 0.2 | 5.0 ± 0.5 | 5.3 ± 0.2 | 7.1 ± 0.7** |
| Blood pressure (mm Hg) | 114.4 ± 2.7 | 111.0 ± 5.2 | 114.7 ± 3.6 | 113.6 ± 3.8 |
| Heart rate (bpm) | 611 ± 14 | 609 ± 6 | 617 ± 6 | 606 ± 16 |
| Left ventricular wall thickness (mm) | ||||
| diastole | 1.57 ± 0.04 | 1.59 ± 0.10 | 1.66 ± 0.09 | 2.14 ± 0.13*†† |
| systole | 1.99 ± 0.03 | 1.90 ± 0.12 | 1.98 ± 0.10 | 2.39 ± 0.13**†† |
| Left ventricular diameter (mm) | ||||
| diastole | 3.03 ± 0.04 | 2.87 ± 0.15 | 3.12 ± 0.15 | 3.23 ± 0.13*†† |
| systole | 1.73 ± 0.06 | 1.55 ± 0.06 | 1.71 ± 0.14 | 1.59 ± 0.06 |
| Septum thickness (mm) | ||||
| diastole | 0.33 ± 0.02 | 0.38 ± 0.02 | 0.33 ± 0.03 | 0.34 ± 0.03 |
| systole | 0.46 ± 0.02 | 0.48 ± 0.01 | 0.39 ± 0.05 | 0.51 ± 0.06 |
| Fraction of shortening (%) | 43.1 ± 1.6 | 45.8 ± 1.0 | 45.7 ± 2.2 | 50.7 ± 1.2**†† |
Statistical significance: *, P < 0.05 and **, P < 0.01 as compared to corresponding vehicle-treated groups; †, P < 0.05 and ††, P < 0.01 as compared to corresponding WT groups.
Notch1 receptor-mediated activation of the Notch pathway in the heart
To study the activation of the Notch pathway at the onset of the hypertrophic response, we used a model of renovascular hypertension, namely the 1K1C (one kidney, one clip) model, which produces hemodynamic overload and induces cardiac remodeling (27). Immunohistochemistry was performed to quantify the number of N1IC-positive cells to evaluate the size of the cardiac cell population with an activated Notch pathway (Fig. 3 A). Approximately 80% of the N1IC-positive cells in the normal adult heart were cardiomyocytes, and this percentage was not strikingly modified under stress. However, hearts from clipped mice showed an increased number of N1IC-positive cells 8 d after surgery. This number decreased to levels observed in sham-operated animals after 20 d. Because the Notch1 receptor was the predominant form expressed in the adult heart, we evaluated the physiological importance of this receptor using conditional Notch1 KOs. Thus, MLC2v-Cre mice with cardiac-specific expression of the Cre recombinase (28) were crossed to Notch1lox/lox animals (29). Mice with cardiac-specific Notch1 deletion (Notch1lox/lox; MLC2v-Cre, thereafter cNotch1 KOs) reached adulthood and appeared grossly normal (unpublished data). To evaluate the heart under stress, work overload was induced using the hypertensive 1K1C model. 8 wk after clipping, WT and cNotch1 KOs developed the same level of hypertension with comparable heart rates (Table II). Heart weight/body weight ratio was similar in both hypertensive groups. However, echographic analysis revealed several differences (Table II). The left ventricular free wall was thicker in mutant mice. In addition, cNotch1 KOs had a larger left ventricular diameter in systole along with a decreased fractional shortening when compared with controls. This paralleled an increase in the expression of cardiac markers of hypertrophy in the hearts of hypertensive KOs (Fig. 3, B and C). Moreover, marked fibrosis was detected in the hypertrophic hearts of mice lacking Notch1 (Fig. 3 D). Quantitation of extracellular matrix accumulation demonstrated an ∼10-fold difference between hearts from hypertensive WT and Notch1-deficient animals (unpublished data). Finally, mice with cardiac-specific Notch1 deletion also showed a higher rate of mortality under conditions of hemodynamic overload than normal littermates (Fig. 3 E). To evaluate whether increased fibrosis and decreased cardiac function were caused by accelerated cardiomyocyte loss, we determined the rate of apoptosis using TUNEL assay. Fig. 4 A shows apoptotic cardiomyocytes in the stressed heart of cNotch1 KOs. Quantitative analysis demonstrated that the number of TUNEL-positive cells was greater in 1K1C mice lacking Notch1 than in WT (Fig. 4 B).
Figure 3.
Notch1 controls the adaptive cardiac response to stress. (A) Quantitation of Notch1 activation in the adult stressed heart. Heart tissue sections from clipped (1K1C) and sham-operated (sham) mice were stained with an antibody against proteolytically activated N1IC at 8 and 20 d after clipping. The N1IC-positive nuclei were counted in representative sections and represented as percentage of total nuclei. n = 5–8 areas per section and 3 mice per group. (B and C) Quantitative RT-PCR analysis of the expression of cardiac marker genes in sham and 1K1C mice. N1Del, mice lacking Notch1 in the heart. n = 5–7 mice per group. Error bars show the mean ± SD. (D) Development of cardiac fibrosis in mice lacking Notch1 in the heart. Heart tissue sections from sham and clipped WT and N1Del mice were subjected to van Gieson staining to reveal fibrosis (red). (E) Mortality rate in clipped and sham-operated WT mice and mice lacking Notch1 in the heart (N1Del). n = 20–25 mice per group. Asterisks in A, B, and C denote significant difference (P < 0.01). Bar, 2 mm.
Table II.
Physiological and echocardiographic parameters in sham-operated and 1K1C mice lacking Notch1 in the heart
| WT
|
cNotch1 KOs
|
|||
|---|---|---|---|---|
| Sham (n = 4–6) | 1K1C (n = 5–6) | Sham (n = 4–6) | 1K1C (n = 5–8) | |
| Heart weight (mg) | 96.3 ± 3.6 | 156.2 ± 11.2 ** | 97.3 ± 3.6 | 144.2 ± 1.3 ** |
| Body weight (g) | 26.0 ± 0.6 | 27.2 ± 0.7 | 25.7 ± 0.7 | 26.2 ± 0.8 |
| Heart weight/body weight ratio (mg/g) | 3.7 ± 0.1 | 5.8 ± 0.5 ** | 3.8 ± 0.1 | 5.5 ± 0.5 ** |
| Blood pressure (mm Hg) | 117.1 ± 4.3 | 149.0 ± 3.2 ** | 120.3 ± 2.3 | 148.0 ± 3.2 ** |
| Heart rate (bpm) | 556 ± 90 | 528 ± 51 | 640 ± 20 | 561 ± 75 |
| Left ventricular wall thickness (mm) | ||||
| diastole | 1.32 ± 0.04 | 1.89 ± 0.04 ** | 1.39 ± 0.04 | 2.11 ± 0.04 **†† |
| systole | 1.75 ± 0.09 | 2.23 ± 0.07 ** | 1.69 ± 0.03 | 2.43 ± 0.05 **† |
| Left ventricular diameter (mm) | ||||
| diastole | 3.18 ± 0.19 | 3.17 ± 0.12 | 2.97 ± 0.07 | 3.17 ± 0.08 |
| systole | 1.75 ± 0.06 | 1.60 ± 0.08 | 1.71 ± 0.07 | 2.01 ± 0.07 **†† |
| Septum thickness (mm) | ||||
| diastole | 0.38 ± 0.04 | 0.39 ± 0.09 | 0.34 ± 0.02 | 0.49 ± 0.04 ** |
| systole | 0.48 ± 0.04 | 0.60 ± 0.13 | 0.46 ± 0.02 | 0.61 ± 0.05 ** |
| Fraction of shortening (%) | 44.8 ± 1.0 | 49.5 ± 1.5 * | 42.4 ± 0.8 | 36.4 ± 0.8 **†† |
Statistical significance: *, P < 0.05 and **, P < 0.01 as compared to corresponding vehicle-treated groups; †, P < 0.05 and ††, P < 0.01 as compared to corresponding WT groups.
Figure 4.
Increased apoptosis in the heart tissue of clipped N1Del mice. (A) Heart tissue sections were processed for detection of apoptotic nuclei using TUNEL assay and for detection of cardiac myocytes using anti-myomesin antibody. Representative photomicrographs of TUNEL-positive nuclei in α-actinin–positive cardiomyocytes and in nonmyocyte cells are shown. (B) Quantitative analysis of apoptosis in the hearts of clipped mice. Heart tissue sections cut at anatomically equivalent locations were stained as in A. The total number of TUNEL-positive nuclei per section was counted under a fluorescence microscope. The bar graph represents the mean number of nuclei in sham operated (n = 4) and clipped (n = 4) WT mice and sham (n = 3) and clipped (n = 5) N1Del mice. The asterisk denotes significant difference (P < 0.05) between clipped WT and N1Del mice. Error bars show the mean ± SD. Bar, 20 μm.
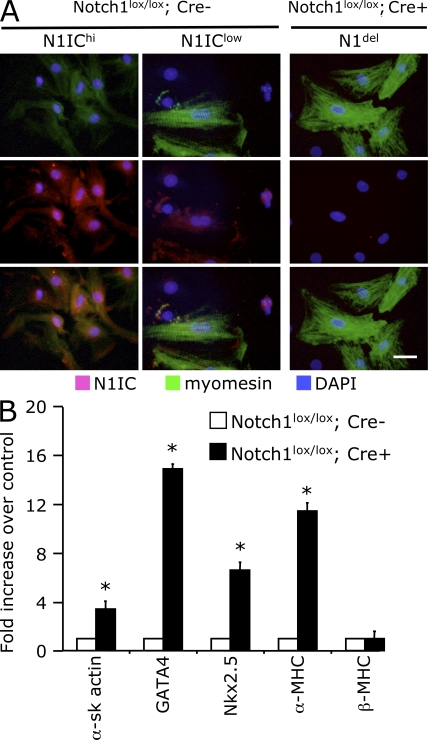
To investigate the role of the Notch pathway in cardiac myocytes, we studied neonatal mouse cardiomyocytes isolated from WT and cNotch1 KOs (Fig. 5 A). When stained for N1IC, cultured WT cardiomyocytes demonstrated two different distribution patterns. Some cells were characterized by intense nuclear N1IC staining. Diffuse myomesin staining suggested that these cells were poorly differentiated or in the process of dedifferentiation. In contrast, mature differentiated cardiomyocytes with organized sarcomeres showed little or no N1IC staining. Notch1-deficient cardiomyocytes presented a state of maturation that appeared to be even more advanced than that of the WT differentiated population. Furthermore, the expression of cardiac genes in myocytes isolated from mutant animals was strikingly up-regulated as compared with WT cells (Fig. 5 B). Interestingly, the expression of the α-MHC gene (a marker of cardiac maturation) was highly stimulated in cardiomyocytes lacking Notch1, whereas β-MHC gene expression (a maker of cardiac hypertrophy) was similar to that measured in WT cells. Finally, we demonstrated that the N1IC staining decreased markedly in phenylephrine-treated WT cardiomyocytes, suggesting that inactivation of Notch1 signaling could also be necessary for the initiation of the hypertrophic response (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20081427/DC1).
Figure 5.
Notch1 activation controls maturation and cardiac gene expression in cardiomyocytes. (A) Analysis of Notch1 activation in cultured cardiomyocytes isolated from neonatal mice carrying floxed Notch1 allele (Notch1lox/lox) without (Cre−) or with (Cre+) Cre recombinase expression under the control of the MLC2v promoter. The cultures were stained for activated Notch1 (N1IC) and myomesin. The Cre− (WT) cardiomyocytes demonstrate either high (N1IChi) or low (N1IClow) N1IC staining concomitant with poor or elaborate myofibrillar structure, respectively. (B) Quantitative RT-PCR analysis demonstrating the up-regulation of cardiac-specific gene expression in cultures of Cre+ (Notch1 deleted) cardiac myocytes relative to Cre− myocyte cultures. Error bars represent the means ± SD. Bar, 20 μm.
The Notch pathway controls cardiogenic differentiation in the cardiac precursor population
Immunostaining analysis revealed that, besides being stimulated in cardiomyocytes, the Notch pathway was also activated in cells belonging to an undifferentiated mesenchymal fraction that was shown previously to contain CPCs (Fig. 1). Therefore, the expression of GATA4 and Nkx2.5 on cardiac nonmyocyte cells was examined in TG1306/1R transgenics to evaluate whether mesenchymal Notch signaling cells were committed to the cardiogenic lineage. The vast majority of undifferentiated N1IC-positive cells coexpressed GATA4 (Fig. 6 A). In addition, cells with an activated Notch pathway expressed Nkx2.5 (Fig. 6 B). N1IC-negative Nkx2.5-positive cells were also found in the same clusters. Because the Notch pathway has been shown to be involved in sustaining proliferation in other organs systems, we investigated whether N1IC-positive CPCs were actively cycling. First, we demonstrated that CPCs with an activated Notch pathway (N1IC positive) and expressing the cell proliferation marker Ki67 could be readily detected in the adult heart (Fig. 6 C). Using laminin staining to identify cell boundaries, we confirmed that small nonmyocyte cells expressed Ki67. In addition, some cardiomyocytes were also Ki67 positive (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20081427/DC1). Furthermore, we quantified BrdU-positive cells in the stressed adult heart using the 1K1C model (Fig. 6, D and E). In WT animals, the percentage of BrdU-positive cells increased in the stressed heart. Interestingly, the increase in the number of labeled cells in the hearts of hypertensive mice lacking Notch1 was significantly reduced as compared with their WT littermates (Fig. 6 E).
Figure 6.
Notch1 activation in proliferating undifferentiated cardiac-committed cells in the adult myocardium. (A) Confocal microscopy of normal heart tissue sections showing activation of Notch1 (N1IC) in GATA4-positive α-actinin–negative cells. Nuclei are stained with DAPI. (B) Staining of cardiac tissue sections, as in A, showing N1IC staining in Nkx2.5-positive nuclei of undifferentiated α-actinin–negative cells. (C) Activation of Notch1 (N1IC) in proliferating Ki67-positive α-actinin–negative cells. (D) Analysis of BrdU incorporation in hearts of clipped (1K1C) and sham-operated (sham) WT mice and mice lacking Notch1 in the heart (N1del). Tissue sections of hearts from mice administered with BrdU for 96 h in drinking water were stained with anti-BrdU antibody. The BrdU+ nuclei are red. (E) Quantitation of BrdU incorporation represented as percentage of BrdU+ nuclei. * and † denote significance at P < 0.01 within groups and between groups, respectively. Error bars show the mean ± SD. Bars: (A, B, and C) 10 μm; (D) 100 μm.
To further analyze the role of Notch in the control of postnatal neocardiogenesis, we isolated the cardiac mesenchymal fraction from WT neonatal mice, which was previously shown to contain CPCs (4–7). Under conditions that favored expansion, CPCs demonstrated nuclear N1IC staining. These proliferating cells also appeared to be committed to the cardiogenic lineage as judged by the colocalization of N1IC and Nkx2.5 staining (Fig. 7 A, left). After the induction of differentiation, single Nkx2.5-positive cells were also found adjacent to double-positive precursors (Fig. 7 A, middle, arrows). At an even later time point, differentiated α-actinin–positive cardiomyocytes with assembled myofibrillar apparatus appeared in the culture. These cells were N1IC negative (Fig. 7 A, right, arrows). Cardiogenic differentiation was also evident by the enhanced expression of late cardiac-specific genes such as cardiac troponin I and α–myosin heavy chain in differentiating cells (Fig. 7 B). Interestingly, a concomitant down-regulation of Jagged1 expression was observed in differentiating cultures. Expression of Hes1, a direct Notch target gene, was also diminished. This occurred in the absence of Notch1 receptor down-regulation. To evaluate whether inhibition of the Notch pathway could stimulate cardiogenesis in isolated CPCs, precursors were cultured in the presence of the γ-secretase inhibitor DAPT. Treated cells demonstrated an acceleration of spontaneous differentiation into cardiomyocytes (Fig. 7 C). Mesenchymal CPCs were also obtained from neonatal cNotch1 KOs. In this population, similar to what we observed in WT cells maintained in the presence of DAPT, the differentiation toward the cardiogenic lineage was significantly advanced.
Figure 7.
Notch signaling controls differentiation of CPCs into mature cardiomyocytes. (A) Analysis of Notch1 activation in growing and differentiating cardiac-committed nonmyocyte cells in vitro. Cultures of nonmyocyte cells isolated from neonatal hearts were maintained in expansion conditions and stained for activated Notch1 (N1IC, green) and Nkx2.5 (red). The cultures were shifted to differentiation conditions and stained either for N1IC (green) and Nkx2.5 (red) in the middle column or for N1IC (green) and α-actinin (red) in the right column. Arrows indicate cardiac-committed or differentiated cardiomyocyte nuclei lacking N1IC staining. (B) Quantitative RT-PCR analysis of expression of markers of cardiac differentiation. Nonmyocyte cells were maintained in differentiation medium or control medium and analyzed by quantitative RT-PCR for the expression of the indicated genes. Down-regulation of Jagged1 and Hes1 expression during nonmyocyte differentiation in vitro is also presented. The expression levels are relative to controls maintained in expansion medium. Results are presented as the mean of three independent experiments. (C) Pharmacological and genetic inhibition of Notch accelerates differentiation in vitro. On the left, nonmyocyte cultures were established from WT neonatal mouse hearts and placed in differentiation medium in presence of vehicle alone (DMSO) or the γ-secretase inhibitor DAPT. On the right, nonmyocyte cultures were established from WT or Notch1lox/lox MLC2vCre+ (N1del) newborn mice and placed in differentiation medium. RNA was isolated from the differentiated cultures and analyzed by semiquantitative RT-PCR for the expression of the indicated genes. Representative results of three independent experiments are shown. Bar, 20 μm.
Jagged1 expression is stimulated in the stressed heart
Because the adult heart predominantly expressed Jagged1, this suggested that it could be the main ligand involved in the activation of the Notch pathway. Therefore, we first determined the nature of the cardiac cells expressing Jagged1. Jagged1 expression was readily detected in cardiomyocytes. The staining pattern, in particular the alignment along the muscle fibers, was compatible with the expected surface expression of Jagged1 (Fig. 8, A and B). Jagged1 on the surface of cardiomyocytes could activate both adjacent Notch-expressing myocytes and CPCs. Indeed, α-actinin–negative Nkx2.5-positive cells can be observed lying adjacent to Jagged1-expressing fibers (Fig. 8 B, arrows). We then investigated cardiac expression of Jagged1 in the stressed heart using hypertensive 1K1C WT as well as TG1306/1R transgenic mice (Fig. 8, C and D). Sham-operated WT animals in the 1K1C model demonstrated a patchy pattern of Jagged1 expression distributed throughout both cardiac ventricles (Fig. 8 C). In hypertensive 1K1C WT mice, no change was noticed during the first 4 d after induction of the hypertrophic response. However, 8 and 12 d after induction, there was a dramatic up-regulation of Jagged1 expression in the myocardium. Similarly, Jagged1 expression was markedly up-regulated in all cardiomyocytes of TG13061R transgenic mice as compared with their WT littermates (Fig. 8 D).
Figure 8.
Up-regulation of Jagged1 expression in the stressed heart. (A) Immunofluorescence analysis of Jagged1 expression in the heart. The micrograph shows Jagged1 expression on the surface of α-actinin–positive cardiomyocytes with an enlarged area shown in the inset. Nuclei are stained with DAPI. (B) Confocal micrograph showing Jagged1 expression on the surface of α-actinin–positive cardiomyocytes. Arrows in the inset indicate undifferentiated Nkx2.5-positive α-actinin–negative CPCs lying between Jagged1-expressing fibers. The nuclei are stained with DAPI. (C) Heart tissue sections from sham-operated (sham) and clipped (1K1C) mice were subjected to alkaline phosphatase immunohistochemistry using an antibody directed against Jagged1. The representative micrographs show Jagged1 expression in the myocardium at 4, 8, and 12 d after surgery. (D) Jagged1 immunostaining in WT and TG1306/1R transgenic mice as in C. Bars: (A and B) 20 μm; (C and D) 100 μm.
DISCUSSION
In this study, we show that the Notch pathway regulates the cardiac response to stress via the Notch1 receptor. Specifically, we demonstrate that: (a) the receptor Notch1 and its ligand Jagged1 are the predominant members of the Notch family expressed in the adult heart; (b) both Notch1 and Jagged1, as well as the Notch target gene Hes1, are up-regulated in the stressed heart; (c) the Notch pathway is activated in the cardiomyocyte as well as in CPCs; (d) the Notch pathway regulates cardiac gene expression in adult cardiomyocytes and controls the extent of the hypertrophic response; (e) the Notch pathway controls proliferation and cardiogenesis in CPCs; and (f) pharmacological and genetic blockade of the Notch pathway in the heart is largely detrimental to the stressed heart. Based on these findings, we propose that Notch exerts a dual role in the adult heart. On the one hand, Notch prevents uncontrolled growth of cardiomyocytes and might contribute to cell survival. On the other hand, Notch blocks cardiac differentiation in the resident precursor population and may therefore allow expansion of a transient amplifying cell compartment. Blockade of the Notch pathway appears to be detrimental to the normal adaptation of the heart. Pharmacological inhibition produces a down-regulation of Notch target genes such as Hes1, Hey1, and Hey2, activates cardiac gene expression, and exacerbates cardiac hypertrophy. Hey genes, Hey2 in particular, were shown to inhibit cardiac gene transcription (30), to block ventricular expression of atrial genes during cardiac development (31) and in the adult heart (32), and to suppress cardiac hypertrophy via direct interaction with the transcription factor GATA4 (30, 33). Experiments performed in embryonic stem cells lacking either Hey-2 or both Hey-1 and Hey-2 also suggest that Hey proteins negatively regulate the transcription of GATA-4 and GATA-6 (31). Therefore, Notch appears to limit the extent of cardiomyocyte hypertrophy by negatively regulating cardiac gene expression. The reexpression of atrial genes, such as the atrial natriuretic factor, is a hallmark of the hypertrophic heart and likely contributes to alteration of cardiac function. Accordingly, cardiomyocytes lacking Notch1 demonstrate a stimulated cardiac gene program and a more mature (hypertrophic) phenotype. Notch activation may, therefore, represent a protective mechanism enabling cell survival. Indeed, mice lacking Notch1 in the heart demonstrate increased apoptotic cell death during the adaptive response to hemodynamic overload. Consistent with this, a recent study suggests that Notch activates protective signaling in the myocardium; specifically, cross talk between hepatocyte growth factor and its cognate receptor c-Met, the Akt–PKB signaling pathway, and Notch regulates survival mechanisms in cardiomyocytes (34).
There is a striking difference between the cardiac phenotype that develops in TG1306/1R transgenic mice receiving a γ-secretase inhibitor (short-term inhibition) and that observed in hypertensive cNotch1 KOs (long-term inhibition). In a chronic model of cardiac hypertrophy and failure (TG1306/1R transgenics), short-term Notch inhibition results in increased cardiac performance, which likely reflects enhanced protein synthesis and sarcomeric assembly in hypertrophic cardiomyocytes. In contrast, long-term inhibition in Notch1-deficient mice with renovascular hypertension (1K1C model) leads to maladaptive remodeling characterized by diminished cardiac function. Notch1-deficient hearts demonstrate severe cardiac fibrosis under conditions of hemodynamic overload associated with decreased animal survival. Tissue fibrosis is a sign of a deficient regenerative capacity in normally self-renewing organs (35). This observation suggests that cardiac dysfunction may result from decreased mobilization of CPCs and inadequate replacement of damaged cardiomyocytes. For instance, zebrafish can regenerate its partially amputated heart via proliferation of immature cardiomyocytes (36). This process relies in part on an activated Notch pathway (37). In mutant fish lacking this regenerative capacity, the failure to form a new compact myocardium leads to fibrosis and scarring (36).
Recently, evidence has accumulated suggesting that cardiac myocytes can be continuously replaced in the heart through a process involving CPC replication and differentiation. Our results demonstrate the existence of Notch-activating CPCs that can give rise to functional cardiomyocytes. N1IC-positive cells coexpress GATA4 and/or Nkx2.5 in vivo and during expansion in vitro suggesting that CPCs are committed to the cardiogenic lineage. Interestingly, CPCs with an activated Notch1 pathway did not appear to express Sca-1, a widely used marker of CPCs in the heart. Whether Notch1 signaling and Sca-1–positive CPCs represent two distinct steps toward cardiomyocyte differentiation or two separate cardiogenic lineages is still unclear. Furthermore, nonmyocyte cells with an activated Notch pathway express the proliferation marker Ki67 suggesting that these cells are actively cycling. In CPCs, down-regulation of Notch appears to be necessary for achieving terminal cardiac differentiation. By preventing cardiogenesis, Notch signaling may therefore facilitate the expansion of transient amplifying cells. In this context, it is interesting to note that the number of BrdU-positive cells in Notch1-deficient hearts is significantly lower than that observed in WT hearts under both basal and hemodynamic stress, indicating that proliferation of cardiac cells relies on a functional Notch1 pathway. The Notch pathway has been described to promote stem cell maintenance and proliferation in several different self-renewing organs (38). For instance, expansion of satellite cells in the skeletal muscle is dependent on an activated Notch pathway (39). In cNotch1 KOs, Notch1 deletion is mediated by expression of the Cre recombinase under the control of the Mlc2v promoter. Therefore, the Notch pathway could act primarily in late committed precursors and/or immature cardiomyocytes in which the MLC2v promoter is activated. In isolated cardiomyocytes, Notch1 activation is associated with an immature phenotype characterized by the absence of organized sarcomeres. Sarcomere disassembly is a prerequisite for cardiomyocyte proliferation (40). The activation of the Notch pathway in cardiac myocytes could either preserve the capacity of incompletely differentiated cells to proliferate and/or facilitate the reentry of differentiated cardiomyocytes into the cell cycle in part by inhibiting sarcomeric protein synthesis. Accordingly, Ki67-positive cardiomyocytes with an activated Notch1 pathway are detected in the stressed heart. These cells could be primarily affected by Notch1 inactivation.
Blocking the Notch pathway promotes the differentiation of CPCs into cardiomyocytes and accelerates the maturation of cardiomyocytes. These findings are supported by several observations. First, CPCs differentiate more readily into cardiomyocytes when the Notch pathway is pharmacologically or genetically inhibited. Second, sarcomeric structures are more elaborate in cardiomyocytes lacking Notch1. Third, inhibition of the Notch pathway results in a striking up-regulation of cardiac gene expression both in vitro and in vivo. These Notch-mediated effects are very similar to those observed in embryonic stem cells, where Notch inhibits mesodermal commitment, cardiac differentiation (41, 42), and cardiomyocyte production (43). In contrast, forced N1IC expression in mesodermal cardiac progenitor cells expressing Mesp1 during development prevents cardiogenesis and results in cardiac abnormalities (44). In cNotch1 KOs, the absence of Notch in transient amplifying cells could therefore induce premature differentiation, leading to a gradual exhaustion of the proliferating precursor pool, and affect the adaptive capacity of the heart.
The activation of the Notch pathway in the heart in response to stress likely depends on Jagged1 expression on the surface of cardiomyocytes. This is indicated by the high level of Jagged1 in the heart of 1K1C hypertensive mice and TG1306/1R transgenic animals. Reciprocal activation of the Notch pathway could then occur between adjacent myocytes. Jagged1 expression is decreased in mice receiving a γ-secretase inhibitor. This suggests that Notch-mediated inductive signaling may take place between neighboring cells to regulate Jagged1 expression. Indeed, a positive-feedback mechanism has been shown to control Jagged1 expression in the developing heart during the onset of endocardial cushion formation (17). Furthermore, Jagged1 on the surface of cardiomyocytes could also convey a signal to Notch1-positive CPCs lying in interstitial spaces. Interestingly, decreasing Jagged1 expression is observed in cultures of differentiating cardiac precursors, resulting in a down-regulation of the Notch pathway.
In CPCs clusters, some cells appear to activate Notch and some do not. Moreover, in pairs of adjacent cells, Notch is often activated only in one cell. Because down-regulation of the Notch pathway favors cardiac differentiation, it would be interesting to evaluate whether N1IC-positive cells are less advanced in their cardiogenic differentiation than N1IC-negative cells. Furthermore, it has yet to be determined whether cardiogenic differentiation during postnatal cardiogenesis requires asymmetrical CPC division, producing daughter cells preferentially expressing the Notch receptor or its ligand Jagged1. In the skeletal muscle, the maintenance and the proliferation of satellite cells are dependent on an activated Notch pathway. Pools of transient activating cells are amplified in the damaged muscle after expression of the Notch ligand Delta-like (39). Furthermore, the mechanism for initiating myogenic differentiation in undifferentiated precursors implicates the Notch antagonist Numb, which is asymmetrically distributed into dividing cells (45). Cells that inherit Numb are committed to the myogenic lineage (46). Interestingly, undifferentiated Numb-positive cells have been found in the adult mouse heart (47). These cells could have undergone asymmetrical division similar to that seen during the regeneration of the skeletal muscle. Finally, it is noteworthy that the Notch-mediated regenerative capacity of skeletal muscle is gradually lost with age because of a decreased capacity to up-regulate Notch ligand expression and to recruit precursor cells (39). It will be important to determine whether a similar regulation exists in the heart as this may provide a mechanism by which cardiac function and the capacity of the left ventricle to adapt to hemodynamic overload decreases with age. If this is true, Notch may represent a novel therapeutic target for the treatment of cardiac diseases.
MATERIALS AND METHODS
Mice.
The transgenic mouse line TG1306/1R expressing angiotensinogen under control of the α-MHC promoter has been previously described (23, 24). The cardiac Notch1 deletion was obtained by crossing mice harboring floxed Notch1 allele (29) with mice expressing the Cre recombinase under control of MLC2v promoter (28). WT C57BL/6 mice were purchased from Charles River Laboratories. Mice were housed under standard conditions. Animal experiments were approved by the Government Veterinary Office (Lausanne, Switzerland) and performed according to the University of Lausanne Medical School institutional guidelines.
Microarray analysis.
Microarray experiments were performed on three different mouse groups: transgenic hypertrophic TG1306, transgenic dilated TG1306, and WT littermates, with the former two groups being distinguished on the basis of heart morphology, cardiac weight index, and expression of cardiac hypertrophy marker genes. Aliquots containing 25 μg of total RNA were reverse transcribed into dye-labeled cDNA with Superscript II reverse transcription (Invitrogen) in presence of Cy3- or Cy5-labeled dCTP (GE Healthcare). SpotReport Alien and Arabidopsis Spike RNAs (Stratagene) were used as controls. The dye-labeled cDNAs were purified using the MinElute PCR Purification kit (QIAGEN) after hydrolysis of the RNA template. Microarray slides (DNA Array Facility in Lausanne, Switzerland) contained the NIA 15K mouse embryonic clone set (National Institute of Aging, National Institutes of Health, Bethesda, MD) to which an additional 1563 custom clone set was added. The slides were hybridized with heat-denatured Cy3 and Cy5 cDNA samples to be compared in 30 μl of hybridization solution containing 0.33 mg/ml poly-A (Sigma-Aldrich), 0.33 mg/ml of yeast transfer RNA (Sigma-Aldrich), 0.66 mg/ml of mouse Cot1 DNA (Invitrogen). Hybridization was performed overnight in the dark in sealed hybridization chambers (TeleChem International, Inc.) at 64°C. After incubation, the array slides were washed in washing solutions containing SSC, SDS, and Triton X-100, spun dry, and stored in a dark desiccated environment until scanning. The slides were scanned using a microarray laser scanner (Agilent Technologies), which acquired two 16-bit grayscale images corresponding to the Cy5 and Cy3 fluorescence emission channels (excited with an argon laser operating at ∼650 and 550 nm, respectively). The images were processed with GenePix Pro 5.1 software (MDS Analytical Technologies) to produce a matrix containing Cy5 (red) and Cy3 (green) intensities for each spot on the microarray. The bulk of quality control and normalization steps are automated and provided by in house–developed web tools using the open source Bioconductor packages from the statistical software R (R Foundation for Statistical Computing).
One kidney, one clip model.
In brief, 12-wk-old mice weighing 22–25 g were anaesthetized using 1.5% halothane in oxygen. The left kidney was exposed and a clip (0.12-mm opening) was inserted on the renal artery isolated by blunt dissection. The contralateral kidney was removed. Sham procedure included the entire surgery with the exception of artery clipping (27).
Physiological measurements.
Blood pressure was measured on conscious animals by the Tail-Cuff method using a BP-2000 apparatus (Visitech Systems, Inc.). Cardiac dimensions and function were assessed in anesthetized mice (90 mg/Kg ketamine and 5 mg/Kg xylazine i.p.) by transthoracic echocardiography using an ultrasound machine (ATL HDI 5000; Philips) equipped with a 12-MHz phase array linear transducer (L12-5). M-mode images were used for the measurement of wall thickness, chamber dimension, and fraction of shortening. Cardiac weight index was defined as the ratio of heart weight/body weight (in milligrams/gram).
γ-Secretase inhibition.
To inhibit the Notch pathway in vivo, γ-secretase inhibitor was administered to 15-wk-old male TG1306 transgenic and WT littermates. Mice were injected every 12 h for 14 d with either 1.5 mg/kg of the γ-secretase inhibitor LY411,575 (provided by H. Jacobsen, F Hoffmann-La Roche Ltd, Basel, Switzerland) in 0.9% NaCl and 0.3% Tween 80 (25) or vehicle alone. On day 10, physiological measurements were performed. The cardiac weight index was determined at sacrifice. Excised hearts were either processed for histological analysis or stored at −80°C until used.
Analysis of cardiac cell proliferation.
To analyze cell proliferation in vivo, 1 mg/ml BrdU was administered to 8-wk-old mice for 96 h in drinking water containing 5% glucose. Mice were killed and hearts were removed and fixed in 4% paraformaldehyde in PBS. The hearts were dehydrated and processed for paraffin sectioning and immunostaining.
Analysis of apoptotic cell death.
To detect apoptotic cells in the myocardium, paraffin or cryosections of heart tissue were analyzed using the In Situ Cell Detection kit (Roche). To simultaneously stain with myomesin, cryosections were first stained with antimyomesin antibody and a secondary Cy3 conjugate anti–mouse antibody, postfixed, and then processed for TUNEL assays. To quantitate apoptosis, sections were subjected to TUNEL assays, and the total number of positive nuclei was determined by counting under a fluorescence microscope.
Cell culture.
To isolate neonatal ventricular cardiomyocytes and mesenchymal nonmyocyte cells, ventricles were separated from the atria and minced. Tissues were digested in a solution containing 0.45 mg/ml collagenase (Worthington Biochemical Corporation) and 1 mg/ml pancreatin (EMD). Cardiomyocytes were separated from mesenchymal cells by two steps of differential plating at 45 min each. Cardiomyocytes were plated on gelatinized plates in complete medium (a 3:1 mixture of DMEM and Medium 199 supplemented with 10% horse serum, 5% fetal calf serum and antibiotics) and allowed to adhere overnight then maintained in a serum-free 3:1 mixture of DMEM and Medium 199 and antibiotics. The mesenchymal nonmyocyte cells were maintained in complete medium. Culture of CPCs present in the mesenchymal fraction and induction of differentiation into functional cardiomyocytes was performed as previously described (7). In brief, differentiation was induced in confluent cultures by maintaining cells in α-MEM, 10% FCS supplemented with 50 μg/ml of ascorbic acid, 10 mM β-glycerol phosphate, and 1 μM dexamethasone. Control cultures were maintained in the same medium without supplements. To inhibit Notch signaling, cells were treated with 1 μM DAPT (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester; EMD) dissolved in DMSO. Control cultures were treated with DMSO alone.
Semiquantitative and quantitative RT-PCR analysis.
RNA was extracted from cultured cells or heart tissue using TRIZOL reagent (Invitrogen). RNA was reverse transcribed into cDNA using oligo dT primers and MMLV reverse transcription according to the manufacturer's instructions (Invitrogen). PCR primers and conditions were essentially as described (7, 42). Quantitative TaqMan analysis was performed using Gene Expression Assays (Applied Biosystems) on an ABI 7500 Thermal Cycler (Applied Biosystems). The relative gene expression levels were calculated using the comparative Ct method and are reported as ratios to those of the housekeeping genes GAPDH or β2 microglobulin as indicated.
Antibodies.
The following primary antibodies were used: rabbit antiactivated N1IC (1:200; Abcam), mouse antimyomesin B4 (1:20; gift from J-C. Perriard, Die Eidgenössische Technische Hochschule Zürich, Zurich, Switzerland), mouse antisarcomeric α-actinin (1:500; Sigma-Aldrich), rabbit anti-Nkx2.5 (1:100; Santa Cruz Biotechnology, Inc.), goat anti-Nkx2.5 (SC12514; 1:200; Santa Cruz Biotechnology, Inc.), goat anti–GATA-4 (SC-1237; 1:100; Santa Cruz Biotechnology, Inc.), goat anti-Jagged1 (1:100; Santa Cruz Biotechnology, Inc.), rat anti-BrdU (1:200; Oxford Biotech), rabbit antilaminin (1:200; Sigma-Aldrich), rat anti–Sca-1 (1:100; BD), and goat anti-Ki67 (1:50; Santa Cruz Biotechnology, Inc.). For double immunostaining, the following combinations of secondary antibodies were used: biotinylated donkey anti–rabbit (1:150; Jackson ImmunoResearch Laboratories) or biotinylated donkey anti–goat (1:150; Jackson ImmunoResearch Laboratories) plus Cy3- or Cy2-conjugated streptavidin (1:900; Jackson ImmunoResearch Laboratories), Cy3-conjugated goat anti–mouse (1:400; Jackson ImmunoResearch Laboratories), or Cy3-conjugated donkey anti–goat (1:200; Jackson ImmunoResearch Laboratories) antibodies. Jagged1 antibody binding was detected with horseradish peroxidase–conjugated anti–mouse or anti–goat antibody and an UltraVision detection system (Thermo Fisher Scientific) or Alexa Fluor 488–conjugated anti–goat antibody. BrdU antibody binding was detected using horseradish peroxidase–conjugated anti–rat antibody (1:200; Invitrogen) using diaminobenzidine as a substrate.
Immunofluorescence detection.
Cultured cells or 5-μm frozen tissue sections were fixed in 2 and 4% paraformaldehyde in PBS, respectively, permeabilized with 0.25% Triton X-100 in PBS, and blocked in PBS containing 0.001% Triton X-100, 1% BSA, and 1% FCS. Specimens were incubated overnight with primary antibodies, which were revealed by incubation for 1 h at room temperature with fluorochrome-conjugated secondary antibodies. Sections were observed with a fluorescence microscope (AxioVision; Carl Zeiss, Inc.). Confocal microscopy was performed on a confocal microscope (SP5; Leica). Stained cells in culture were observed under an inverted fluorescence microscope (LSR Eclipse TE2000S; Nikon).
Histology.
For histological analysis, hearts were dissected free from atria and vessels and fixed in 4% paraformaldehyde in PBS or 10% formalin. Staining included standard hematoxylin and eosin or Van Gieson staining. Fibrosis was quantified by computer-assisted densitometric morphometry (Image Pro Plus; Media Cybernetics). Collagen deposition was expressed as a percentage of total cross-sectional area.
Online supplemental material.
Fig. S1 shows that the Sca-1+ cell population in WT adult and neonatal hearts, as well as in adult TG1306/1R transgenic hearts, does not generally activate the Notch1 signaling pathway. Fig. S2 demonstrates the efficacy of the γ-secretase inhibitor treatment in vivo by quantitating the disappearance of the Notch-dependent MZB cell population in the spleen of treated mice. Fig. S3 depicts the down-regulation of the Notch1 signaling pathway in cardiomyocytes in the presence of the hypertrophic agent phenylephrine. Fig. S4 demonstrates that Notch-signaling cardiomyocytes and nonmyocyte cells are actively cycling in the heart of TG1306/1R transgenic mice as shown by Ki67 staining. Table S1 describes the TG1306/1R transgenic mice with either cardiac hypertrophy or dilated cardiomyopathy used in microarray analyses. Tables S2–S7 report microarray analysis data obtained by comparing gene expression of WT hearts and either hypertrophic or dilated hearts from TG1306/1r transgenic mice. The data have been deposited in National Center for Biotechnology Information's Gene Expression Omnibus, and are accessible through GEO Series accession no. GSE13336 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13336). Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081427/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Alexis Dumortier and Ms. Catherine Roger (Swiss Institute for Experimental Cancer Research, Epalinges, Switzerland) for helping with FACS analysis and histology. The expert technical assistance of Jean-François Aubert, Nathalie Jordan, Steve Cruchet, and Sandy Ogay is gratefully acknowledged. We are also grateful to Dr. Keith Harshman and his collaborators at the DNA array facility (University of Lausanne) and Dr. Yannick Krempp at the Cellular Imaging Facility (University of Lausanne) for their contribution to this work. We thank Dr. Allison Felley for critical reading of the manuscript. We thank Dr. Helmut Jacobsen (F Hoffmann-La Roche Ltd, Basel, Switzerland) for providing us with the γ-secretase inhibitor LY411,575.
We are extremely grateful to the Leenaard Foundation (Lausanne, Switzerland) for their important financial support. This work is funded in part by the Swiss National Science Foundation (grant 320000-114026 to T. Pedrazzini).
The authors have no conflicting financial interests.
Abbreviations used: BNP, brain natriuretic peptide; CPC, cardiac precursor cell; Hes, Hairy/enhancer of split; MZB, marginal zone B; Sca, stem cell antigen.
A. Croquelois, A.A. Domenighetti, and M. Nemir contributed equally to this paper.
References
- 1.Lopez, A.D., and C.C. Murray. 1998. The global burden of disease, 1990-2020. Nat. Med. 4:1241–1243. [DOI] [PubMed] [Google Scholar]
- 2.Towbin, J.A., and N.E. Bowles. 2002. The failing heart. Nature. 415:227–233. [DOI] [PubMed] [Google Scholar]
- 3.Chien, K.R. 1999. Stress pathways and heart failure. Cell. 98:555–558. [DOI] [PubMed] [Google Scholar]
- 4.Beltrami, A.P., L. Barlucchi, D. Torella, M. Baker, F. Limana, S. Chimenti, H. Kasahara, M. Rota, E. Musso, K. Urbanek, et al. 2003. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 114:763–776. [DOI] [PubMed] [Google Scholar]
- 5.Oh, H., S.B. Bradfute, T.D. Gallardo, T. Nakamura, V. Gaussin, Y. Mishina, J. Pocius, L.H. Michael, R.R. Behringer, D.J. Garry, et al. 2003. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc. Natl. Acad. Sci. USA. 100:12313–12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laugwitz, K.L., A. Moretti, J. Lam, P. Gruber, Y. Chen, S. Woodard, L.Z. Lin, C.L. Cai, M.M. Lu, M. Reth, et al. 2005. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 433:647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenblatt-Velin, N., M.G. Lepore, C. Cartoni, F. Beermann, and T. Pedrazzini. 2005. FGF-2 controls the differentiation of resident cardiac precursors into functional cardiomyocytes. J. Clin. Invest. 115:1724–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Artavanis-Tsakonas, S., K. Matsuno, and M.E. Fortini. 1995. Notch signaling. Science. 268:225–232. [DOI] [PubMed] [Google Scholar]
- 9.Radtke, F., A. Wilson, S.J. Mancini, and H.R. Macdonald. 2004. Notch regulation of lymphocyte development and function. Nat. Immunol. 5:247–253. [DOI] [PubMed] [Google Scholar]
- 10.Wilson, A., and F. Radtke. 2006. Multiple functions of Notch signaling in self-renewing organs and cancer. FEBS Lett. 580:2860–2868. [DOI] [PubMed] [Google Scholar]
- 11.Bray, S.J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678–689. [DOI] [PubMed] [Google Scholar]
- 12.Iso, T., L. Kedes, and Y. Hamamori. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194:237–255. [DOI] [PubMed] [Google Scholar]
- 13.Radtke, F., A. Wilson, and H.R. Macdonald. 2005. Notch signaling in hematopoiesis and lymphopoiesis: lessons from Drosophila. Bioessays. 27:1117–1128. [DOI] [PubMed] [Google Scholar]
- 14.Gridley, T. 2007. Notch signaling in vascular development and physiology. Development. 134:2709–2718. [DOI] [PubMed] [Google Scholar]
- 15.Pedrazzini, T. 2007. Control of cardiogenesis by the notch pathway. Trends Cardiovasc. Med. 17:83–90. [DOI] [PubMed] [Google Scholar]
- 16.High, F.A., and J.A. Epstein. 2008. The multifaceted role of Notch in cardiac development and disease. Nat. Rev. Genet. 9:49–61. [DOI] [PubMed] [Google Scholar]
- 17.Timmerman, L.A., J. Grego-Bessa, A. Raya, E. Bertran, J.M. Perez-Pomares, J. Diez, S. Aranda, S. Palomo, F. McCormick, J.C. Izpisua-Belmonte, and J.L. De La Pompa. 2004. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grego-Bessa, J., J. Diez, L. Timmerman, and J.L. De La Pompa. 2004. Notch and epithelial-mesenchyme transition in development and tumor progression: another turn of the screw. Cell Cycle. 3:718–721. [PubMed] [Google Scholar]
- 19.Loomes, K.M., D.B. Taichman, C.L. Glover, P.T. Williams, J.E. Markowitz, D.A. Piccoli, H.S. Baldwin, and R.J. Oakey. 2002. Characterization of Notch receptor expression in the developing mammalian heart and liver. Am. J. Med. Genet. 112:181–189. [DOI] [PubMed] [Google Scholar]
- 20.Garg, V., A.N. Muth, J.F. Ransom, M.K. Schluterman, R. Barnes, I.N. King, P.D. Grossfeld, and D. Srivastava. 2005. Mutations in NOTCH1 cause aortic valve disease. Nature. 437:270–274. [DOI] [PubMed] [Google Scholar]
- 21.Oda, T., A.G. Elkahloun, B.L. Pike, K. Okajima, I.D. Krantz, A. Genin, D.A. Piccoli, P.S. Meltzer, N.B. Spinner, F.S. Collins, and S.C. Chandrasekharappa. 1997. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 16:235–242. [DOI] [PubMed] [Google Scholar]
- 22.Boyer, J., C. Crosnier, C. Driancourt, N. Raynaud, M. Gonzales, M. Hadchouel, and M. Meunier-Rotival. 2005. Expression of mutant JAGGED1 alleles in patients with Alagille syndrome. Hum. Genet. 116:445–453. [DOI] [PubMed] [Google Scholar]
- 23.Mazzolai, L., J. Nussberger, J.F. Aubert, D.B. Brunner, G. Gabbiani, H.R. Brunner, and T. Pedrazzini. 1998. Blood pressure-independent cardiac hypertrophy induced by locally activated renin-angiotensin system. Hypertension. 31:1324–1330. [DOI] [PubMed] [Google Scholar]
- 24.Domenighetti, A.A., Q. Wang, M. Egger, S.M. Richards, T. Pedrazzini, and L.M. Delbridge. 2005. Angiotensin II-mediated phenotypic cardiomyocyte remodeling leads to age-dependent cardiac dysfunction and failure. Hypertension. 46:426–432. [DOI] [PubMed] [Google Scholar]
- 25.Hyde, L.A., N.A. McHugh, J. Chen, Q. Zhang, D. Manfra, A.A. Nomeir, H. Josien, T. Bara, J.W. Clader, L. Zhang, et al. 2006. Studies to investigate the in vivo therapeutic window of the gamma-secretase inhibitor N2-[(2S)-2-(3,5-difluorophenyl)-2-hydroxyethanoyl]-N1-[(7S)-5-methyl-6-oxo -6,7-dihydro-5H-dibenzo[b,d]azepin-7-yl]-L-alaninamide (LY411,575) in the CRND8 mouse. J. Pharmacol. Exp. Ther. 319:1133–1143. [DOI] [PubMed] [Google Scholar]
- 26.Tanigaki, K., H. Han, N. Yamamoto, K. Tashiro, M. Ikegawa, K. Kuroda, A. Suzuki, T. Nakano, and T. Honjo. 2002. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 3:443–450. [DOI] [PubMed] [Google Scholar]
- 27.Wiesel, P., L. Mazzolai, J. Nussberger, and T. Pedrazzini. 1997. Two-kidney, one clip and one-kidney, one clip hypertension in mice. Hypertension. 29:1025–1030. [DOI] [PubMed] [Google Scholar]
- 28.Chen, J., S.W. Kubalak, and K.R. Chien. 1998. Ventricular muscle-restricted targeting of the RXRalpha gene reveals a non-cell-autonomous requirement in cardiac chamber morphogenesis. Development. 125:1943–1949. [DOI] [PubMed] [Google Scholar]
- 29.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H.R. Macdonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558. [DOI] [PubMed] [Google Scholar]
- 30.Kathiriya, I.S., I.N. King, M. Murakami, M. Nakagawa, J.M. Astle, K.A. Gardner, R.D. Gerard, E.N. Olson, D. Srivastava, and O. Nakagawa. 2004. Hairy-related transcription factors inhibit GATA-dependent cardiac gene expression through a signal-responsive mechanism. J. Biol. Chem. 279:54937–54943. [DOI] [PubMed] [Google Scholar]
- 31.Fischer, A., J. Klattig, B. Kneitz, H. Diez, M. Maier, B. Holtmann, C. Englert, and M. Gessler. 2005. Hey basic helix-loop-helix transcription factors are repressors of GATA4 and GATA6 and restrict expression of the GATA target gene ANF in fetal hearts. Mol. Cell. Biol. 25:8960–8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xin, M., E.M. Small, E. van Rooij, X. Qi, J.A. Richardson, D. Srivastava, O. Nakagawa, and E.N. Olson. 2007. Essential roles of the bHLH transcription factor Hrt2 in repression of atrial gene expression and maintenance of postnatal cardiac function. Proc. Natl. Acad. Sci. USA. 104:7975–7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang, F., Y. Sakata, L. Cui, J.M. Youngblood, H. Nakagami, J.K. Liao, R. Liao, and M.T. Chin. 2006. Transcription factor CHF1/Hey2 suppresses cardiac hypertrophy through an inhibitory interaction with GATA4. Am. J. Physiol. Heart Circ. Physiol. 290:H1997–H2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gude, N.A., G. Emmanuel, W. Wu, C.T. Cottage, K. Fischer, P. Quijada, J.A. Muraski, R. Alvarez, M. Rubio, E. Schaefer, and M.A. Sussman. 2008. Activation of Notch-mediated protective signaling in the myocardium. Circ. Res. 102:1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borchardt, T., and T. Braun. 2007. Cardiovascular regeneration in non-mammalian model systems: what are the differences between newts and man? Thromb. Haemost. 98:311–318. [PubMed] [Google Scholar]
- 36.Poss, K.D., L.G. Wilson, and M.T. Keating. 2002. Heart regeneration in zebrafish. Science. 298:2188–2190. [DOI] [PubMed] [Google Scholar]
- 37.Raya, A., C.M. Koth, D. Buscher, Y. Kawakami, T. Itoh, R.M. Raya, G. Sternik, H.J. Tsai, C. Rodriguez-Esteban, and J.C. Izpisua-Belmonte. 2003. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc. Natl. Acad. Sci. USA. 100:11889–11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crosnier, C., D. Stamataki, and J. Lewis. 2006. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat. Rev. Genet. 7:349–359. [DOI] [PubMed] [Google Scholar]
- 39.Conboy, I.M., M.J. Conboy, G.M. Smythe, and T.A. Rando. 2003. Notch-mediated restoration of regenerative potential to aged muscle. Science. 302:1575–1577. [DOI] [PubMed] [Google Scholar]
- 40.Ahuja, P., E. Perriard, J.C. Perriard, and E. Ehler. 2004. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. J. Cell Sci. 117:3295–3306. [DOI] [PubMed] [Google Scholar]
- 41.Schroeder, T., S.T. Fraser, M. Ogawa, S. Nishikawa, C. Oka, G.W. Bornkamm, S. Nishikawa, T. Honjo, and U. Just. 2003. Recombination signal sequence-binding protein Jkappa alters mesodermal cell fate decisions by suppressing cardiomyogenesis. Proc. Natl. Acad. Sci. USA. 100:4018–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nemir, M., A. Croquelois, T. Pedrazzini, and F. Radtke. 2006. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ. Res. 98:1471–1478. [DOI] [PubMed] [Google Scholar]
- 43.Schroeder, T., F. Meier-Stiegen, R. Schwanbeck, H. Eilken, S. Nishikawa, R. Hasler, S. Schreiber, G.W. Bornkamm, S. Nishikawa, and U. Just. 2006. Activated Notch1 alters differentiation of embryonic stem cells into mesodermal cell lineages at multiple stages of development. Mech. Dev. 123:570–579. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe, Y., H. Kokubo, S. Miyagawa-Tomita, M. Endo, K. Igarashi, K. Aisaki, J. Kanno, and Y. Saga. 2006. Activation of Notch1 signaling in cardiogenic mesoderm induces abnormal heart morphogenesis in mouse. Development. 133:1625–1634. [DOI] [PubMed] [Google Scholar]
- 45.Conboy, I.M., and T.A. Rando. 2002. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 3:397–409. [DOI] [PubMed] [Google Scholar]
- 46.Shinin, V., B. Gayraud-Morel, D. Gomes, and S. Tajbakhsh. 2006. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat. Cell Biol. 8:677–687. [DOI] [PubMed] [Google Scholar]
- 47.Urbanek, K., D. Cesselli, M. Rota, A. Nascimbene, A.A. De, T. Hosoda, C. Bearzi, A. Boni, R. Bolli, J. Kajstura, et al. 2006. Stem cell niches in the adult mouse heart. Proc. Natl. Acad. Sci. USA. 103:9226–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.