Abstract
B cell receptor (BCR) signaling contributes to the pathogenesis of B cell malignancies, and most B cell lymphomas depend on BCR signals for survival. Identification of genes that restrain BCR-mediated proliferation is therefore an important goal toward improving the therapy of B cell lymphoma. Here, we identify Ptger4 as a negative feedback regulator of proliferation in response to BCR signals and show that its encoded EP4 receptor is a principal molecule conveying the growth-suppressive effect of prostaglandin E2 (PGE2). Stable knockdown of Ptger4 in B cell lymphoma markedly accelerated tumor spread in mice, whereas Ptger4 overexpression yielded significant protection. Mechanistically, we show that the intrinsic activity of Ptger4 and PGE2–EP4 signaling target a similar set of activating genes, and find Ptger4 to be significantly down-regulated in human B cell lymphoma. We postulate that Ptger4 functions in B cells as a candidate tumor suppressor whose activity is regulated by PGE2 in the microenvironment. These findings suggest that targeting EP4 receptor for prostaglandin may present a novel strategy for treatment of B cell malignancies.
Recognition of foreign antigens by BCRs expressed on the surface of mature naive B cells triggers their massive proliferation, which is critically important for the effective defense of the organism against invading pathogens (1). Antigen-activated B cells undergo clonal expansion in dynamic structures called germinal centers (GCs) where much of the diversity of the Ig genes is generated by somatic hypermutation and class-switch recombination (2). These molecular processes require frequent DNA strand breaks that can, when deregulated, provide a rich source for the genesis of B cell lymphomas (3). For instance, the hallmark of Burkitt's lymphoma and some cases of diffuse large B cell lymphoma (DLBCL) is a reciprocal chromosomal translocation where the MYC protooncogene comes under the control of an active Ig locus, resulting in constitutive expression of MYC (4). However, although this and other translocations are thought to cause many types of B cell lymphoma, additional transforming events that override the normal mechanisms controlling B cell proliferation are needed for malignant transformation. Indeed, mutations affecting the expression level or the activity of tumor suppressor genes, as well as genomic amplifications and hypermutations of multiple protooncogenes have also been implicated in the pathogenesis of B cell lymphomas (for review see reference [3]).
Despite the aggressive behavior of several types of B cell lymphoma, data gathered over the past few years have demonstrated that BCR signaling is essential for the survival of neoplastic B lymphoma cells, which also holds true for their nonmalignant counterparts (5–8). Observations that the majority of non-Hodgkin's lymphomas persistently express BCR and that IgH translocations almost never affect the functionality of Ig alleles, as well as the discovery of autoreactive BCR in certain neoplasms, indirectly suggests the need for BCR-derived survival and proliferation signals (3). Correspondingly, BCR signaling was found to promote growth of DLBCL and chronic lymphocytic leukemia B cells (5, 7), whereas follicular lymphoma B cells displayed potentiated BCR signaling versus tumor-infiltrating normal B cells (6). Most recently, a direct link between BCR signaling and B lymphomagenesis was established by demonstrating that PAX5 promotes neoplastic growth by activating BCR signaling (8). These findings, together with the recognition that the survival of many B cell tumors depends on signals provided by their microenvironment, might lead to novel treatment options for B cell lymphoma.
More than three decades have passed since the first description of macrophage-derived PGE2 as a potent suppressor of splenic B cell colony formation (9), and it was later demonstrated that the inhibitory effect of PGE2 was caused by its direct influence on B cell proliferation (10). Further studies identified tingible body macrophages as a major source of prostaglandins in the GC microenvironment, and proposed that these scavengers of apoptotic lymphocytes may use prostaglandin to down-regulate the GC reaction (11). However, since these seminal observations, no further insight into the mechanism or relevance of the attenuating effect of PGE2 on B cell proliferation has been provided. In this study, we investigated how, mechanistically, PGE2 and its receptors may regulate B cell proliferation triggered by BCR signaling, and evaluated the potential contribution to B cell lymphoma growth in mice and humans.
RESULTS
Ptger4 is a delayed-early gene that inhibits B cell proliferation
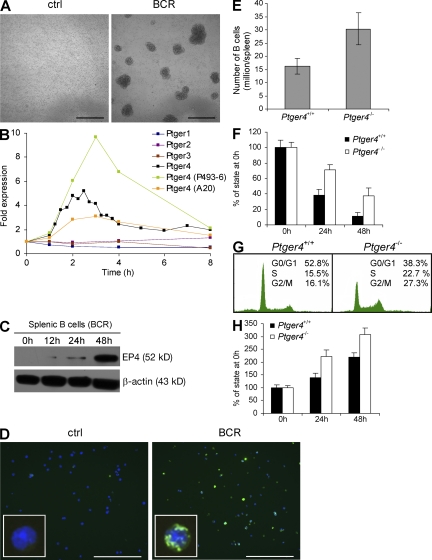
BCR-triggered activation of mature B cells leads to transcriptional reprogramming, which can be observed by a strong induction of immediate-early genes within minutes after mitogenic stimulation, preparing a cell to quickly respond with proliferation (Fig. 1 A; see also Fig. 2 C) (12). In an attempt to identify negative regulators of B cell proliferation, we performed transcriptome analysis at 2 h after BCR stimulation and looked for putative delayed-early genes, which are generally known to function as negative feedback regulators of growth factor signaling (GEO database accession no. GSE9215) (13). Among the most strongly induced genes, we found Ptger4, whose expression kinetics appeared similar to a wave of delayed-early genes (Fig. 1 B). In addition to its high induction upon BCR cross-linking, Ptger4 also seemed interesting as a candidate regulator of B cell activation because it encodes an EP4 subtype of receptor for prostaglandin E2 (PGE2) (14), and PGE2 is known for its potent yet poorly understood immunosuppressive role (15). Notably, none of the other genes coding for PGE2 receptors showed altered expression after BCR triggering (Fig. 1 B). We found the induction of Ptger4 to be conserved in mouse A20 and human P493-6 B cell lines, and confirmed that it resulted in protein expression by Western blotting and immunofluorescence (Fig. 1, B–D and Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20081163/DC1; see also Fig. S2).
Figure 1.
Ptger4 is a delayed-early gene that inhibits B cell proliferation. (A) Mouse splenic B cells stimulated with (BCR) or without (ctrl) anti-IgM F(ab′)2 for 72 h in culture. (B) BCR-induced expression of Ptger4 in mouse and human B cells as measured by qPCR at the indicated times. The relative abundances of each Ptger mRNA compared with Hprt transcripts in primary mouse B cells at time 0 h were as follows: Ptger4 (0.0068; 100%), Ptger2 (0.0013; 19.1%), Ptger1 (0.0009; 13.2%), and Ptger3 (2.2 × 10−6; 0.0003%). Western blotting (C) and immunofluorescence (D; at 48 h) of EP4 in BCR-stimulated mouse B cells. See Fig. S1 for higher protein loading and longer exposure immunoblot analysis, and Fig. S6 for higher quality immunofluorescence images. (E) Numbers of B cells isolated from spleen of Ptger4+/+ and Ptger4−/− mice. (F) ATP content of unstimulated and (H) BCR-stimulated mouse B cells in culture. (G) Cell cycle analysis at 60 h after BCR-triggering of mouse B cells of the indicated genotypes. Error bars are the SD of six to eight independent experiments. Bars, (A) 200 μm; (D) 100 μm. Figs. S1 and S2 are available at http://www.jem.org/cgi/content/full/jem.20081163/DC1.
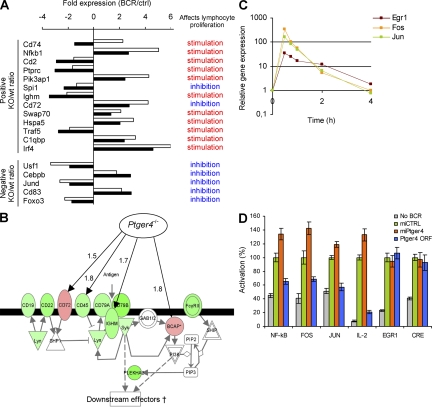
Figure 2.
Ptger4 is a negative feedback regulator of BCR-triggered proliferation. (A) Relative expression values and functions of genes known to affect lymphocyte proliferation, according to Ingenuity Pathway Analysis database (IPA; see URLs in Materials and methods). For each gene, the expression ratio in BCR-stimulated (BCR) versus control (ctrl) sample at 2 h is shown (filled bars, wild-type [wt] B cells; open bars, Ptger4−/− [KO] B cells). Genes were ranked according to their absolute ratios in descending order. (B) Proximal part of the BCR-signaling canonical pathway. The molecules whose encoding genes showed at least a twofold change in expression upon BCR-triggering of wild-type B cells are colored (green, down-regulation; red, up-regulation). Black arrows indicate genes that showed at least a 50% difference in expression between Ptger4−/− and wild-type B cells. Numbers indicate fold induction caused by the lack of Ptger4. (C) BCR-induced expression of the immediate-early genes Egr1, Fos, and Jun in A20 cell line. (D) A20 cells were cotransfected with plasmids encoding GFP cocistronic with nontargeting miRNA (miCTRL) or miRNA targeting Ptger4 (miPtger4b) or the Ptger4 ORF, together with the indicated luciferase reporter plasmid. The knockdown efficiency was similar to that achieved in stable cell lines (Fig. 3 A). Luminescence was determined after BCR stimulation as detailed in Materials and methods. Error bars are the SD of triplicates. To exclude the possibility of saturation, the assay for EGR1 and CRE activity was also performed at a lower concentration (5 μg/ml) of anti-IgG F(ab′)2 (Fig. S4). Fig. S4 is available at http://www.jem.org/cgi/content/full/jem.20081163/DC1.
To investigate the role of Ptger4 in peripheral B cell activation, we first examined the gross phenotypes of B cells extracted from spleen of Ptger4−/− mice (16). We observed that knockout mice consistently harboured significantly increased numbers of B cells that showed greater resistance to spontaneous cell death in vitro compared with wild-type controls (Fig. 1, E and F). In addition, cell cycle analysis and ATP content measurements revealed an augmented mitogenic response of Ptger4-deficient B cells upon BCR ligation, indicating that the early induction of Ptger4 may serve to counterbalance antigen-induced proliferation in mouse B cells (Fig. 1, G and H). To check whether the endogenously produced PGE2 may have influenced the growth of BCR-activated B cells, we examined the levels of PGE2 in culture supernatants and saw that these remained below the detection limit of 15 pg/ml during the course of the experiment (unpublished data). In addition, cotreatment with COX inhibitors indomethacin or NS-398 had no significant effect on proliferation or viability of BCR-stimulated cultures, suggesting that de novo–produced PGE2 may not be a major factor in suppressing the growth of activated wild-type compared with Ptger4-deficient B cells in culture (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20081163/DC1). In summary, these observations suggested that Ptger4 functions as a delayed-early gene upon BCR-triggering, and prompted us to further investigate the mechanism used by Ptger4 to attenuate BCR-signaled proliferation.
Ptger4 acts as a negative feedback regulator of BCR-signaling
First, to understand how lack of Ptger4 may have enhanced BCR-driven proliferation, we examined the early transcriptional response of both Ptger4-deficient and wild-type mouse B cells to stimulation with anti-IgM F(ab′)2 in culture. We were surprised by the regulation of genes known to affect lymphocyte proliferation, which showed two functionally coherent groups of genes defined by the presence or absence of Ptger4 (Fig. 2 A). In line with the aforementioned observations, 11 out of 13 genes (85%) that had higher expression levels in knockout relative to wild-type B cells were found to promote proliferation, whereas all (5 out of 5) genes overexpressed in wild-types had an inhibitory role. Surprisingly, signaling through the BCR in wild-type cells resulted in a profound down-regulation of most genes encoding components of the proximal BCR-signaling cascade itself, suggesting the existence of a potent negative feedback mechanism (Fig. 2 B). Although triggering of Ptger4-deficient B cells also repressed the majority of these genes, several crucial genes of the upper BCR pathway, including Ptprc coding for CD45 (also known as B220) (17) and Ighm coding for the immunoglobulin heavy chain of the B cell receptor itself, were induced nearly twofold in Ptger4−/− cells compared with wild-type controls (Fig. 2 B). This indicates that by maintaining the relatively high expression of certain signaling components, the lack of Ptger4 may prolong the duration of signaling via the BCR, cumulatively delivering a stronger proliferative stimulus.
Among the genes most significantly up-regulated in the absence of Ptger4, we found three potent cell cycle–regulating genes, Cdk4, Plagl2, and Nfkb1, which are all known to promote G1 to S phase transition (18–20), as well as Cd74 and H2-DMa, which are both members of the MHC class II family (Table S1, available at http://www.jem.org/cgi/content/full/jem.20081163/DC1). Notably, CD74 has been shown to promote B cell lymphoma progression, and an anti-CD74 antibody is currently being clinically evaluated for the therapy of human B cell malignancies (21, 22).
Because B cells use antigen-presenting MHC class II molecules to attract activating help from helper T cells (23), endogenous control of the MHC-encoding genes by Ptger4 may thus provide another means to keep B cell activation in check. Indeed, as we demonstrate in this study, signaling via the EP4 receptor potently repressed the expression of multiple MHC components essential for effective antigen presentation by B cells (see also Fig. 5 B). Interestingly, it has been reported that PGE2 and EP2/EP4 agonists significantly inhibit the expression of MHC class II molecules in B lymphocytes stimulated with IL-4 and/or lipopolysaccharide (24, 25), suggesting that the inhibition of MHC class II by EP4 may occur independently of the stimulus used to activate B cells.
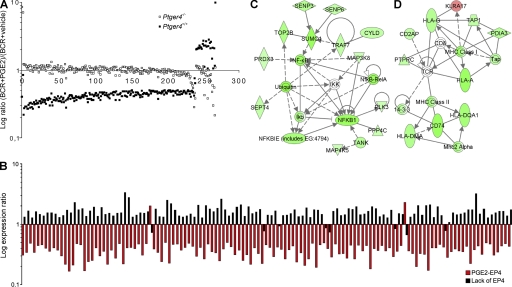
Figure 5.
PGE2–EP4 signaling results in general repression of activating genes. (A) Genes regulated at least twofold in BCR-triggered Ptger4+/+ and/or Ptger4−/− B cells by 1 nM PGE2 at 24 h. For each genetic background, log expression ratios of PGE2-treated (BCR+PGE2) versus vehicle-treated (BCR+vehicle) activated B cells are shown for each gene. The x axis denotes number of genes. (B) Genes that are induced when EP4 is absent are repressed when EP4 is abundant and activated. Shown are the log expression ratios of 128 genes commonly regulated in conditions designated as “lack of EP4” (Ptger4−/− versus Ptger4+/+ BCR-triggered B cells at 2 h) and “PGE2–EP4” (Ptger4+/+ versus Ptger4−/− BCR-triggered and 1 nM PGE2 cotreated B cells at 24 h). The identity of genes and their values are listed in Table S2. (C, D) Genes from the “PGE2–EP4” condition in (B) were connected using the IPA knowledge database. Two most significant and coherent networks consisted of genes associated with the NF-kB pathway (C) and MHC class I and II molecules (D). Green, down-regulated genes; red, up-regulated genes.
To verify the regulatory function of Ptger4 in BCR-driven gene transcription, we focused on a set of immediate-early genes induced by antigen-receptor signaling (Fig. 2, C and D). Transient micro RNA (miRNA)–mediated knockdown of Ptger4 in the A20 B cell lymphoma line resulted in increased transcriptional activity of NF-κB, in line with the observation from primary Ptger4−/− B cells, where genes coding for essential components of the NF-κB signaling pathway, including Traf5, Nfkb1, Nfkbie, and Fbi1, have been found significantly induced over controls (GEO accession no. GSE9847) (20, 26). In contrast, ectopic expression of the Ptger4 ORF region repressed BCR-driven transcription through NF-κB, complementing the results from loss-of-function studies. Ptger4 exerted a similar inhibitory effect on transcriptional activity of the AP-1 components FOS and JUN, and potently decreased expression from IL-2 promoter known to contain critically important AP-1 and NF-κB sites (Fig. 2 D) (27). On the other hand, neither silencing nor overexpression of Ptger4 influenced transcriptional activity of EGR1 or transcription through cAMP-response element (CRE), both of which greatly increase upon BCR ligation (Fig. 2 D and Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20081163/DC1) (28, 29). Together, these data indicate that Ptger4 negatively regulates BCR-mediated gene expression and B cell proliferation, primarily by inhibiting NF-κB and AP-1 transcriptional activity.
Ptger4 is a candidate tumor suppressor in mouse B cell lymphoma
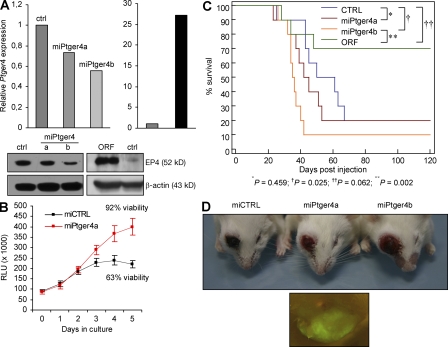
As primary Ptger4−/− B cells resisted rapid cell death in vitro in the absence of activating stimuli, we next set out to determine how altering the Ptger4 gene dosage would affect B cell growth and survival. To this end, we designed lentiviral vectors containing either GFP cocistronic with miRNA targeting Ptger4 (miPtger4) or the cloned Ptger4 ORF region to generate stable A20 B cell lines. Quantitative PCR (qPCR) and immunoblotting confirmed the efficiency of the stably integrated Ptger4 knockdown and overexpressing cassettes (Fig. 3 A). Interestingly, although the introduced changes did not seem to influence the in vitro growth of A20 cells, which have a doubling time of <12 h, the slower growing P493-6 human B cell line was significantly impacted by a silencing construct that targeted both mouse and human Ptger4 and achieved comparable gene knockdown levels in both cell lines (Fig. 3 B and unpublished data). The modified human B cells showed a similar rate of proliferation at low cell concentrations compared with controls, but later resisted overgrowth and reached about twice the density of viable control cells in the medium. This is in agreement with increased ex vivo survival of Ptger4−/− cells and indicates that reducing the levels of Ptger4 enhances survival of mouse and human B cells.
Figure 3.
Ptger4 is a candidate tumor suppressor in B cells. (A) Stable A20 cell lines were generated by lentiviral infection as described in Materials and methods. Ptger4 expression was determined by qPCR and confirmed by Western blotting. (B) ATP content and viability of stable P493-6 cell lines in culture. RLU, relative luminescence units. Error bars are the SD of triplicates. (C) Survival of BALB/c mice injected with 2 × 105 A20 cells stably expressing the indicated constructs. 10 mice were used to derive each survival curve. Logrank test comparing two survival curves was used to derive P values, as indicated. (D) Tumor formation at the site of injection of the indicated stable A20 cell line at 20 d after injection (5 × 105 cells were injected per mouse; top), and a fluorescence micrograph showing GFP-expressing tumor tissue in the eye region (bottom).
We then asked whether vigorous in vitro proliferation of A20 cells could have obscured the effect of over- or underexpressed Ptger4 by setting up a more sensitive in vivo system. Because A20 cells are known to induce tumor formation when introduced into syngeneic mice, we injected 200,000 viable cells from each of the generated A20 lines in the BALB/c strain and monitored tumor development over time (Fig. 3 C). Stable coexpression of GFP in the injected cells allowed us to easily track tumor origin by fluorescence microscopy of the dissected or intact tumor tissue. Stunningly, although most of the control mice injected with nontargeting miRNA (miCTRL)-carrying cells succumbed to disseminated B-lymphoma development over the first two-month period, as expected, overexpression of Ptger4 yielded significant protection against lymphoma spread, and the majority (70%) of mice survived beyond the monitoring period with no adverse symptoms (Fig. 3 C). In contrast, A20 cells with stably silenced Ptger4 potently aggravated tumor burden, which frequently manifested as liver and neck region metastasis, as well as hind limb paralysis, and the mice quickly became terminally ill. Additionally, the more efficient miRNA, miPtger4b, triggered a more severe phenotype than the less efficient miPtger4a, as was also evident from a separate experiment in which a higher number of injected cells induced tumor formation at the site of injection in the eye, with the size of the tumor corresponding to the Ptger4 knockdown efficiency (Fig. 3 D). Collectively, these data demonstrate that Ptger4 negatively regulates B cell survival and proliferation, and suggest that Ptger4 may function as a tumor suppressor in B cells.
PGE2 suppresses B cell proliferation by targeting the EP4 receptor
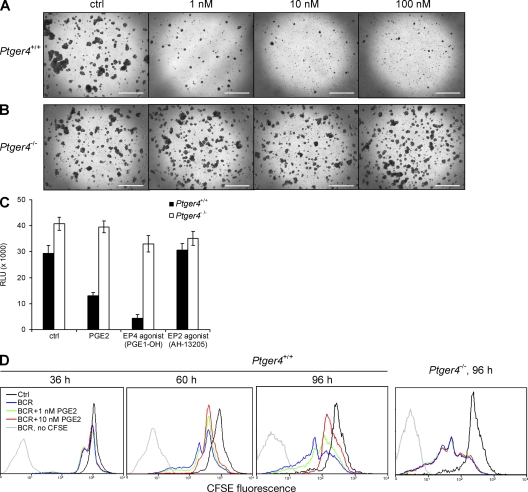
We next examined the potential physiological relevance of the inhibitory role of Ptger4 on B cell growth by focusing on the effects of the only known but extremely potent endogenous ligand for EP4 receptor, prostaglandin E2 (PGE2). Several reports have documented a repressive role of PGE2 on immune responses and B cells in particular (10, 30–32), but no study to date has identified the exact mechanism by which this inhibition occurs. Because we identified Ptger4 as a delayed-early gene and the only BCR-induced gene of the Ptger series (Fig. 1 B), we speculated that the EP4 receptor may be important for conveying the growth-regulatory actions of PGE2 in B cells. We first checked the influence of PGE2 on BCR-triggered proliferation of wild-type cells in vitro and saw that PGE2 at a concentration as low as 1 nM effectively prevented B cell cluster formation and proliferation (Fig. 4 A). In contrast, EP4-null B cells were completely insensitive to the inhibition by PGE2, forming massive clusters of proliferating cells similar to the vehicle-treated controls, even at high doses of PGE2 (Fig. 4 B). This strongly suggested that EP4 was the major, if not the only receptor transmitting the PGE2-delivered signal to block B cell growth. In addition, treatment with the EP4-specific agonist PGE1-OH faithfully mimicked the inhibitory activity of PGE2 in wild-type, but not EP4-null, cells, as opposed to stimulating the closest EP4-related receptor, EP2, which did not influence proliferation of either cell type (Fig. 4 C).
Figure 4.
The suppressive effect of PGE2 on B cell proliferation is mediated by the EP4 receptor. Ptger4+/+ (A) and Ptger4−/− (B) splenic B cells were incubated in vitro for 60 h with anti-IgM F(ab′)2 and cotreated with PGE2 at the concentrations as indicated (ctrl, vehicle-treated control). Bar, 200 μm. (C) BCR-stimulated splenic B cells were cotreated as indicated for 60 h, followed by determination of ATP content. Final concentrations of PGE1-OH and AH-13205 were adjusted to compensate for the differences between inhibitory constants for the mouse EP4 receptor (56). RLU, relative luminescence units. Error bars are the SD of triplicates. (D) The CFSE staining method was used to follow proliferation of Ptger4+/+ and Ptger4−/− mouse B cells. Flow cytometry analysis of viable cells is shown (see Materials and methods).
To more closely examine how triggering of EP4 by PGE2-affected B cell activation, we used time-lapse microscopy to follow cell growth and cluster development of BCR-triggered Ptger4+/+ and Ptger4−/− B cells in the presence of PGE2 over several days. Although the two cell populations did not differ during the initial phase (the first 18–24 h) of cluster formation, the effect of PGE2 became progressively more apparent, as the wild-type cells were less efficient at forming bigger groups of cells. Once Ptger4−/− cells entered a cluster, they stayed associated with it until the activation reaction was terminated, as opposed to wild-type cells, which dynamically shuttled in and out of clusters, resulting in a smaller net size of the clusters and in more cells found dispersed throughout the culture plate (Videos S1 and S2, available at http://www.jem.org/cgi/content/full/jem.20081163/DC1). Notably, this time-dependent activity of PGE2 was in line with the kinetics of BCR-induced EP4 expression, as revealed by immunoblotting (Fig. 1 C). To discern the impact of signaling through EP4 on cell proliferation, we used the CFSE method to follow cell division of BCR-activated B cells (Fig. 4 D). In agreement with a previous study (10), PGE2 allowed wild-type B cells to undergo only one round of cell division, but significantly inhibited further proliferation of daughter B cells. In contrast, Ptger4-deficient cells displayed complete protection against the proliferation-limiting effects of PGE2, confirming the suggestion that EP4 is the primary molecule mediating the negative influence of PGE2 on B cell activation. The addition of PGE2 diminished the viability of activated wild-type B cells in a time- and dose-dependent manner (Fig. S5), suggesting that PGE2 signaling via the EP4 receptor promoted the apoptosis of replicating progeny. The greater proportion of undivided cells at 96 h compared with 60 h in cultures supplemented with 10 nM PGE2 (Fig. 2 D) is thus a likely consequence of a preferential death of the divided population at the later time point.
PGE2–EP4 signaling results in general repression of activating genes
We next asked how, mechanistically, PGE2–EP4 signaling antagonizes the proliferative burst of B cells triggered by antigen receptor cross-linking. Therefore, we inspected the transcriptional status of activated wild-type B cells incubated with or without PGE2 in comparison to EP4-null cells at 24 h after BCR triggering, a time point at which the inhibited phenotype started to become apparent and when EP4 protein was first readily detected by immunoblotting. Interestingly, of the 263 genes whose expression was altered by more than twofold in wild-type B cells because of the presence of PGE2, 91% (240 genes) were down-regulated, indicating a general repressive effect of PGE2 on gene transcription in activated B cells (Fig. 5 A). To the contrary, only 10 of the ∼22,000 genes examined were either induced or repressed in knockout cells, suggesting that EP4 was indeed the primary receptor through which PGE2 exerted its attenuation of the BCR-induced response. This finding was confirmed by an independent direct comparison of activated and PGE2-treated wild-type and Ptger4−/− B cells, which revealed EP4-mediated 85% (527 out of 623 genes affected) attenuation of gene transcription (unpublished data).
We then examined the identity of the genes regulated by the PGE2–EP4 pathway, and observed that a significant proportion (∼21%) of these genes were also impacted in Ptger4−/− B cells early after triggering of the BCR (Fig. 5 B and Table S2, available at http://www.jem.org/cgi/content/full/jem.20081163/DC1). However, genes that changed expression in both models had a reciprocal pattern of change; genes that were up-regulated because of the lack of EP4 were down-regulated in conditions where EP4 was abundant and stimulated by PGE2, confirming that this set of genes was specifically controlled by the EP4 receptor. PGE2–EP4 signaling most profoundly attenuated the expression of several genes coding for molecules of the NF-κB pathway, ribosomal proteins, translation-initiation factors, and ATP-producing molecules, as well as components of the MHC and (immuno)proteasome, most of which are known to be mandatory for efficient B cell activation (Fig. 5, C and D, and Table S2). These data validate and complement the observations from Ptger4-deficient B cells, indicating that EP4 functions to prime the inhibitory activity of its endogenous ligand PGE2 by repressing transcription of a functionally coherent set of activating genes, eventually leading to a systemic attenuation of B cell proliferation.
Ptger4 antagonizes GC B cell development
To verify the implications of our findings beyond the model of in vitro BCR-stimulated cells, we first screened the published data for potential differences in Ptger4 expression levels between naive follicular B cells and activated GC B cells in mice and humans (33, 34). In accord with the expectations, GC B cells exhibited profoundly reduced levels of Ptger4 compared with naive controls, confirming the in vitro observation that low Ptger4 expression favors B cell proliferation (Fig. S6, A and B, available at http://www.jem.org/cgi/content/full/jem.20081163/DC1). We then examined the expression of EP4 protein and saw that GC B cells expressed it at a much lower level compared with naive B cells, supporting the data from the aforementioned transcriptional analyses (Fig. S6 C). Furthermore, the analysis of GC B cell content in Ptger4−/− mice immunized with a T cell–dependent antigen found a significant increase compared with wild-type controls, indicating that the lack of Ptger4 was causative for the enhanced activation of B cells in vivo (Fig. S6 D). The importance of these findings is underscored by the fact that many B cell malignancies in humans show expression patterns of differentiation markers and global gene-expression profiles characteristic of GC B cells (35), thus most types of B cell lymphoma are thought to originate from GC B cells. It should be noted, however, that the higher proportion of GC B cells in Ptger4−/− mice may have resulted, at least in part, as a consequence of altered functioning of other EP4-expressing cell types, such as T cells (31) and dendritic cells (36), which also participate in T cell–dependent immune responses.
The protooncogene MYC down-regulates PTGER4 in human B cell lymphomas
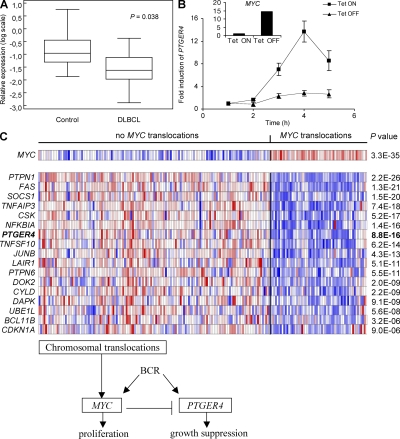
To investigate the relevance of PTGER4 to human disease, we first looked for altered expression of PTGER4 in normal versus malignant B cell populations by mining the GEO expression database (37). Interestingly, the analysis showed that PTGER4 is markedly down-regulated in the most commonly occurring, aggressive form of B cell malignancy, DLBCL, indicating that the reduction of proliferation-attenuating function of PTGER4 may indeed contribute to human B cell lymphoma growth (Fig. 6 A). We then asked about the possible mechanism leading to the observed decrease in PTGER4 expression in human neoplasia by focusing on the MYC gene. Because of chromosomal breakpoints at the MYC locus, increased transcription of MYC is a frequent cause of B lymphomagenesis, and has been associated with an adverse clinical outcome in several cases of aggressive B cell lymphoma (38). To that end, we used a human B cell line (P493-6) carrying a conditional tetracycline (Tet)-regulatable MYC allele that allows for abundant expression of MYC in the absence of Tet, but only weak MYC expression when Tet is present (39). We saw that the presence of Tet markedly influenced BCR-induced expression of PTGER4; in the high-MYC condition, the induction of PTGER4 was moderate, whereas low amounts of MYC transcripts greatly facilitated PTGER4 up-regulation, suggesting that MYC represses PTGER4 expression (Fig. 6 B). To examine whether the association between MYC and PTGER4 is relevant in vivo, we selected PTGER4 and several known tumor suppressors with established roles in human B cell lymphoma, and examined their expression in clinical cases of Burkitt's lymphoma, DLBCL, and some other types of mature aggressive B cell lymphomas with or without MYC translocations (38). We observed that this set of tumor-suppressor genes, including PTGER4, was coordinately down-regulated in cases with MYC translocations compared with cases without MYC translocations, irrespective of morphological, clinical, or immunohistochemical characteristics of the investigated B cell lymphomas (Fig. 6 C). These findings are consistent with a tumor-suppressive role of PTGER4 in human B cell cancer, and implicate the MYC oncogene as a potent negative regulator of PTGER4 expression. Although our data allow for a causative correlation, the extent to which the reduced PTGER4 expression contributes to the origin of these malignancies remains speculative, and further studies will be required to evaluate the potential of PTGER4 and its encoded EP4 receptor as a target for the therapy of human B cell lymphoma.
Figure 6.
MYC down-regulates the expression of PTGER4 in human B cell lymphoma. (A) PTGER4 is significantly down-regulated in human DLBCL cells relative to normal B cells. Data were analyzed from reference 30. The P value from Wilcoxon rank sum test is shown. (B) MYC inhibits BCR-triggered induction of PTGER4 in the human P493-6 B cell line. Cells were incubated with (Tet ON) or without Tet (Tet OFF) for 3 d, and the expression of PTGER4 after BCR-triggering was determined by qPCR. Tet-dependent MYC expression is shown in the insert. Error bars are s.d. of triplicates. (C) PTGER4 and several known tumor suppressors are coordinately down-regulated in multiple human B cell lymphomas with MYC translocation (38). Data were analyzed from a comprehensive study, including samples of 220 mature aggressive B cell lymphomas using the algorithm of the human cancer-profiling database Oncomine (see URLs in Materials and methods) (57). Shown is the heat map comprising the microarray results, where red indicates induction of genes and blue indicates repression of genes. Samples were classified according to the presence (or absence) of MYC translocations, as indicated. Genes showing the lowest P values were selected and ranked in ascending order. The selected genes, except PTGER4, have been described as tumor suppressors in B cell lymphoma according to Pubmed (see URLs in Materials and methods). The scheme om the bottom presents the interaction between MYC and PTGER4 in a lymphoma-relevant context.
DISCUSSION
Ptger series of genes have traditionally been viewed and studied in light of the interactions of their encoded receptors with PGE2 as the natural ligand, whereas kinetics of gene expression or the intrinsic functioning of EP receptor subtypes has received little attention. In this study, we identify Ptger4 as a typical delayed-early gene negatively regulating the robust proliferative response of B cells to antigen receptor triggering and use transcriptome analysis to delineate the attenuating mechanism used by Ptger4.
The observation that Ptger4 knockout B cells display a more activated genotype compared with wild-type controls early after BCR-stimulation in vitro suggests an inherent growth-suppressing activity of Ptger4 that is independent of PGE2–EP4 interactions. This is supported by the finding that BCR-stimulated mouse B cells do not secrete any substantial amounts of PGE2 in culture and that incubation of wild-type B cells with COX inhibitors to block the potential PGE2 biosynthesis does not augment proliferation of activated B cells, excluding the possible influence of de novo–produced PGE2. In addition, stable knockdown of Ptger4 promoted growth of a human B cell line in culture, suggesting that the intrinsic growth-limiting role of Ptger4 is conserved between mouse and human B cells. Although there is evidence for the secretion of relatively high amounts of PGE2 by quiescent and activated human B cells (40, 41), we are not aware of any studies supporting the production of PGE2 by mouse B lymphocytes, with the exception of a study by Graf et al. (42) showing that a biphenotypic B/macrophage population of cells produces substantial amounts of PGE2 upon stimulation with lipopolysaccharide, CD40, or BCR ligand. In contrast, it has been noted that the expression of COX-1 and -2 is not observed in mouse B cells (41, 42), and the lack of induction of COX-2 is confirmed by our microarray analysis of mouse B cells after BCR engagement (GSE9215 and GSE9847). Interestingly, although controversy exists regarding the net effect of PGE2 on proliferative responses of activated human B cells (10, 40, 41, 43–45), there appears to be a consensus on the inhibitory function of PGE2 on mouse B cell proliferation (9, 24, 46, 47). Depending on the mitogenic stimulus studied, reports have documented inhibition (10), no influence (43), stimulation (45), or concentration-dependent effect (41) of PGE2 on proliferation of human B cells. It is possible that the various stimuli differently regulate the expression of or signaling through each of the four distinct PGE2 receptors, some of them with opposing functions, thus critically influencing the net result of PGE2 action on humans B cells. Further studies will be required to show whether PGE2–EP4 signaling is inhibitory in each of these B cell–activating conditions.
Two main observations lead us to assume that Ptger4 acts as a feedback regulator to attenuate BCR-signaling pathway. First, Ptger4 counteracted the transcriptional activity of early-induced NF-κB and AP-1 complexes, which are both essential for propagating B cell growth and proliferation, and, second, it facilitated down-regulation of several proximal BCR-signaling components. Given the “natural” tendency of the BCR to undergo autoinactivation upon signal transmission, the negative feedback regulation by Ptger4 may thus shorten the activation period of the BCR, ultimately resulting in a weaker proliferative response of wild-type compared with Ptger4-deficient B cells.
A striking finding of our study is the reduced in vivo tumorigenic potential of the aggressive A20 B-lymphoma cells overexpressing Ptger4. Regardless of whether the diminished tumorigenicity resulted from the enhanced cell-intrinsic effects of Ptger4, increased PGE2–EP4 signaling, or both, the over-present Ptger4 was clearly causative for protection of mice against lymphoma spread. Conversely, the aggravated symptoms caused by tumor development and the increased mortality imposed by the ∼40–45% silenced expression of Ptger4 demonstrate the inhibitory potential of Ptger4 and indicate that its deficit was, indeed, the primary genetic lesion leading to the elevated rate of tumor invasion. We were therefore not surprised to find PTGER4 significantly down-regulated in DLBCL, the most common subtype of non-Hodgkin's lymphoma that accounts for 30–40% of all lymphoma cases and that frequently shows a poor prognosis in humans (3, 38). Together, these data further establish Ptger4 as a negative regulator of BCR-signaling and identify it as a candidate tumor suppressor in mouse and human B cells.
Consistent with previous reports, PGE2 potently inhibited B cell activation, and we for the first time provide definitive proof that the EP4 receptor is the principal molecule responsible for the repressive activity of PGE2 in B cells. This finding indicates that PGE2 potentiates the existing intrinsic inhibition by EP4, which is supported by the progressively greater negative influence of PGE2 on B cell proliferation and viability correlating with the expression level of EP4, and, importantly, by the reciprocal EP4-related gene regulation between activated Ptger4−/− and PGE2-treated wild-type B cells. Transcriptional profiling of PGE2–EP4 signaling uncovered a systemic down-regulation of numerous prosurvival genes that likely accounted for the impacted phenotype. Several of these genes mapped to the most significantly affected NF-κB pathway, indicating that the latter could play a key role in the EP4–phenotype correlation. Importantly, genetic studies in mice have confirmed the essential role of NF-κB in promoting B cell activation (18), and activated B cell–like DLBCL cells depend on constitutive NF-κB activity for survival (48). The EP4-attenuated expression of cell-cycling genes, as well as genes coding for components of the energy-producing and protein-translation machinery might have, at least in part, come as a consequence of the aberrant upstream NF-κB activity, together leading to a proliferation shutdown.
Another functionally coherent network consisted of MHC class I and II members, which B cells use to present antigens to and thus stimulate T cells, thereby ensuring appropriate immune response to foreign antigens. This suggests that EP4 not only functions to negatively regulate the activation of the B cell in which it is expressed, but may also limit the responses of other immune cells in its vicinity. Nevertheless, one of the genes most significantly up-regulated by the lack of Ptger4 and repressed by PGE2–EP4 signaling, the MHC class II chaperone CD74, was found to directly stimulate B cell survival and promote malignant B cell proliferation (21). The expression of PTGER4 may thus be relevant to the efficiency of the anti-CD74 monoclonal antibody LL1, which is currently under evaluation as a novel therapeutic agent to treat B cell lymphoma (22).
Activation of the MYC protooncogene by chromosomal translocations or hypermutation is a key event in the pathogenesis of many B cell lymphomas, and MYC breakpoints in lymphomas with a high chromosomal complexity score strongly correlate with a poor survival rate (38). Given that the critical mechanisms by which MYC contributes to malignant transformation are not clear, it is tempting to speculate that the coordinate down-regulation of multiple tumor suppressor genes, including PTGER4, plays an important role in MYC-induced lymphomagenesis. It is noteworthy that PTGER4 has also been identified as one of a few genes targeted by the transcription factor and candidate tumor suppressor ETV6, deletions of which are observed in up to 47% of childhood pre–B acute lymphoblastic leukemia (49), implying a different mechanism of PTGER4 down-regulation in B cell malignancy. Regardless of the primary genetic lesion leading to reduced expression of PTGER4, our findings support the possibility that regulation of B cell proliferation is a contributing mechanism by which PTGER4 could hold human B cell lymphoma formation in check.
Collectively, our data postulate Ptger4 as a delayed-early gene and a negative feedback regulator of BCR-signaled activation of B cells. This gene functions via its encoded EP4 receptor, which is also the principal molecule transmitting the inhibitory message of PGE2 to proliferating B cells. Regulation of Ptger4 inversely correlates with the expression of many genes that promote growth and survival of B cell lymphoma, such as MYC, members of the NF-kB pathway, and Cd74, but appears to be coordinated with several tumor suppressors. Varying the Ptger4 expression level significantly affects B cell lymphoma spread in mice and we find it down-regulated in human B cell malignancy, indicating that Ptger4 is a tumor suppressor in B cells. Finally, because EP4 is expressed as an easily accessible membrane receptor, interfering with the PGE2–EP4 signaling pathway may present a clinically useful alternative or supplement to chemotherapy in the treatment of B cell lymphoma.
MATERIALS AND METHODS
Mice and reagents.
We obtained female C57BL/6 and BALB/c mice from The Jackson Laboratory. Female Ptger4−/− mice and Ptger4+/+ littermate controls were generated as previously described (16). All mice were housed in the Exploration and Experimental Functional Research Center (CERFE; Evry, France), and all experiments and protocols using animals were approved by the CERFE direction committee and the Institut de Radiobiologie Cellulaire et Moléculaire (CEA) animal research committee. Mice used for experiments were 8–12 wk old. PGE2, AH-13205, indomethacin, and tetracycline were purchased from Sigma-Aldrich. Prostaglandin E1 alcohol (PGE1-OH) and NS-398 were obtained from Cayman Chemical. FITC-, PE-, APC-, or biotin-conjugated antibodies to CD19, CD23, CD38, CD93, CD95, or B220 were obtained from eBioscience. F(ab′)2 anti–human IgM was purchased from SouthernBiotech, F(ab′)2 anti–mouse IgM was purchased from Jackson ImmunoResearch Laboratories, and F(ab′)2 anti–mouse IgG was purchased from Caltag Laboratories.
Cells, cell sorting, and culture.
Single-cell suspensions were prepared from spleen of mice according to standard protocols. Negatively depleted B cells were recovered using B cell Isolation kit (Miltenyi Biotec) and mature B cells were further sorted as CD93−CD23+ by FACS using a MoFlo cell sorter (Dako) (50). Sorted cells were ≥95% pure. B cells were cultured at a density of 2 × 106 cells/ml in IMDM supplemented with 5% heat-inactivated FCS, 100 U/ml of penicillin, 100 μg/ml of streptomycin, 2 mM l-glutamine, and 50 μM β-mercaptoethanol. To trigger the BCR, cells were stimulated with F(ab′)2 anti–mouse IgM at 20 μg/ml. A20, a mature B lymphoma cell line of BALB/c origin (American Type Culture Collection [ATCC]), and P493-6, a human B lymphoma cell line containing a Tet-regulatable MYC gene (provided by G.W. Bornkamm, GSF Research Centre, Neuherberg, Germany) were cultivated in RPMI-1640 supplemented with 10% heat-inactivated FCS, 100 U/ml of penicillin, 100 μg/ml of streptomycin, 2 mM l-glutamine and 50 μM β-mercaptoethanol. To inhibit the expression of MYC, P493-6 cells were incubated with 0.1 μg/ml Tet for 3 d. For BCR triggering, A20 cells were incubated with F(ab′)2 anti–mouse IgG and P493-6 cells with F(ab′)2 anti–human IgM, both at 20 μg/ml. NIH3T3 cells were cultivated as recommended by the ATCC. All cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Proliferation and viability assays.
The ATP content of B cells was measured using a bioluminescence assay (ViaLight Plus; Cambrex). Tests were performed in 96-well plates at 105 cells/well according to the manufacturer's instructions. Cell cycle was analyzed using flow cytometry according to standard protocols. The CFSE method to follow B cell proliferation over time was performed according to a previously published protocol (51). Cell viability was monitored by the exclusion of 0.4% trypan blue solution visualized by light microscopy.
Transfection and luciferase reporter assays.
A20 cells were transfected by electroporation using Nucleofector II (Amaxa Biosystems) according to the manufacturer's instructions. NF-κB, FOS, JUN, IL-2, EGR1, and CRE activities were measured after electroporation of cells with plasmids (2 μg) encoding the indicated response element or promoter fused to a firefly luciferase reporter gene together with the indicated Ptger4-expression vectors (2 μg) and pRL-TK as an internal control (200 ng; Promega). 48 h after transfection, cells were incubated with 20 μg/ml F(ab′)2 anti–mouse IgG for 8 h, and luminescence was determined using Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. The luciferase reporter plasmids were gifts, as follows: NF-κB reporter plasmid from K.A. Reedquist (University of Amsterdam, Amsterdam, Netherlands), FOS and JUN reporter plasmids from H. van Dam (Leiden University, Leiden, Netherlands), IL-2 reporter plasmid from G.R. Crabtree (Stanford University, Stanford, CA), EGR1 reporter plasmid from J. Milbrandt (Washington University School of Medicine, St. Louis, MO), and CRE reporter plasmid from V.C. Manganiello (National Institutes of Health, Bethesda, MD).
Expression vectors, lentiviruses, and stable cell lines.
Expression vectors to silence or overexpress mouse Ptger4 were constructed using Gateway technology and Block-iT Lentiviral Pol II miR RNAi Expression System (Invitrogen). For the silencing vectors, seven different premiRNAs targeting Ptger4 were designed using the manufacturer's RNAi Designer program, cloned into pcDNA 6.2-GW/EmGFP-miR vectors, and tested by qPCR for Ptger4 knock-down efficiency upon transfection into NIH3T3 cells with Lipofectamine 2000 (Invitrogen). The most efficient premiRNA expression cassette (miPtger4b) a less efficient cassette targeting both mouse and human Ptger4 (miPtger4a), and a nontargeting control cassette (miCTRL) were selected and recombined into pLenti6/V5-DEST destination vector. For the overexpression construct, the ORF region of mouse Ptger4 was amplified by PCR from total cDNA of A20 cells and transferred into pLenti6/V5-DEST vector. Lentiviruses were generated by cotransfecting each recombined destination vector with ViraPower packaging mix (Invitrogen) into 293FT cells as recommended by the manufacturer. A20 and P493-6 cell lines stably expressing silencing or control cassettes were generated by in vitro transduction with the appropriate lentiviral particles, followed by selection of GFP+ cells by FACS at 12 d after transduction. Similarly, A20 cells with stably integrated overexpressing cassette were generated using the blasticidin-selection method. The expression of Ptger4 in stable cells lines was verified by qPCR and immunoblotting. All synthetic oligonucleotide sequences used are listed in Table S3 (available at http://www.jem.org/cgi/content/full/jem.20081163/DC1).
Lymphoma injection and immunization of mice.
Cells of each generated A20 stable cell line were intravenously injected into BALB/c mice, and tumor development was followed over a period of 120 d. The mice were killed when they showed excessive tumor growth. GFP expression analysis by fluorescence microscopy was used to validate the mutated A20-origin of tumors. The results are presented as Kaplan-Meier survival curves. To analyze GC B cell development in response to a T cell–dependent antigen, Ptger4−/− mice and wild-type controls were injected intraperitoneally with 100 μg of 4-hydroxy-3-nitrophenylacetyl chicken γ-globulin (NP-CGG; Biosearch Technologies) mixed with Imject Alum (Thermo Fisher Scientific) as an adjuvant. At day 14 after immunization, mice were killed and GC B cell content in splenic single-cell suspensions was determined by flow cytometry as the percentage of CD19+CD38lowCD95high cells (52). This strategy was also used to sort wild-type GC and naive B cells for the immunoblot analysis.
Microarray analysis.
For the initial screen to identify negative regulators of B cell proliferation, we used microarrays containing 15,247 cDNA clones for the analysis of ∼11,000 mouse genes (53). RNA was isolated from BCR-stimulated or vehicle-treated mature B cells from pooled spleens of 10 C57BL/6 mice using Trizol reagent (Invitrogen), followed by one round of amplification with MessageAmp kit (Ambion). The amplified RNA was reverse-transcribed, labeled, and hybridized to the arrays as previously described (54). Each sample was hybridized to the arrays in 6 replicates using a dye-swap strategy. Slides were scanned with Genepix 4000B array scanner and feature extraction was performed with Genepix Pro 6.0 software (Axon Instruments). For transcriptome analysis of Ptger4−/− and Ptger4+/+ control B cells, microarrays with 24,109 spotted mouse oligonucleotides were used (55). RNA was isolated using RNeasy Mini kit (QIAGEN), amplified, labeled, and hybridized following a published protocol (55). Oligonucleotide arrays were scanned with Agilent G2565AA Microarray Scanner (Agilent Technologies). Data analysis, including intensity-dependent Lowess normalization of raw data, was performed with GeneSpring 7.0 software (Silicon Genetics). Ingenuity Pathway Analysis (Ingenuity Systems) was used for gene ontology and network analysis.
Real-time quantitative PCR.
Total RNA was isolated using RNeasy Mini kit, and reverse-transcribed by Superscript II Reverse transcription (Invitrogen) according to the manufacturers' protocols. Samples were processed on ABI PRISM 7700 Sequence Detection System (Applied Biosystems) using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich). All experiments were performed in triplicate, and results were normalized to Hprt1 expression levels. Primer sequences are listed in Table S3.
Immunoblot analysis and immunofluorescence.
Total cell protein extracts were prepared from B cells by lysis in RIPA Buffer supplemented with Protease Inhibitor Cocktail (both from Sigma-Aldrich). Proteins were separated by SDS-PAGE and immunoblots were performed using anti-EP4 (C-term; Cayman Chemical) and anti-actin (Sigma-Aldrich) antibodies. Immunoblots were developed with SuperSignal ECL reagent (Thermo Fisher Scientific). Immunofluorescence of BCR-stimulated mouse B cells was done using anti-EP4 antibody (C-18; Santa Cruz Biotechnology) following the manufacturer's protocol. To assure specificity of staining, anti-EP4 antibody was preincubated with fivefold (by weight) excess of the specific EP4 blocking peptide (Santa Cruz Biotechnology) for 1 h at room temperature. Indirect detection of EP4 was performed using secondary FITC-conjugated antibody.
Measurement of PGE2 production.
Mouse B lymphocyte culture supernatants were harvested at 0, 2, and 4 d of coincubation with anti-IgM F(ab′)2 with or without indomethacin (20 μM) or NS-398 (10 μM), and analyzed by a highly specific competitive enzyme immunoassay (Prostaglandin E2 EIA kit; Cayman Chemical) to determine the concentration of PGE2. The levels of PGE2 in all samples of culture supernatants were below the detection limit of the assay (15 pg/ml). Three independent experiments were performed.
Statistical analysis.
Results are mean values ± SD. Statistical analyses were performed using the logrank test, Wilcoxon rank sum test, or Student's t test (Oncomine), as indicated. P values of 0.05 or less were considered significant, and exact P values are shown in the figures.
Accession nos.
The complete microarray datasets are available from the Gene Expression Omnibus (GSE9215 and GSE9847).
URLs.
Ingenuity Pathway Analysis: http://www.ingenuity.com/; Oncomine database: http://www.oncomine.org/; Pubmed database: http://www.ncbi.nlm.nih.gov/pubmed/.
Online supplemental material.
Fig. S1 shows that EP4 protein is expressed at low levels in quiescent B cells and is induced upon BCR triggering. Fig. S2 shows that BCR-triggering induces expression of the EP4 receptor on the plasma membrane of B cells. Fig. S3 shows that COX inhibition has no effect on proliferation or viability of BCR-stimulated mouse B cells. Fig. S4 shows that Ptger4 expression does not affect the BCR-induced transcriptional activity of EGR1 or transcription through CRE. Fig. S5 shows that PGE2 diminishes the viability of BCR-activated B cells in culture. Fig. S6 shows that Ptger4 antagonizes GC B cell development in mice. Table S1 shows the lack of Ptger4 facilitates BCR-triggered induction of activating genes. Tabe S2 shows genes that are induced when EP4 is absent are repressed when EP4 is abundant and activated. Table S3 shows synthetic oligonucleotide sequences used for qPCR, amplification of Ptger4 ORF region, and knockdown of Ptger4 expression. The videos show freshly isolated Ptger4−/− (Video S1) and Ptger4+/+ (Video S2) splenic B cells cotreated with 20 μg/ml F(ab′)2 anti-IgM and 10 nM PGE2 in culture. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081163/DC1.
Acknowledgments
We thank P. Vaigot for help with FACS analyses and cell sorting; M.A. Zanta-Bousiff for lentivirus production; K.A. Reedquist, H. van Dam, J. Milbrandt, G.R. Crabtree, and V.C. Manganiello for reporter plasmids; G.W. Bornkamm for the P493-6 cell line, and P. Delis for animal care.
J. Muen was supported by EGIDE.
The authors have no conflicting financial interests.
Abbreviations used: BCR, B cell receptor; DLBCL, diffuse large B-cell lymphoma; GC, germinal center; miRNA, micro RNA; PGE2, prostaglandin E2; qPCR, quantitative real-time PCR; Tet, tetracycline.
References
- 1.Niiro, H., and E.A. Clark. 2002. Regulation of B-cell fate by antigen-receptor signals. Nat. Rev. Immunol. 2:945–956. [DOI] [PubMed] [Google Scholar]
- 2.Allen, C.D., T. Okada, and J.G. Cyster. 2007. Germinal-center organization and cellular dynamics. Immunity. 27:190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuppers, R. 2005. Mechanisms of B-cell lymphoma pathogenesis. Nat. Rev. Cancer. 5:251–262. [DOI] [PubMed] [Google Scholar]
- 4.Hecht, J.L., and J.C. Aster. 2000. Molecular biology of Burkitt's lymphoma. J. Clin. Oncol. 18:3707–3721. [DOI] [PubMed] [Google Scholar]
- 5.Gururajan, M., C.D. Jennings, and S. Bondada. 2006. Cutting edge: constitutive B cell receptor signaling is critical for basal growth of B lymphoma. J. Immunol. 176:5715–5719. [DOI] [PubMed] [Google Scholar]
- 6.Irish, J.M., D.K. Czerwinski, G.P. Nolan, and R. Levy. 2006. Altered B-cell receptor signaling kinetics distinguish human follicular lymphoma B cells from tumor-infiltrating nonmalignant B cells. Blood. 108:3135–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petlickovski, A., L. Laurenti, X. Li, S. Marietti, P. Chiusolo, S. Sica, G. Leone, and D.G. Efremov. 2005. Sustained signaling through the B-cell receptor induces Mcl-1 and promotes survival of chronic lymphocytic leukemia B cells. Blood. 105:4820–4827. [DOI] [PubMed] [Google Scholar]
- 8.Cozma, D., D. Yu, S. Hodawadekar, A. Azvolinsky, S. Grande, J.W. Tobias, M.H. Metzgar, J. Paterson, J. Erikson, T. Marafioti, et al. 2007. B cell activator PAX5 promotes lymphomagenesis through stimulation of B cell receptor signaling. J. Clin. Invest. 117:2602–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurland, J.I., P.W. Kincade, and M.A. Moore. 1977. Regulation of B-lymphocyte clonal proliferation by stimulatory and inhibitory macrophage-derived factors. J. Exp. Med. 146:1420–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simkin, N.J., D.F. Jelinek, and P.E. Lipsky. 1987. Inhibition of human B cell responsiveness by prostaglandin E2. J. Immunol. 138:1074–1081. [PubMed] [Google Scholar]
- 11.Smith, J.P., G.F. Burton, J.G. Tew, and A.K. Szakal. 1998. Tingible body macrophages in regulation of germinal center reactions. Dev. Immunol. 6:285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurosaki, T. 1999. Genetic analysis of B cell antigen receptor signaling. Annu. Rev. Immunol. 17:555–592. [DOI] [PubMed] [Google Scholar]
- 13.Amit, I., A. Citri, T. Shay, Y. Lu, M. Katz, F. Zhang, G. Tarcic, D. Siwak, J. Lahad, J. Jacob-Hirsch, et al. 2007. A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 39:503–512. [DOI] [PubMed] [Google Scholar]
- 14.Narumiya, S., Y. Sugimoto, and F. Ushikubi. 1999. Prostanoid receptors: structures, properties, and functions. Physiol. Rev. 79:1193–1226. [DOI] [PubMed] [Google Scholar]
- 15.Harris, S.G., J. Padilla, L. Koumas, D. Ray, and R.P. Phipps. 2002. Prostaglandins as modulators of immunity. Trends Immunol. 23:144–150. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen, M., T. Camenisch, J.N. Snouwaert, E. Hicks, T.M. Coffman, P.A. Anderson, N.N. Malouf, and B.H. Koller. 1997. The prostaglandin receptor EP4 triggers remodelling of the cardiovascular system at birth. Nature. 390:78–81. [DOI] [PubMed] [Google Scholar]
- 17.Huntington, N.D., Y. Xu, H. Puthalakath, A. Light, S.N. Willis, A. Strasser, and D.M. Tarlinton. 2006. CD45 links the B cell receptor with cell survival and is required for the persistence of germinal centers. Nat. Immunol. 7:190–198. [DOI] [PubMed] [Google Scholar]
- 18.Jost, P.J., and J. Ruland. 2007. Aberrant NF-kappaB signaling in lymphoma: mechanisms, consequences, and therapeutic implications. Blood. 109:2700–2707. [DOI] [PubMed] [Google Scholar]
- 19.Landrette, S.F., Y.H. Kuo, K. Hensen, S. Barjesteh van Waalwijk van Doorn-Khosrovani, P.N. Perrat, W.J. Van de Ven, R. Delwel, and L.H. Castilla. 2005. Plag1 and Plagl2 are oncogenes that induce acute myeloid leukemia in cooperation with Cbfb-MYH11. Blood. 105:2900–2907. [DOI] [PubMed] [Google Scholar]
- 20.Schulze-Luehrmann, J., and S. Ghosh. 2006. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 25:701–715. [DOI] [PubMed] [Google Scholar]
- 21.Binsky, I., M. Haran, D. Starlets, Y. Gore, F. Lantner, N. Harpaz, L. Leng, D.M. Goldenberg, L. Shvidel, A. Berrebi, et al. 2007. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc. Natl. Acad. Sci. USA. 104:13408–13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein, R., M.J. Mattes, T.M. Cardillo, H.J. Hansen, C.H. Chang, J. Burton, S. Govindan, and D.M. Goldenberg. 2007. CD74: a new candidate target for the immunotherapy of B-cell neoplasms. Clin. Cancer Res. 13:5556s–5563s. [DOI] [PubMed] [Google Scholar]
- 23.Lanzavecchia, A. 1985. Antigen-specific interaction between T and B cells. Nature. 314:537–539. [DOI] [PubMed] [Google Scholar]
- 24.Roper, R.L., and R.P. Phipps. 1992. Prostaglandin E2 and cAMP inhibit B lymphocyte activation and simultaneously promote IgE and IgG1 synthesis. J. Immunol. 149:2984–2991. [PubMed] [Google Scholar]
- 25.Fedyk, E.R., and R.P. Phipps. 1996. Prostaglandin E2 receptors of the EP2 and EP4 subtypes regulate activation and differentiation of mouse B lymphocytes to IgE-secreting cells. Proc. Natl. Acad. Sci. USA. 93:10978–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, D.K., J.E. Kang, H.J. Park, M.H. Kim, T.H. Yim, J.M. Kim, M.K. Heo, K.Y. Kim, H.J. Kwon, and M.W. Hur. 2005. FBI-1 enhances transcription of the nuclear factor-kappaB (NF-kappaB)-responsive E-selectin gene by nuclear localization of the p65 subunit of NF-kappaB. J. Biol. Chem. 280:27783–27791. [DOI] [PubMed] [Google Scholar]
- 27.Chen, D., and E.V. Rothenberg. 1994. Interleukin 2 transcription factors as molecular targets of cAMP inhibition: delayed inhibition kinetics and combinatorial transcription roles. J. Exp. Med. 179:931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu, W., T. Fukuyama, P.A. Ney, D. Wang, J. Rehg, K. Boyd, J.M. van Deursen, and P.K. Brindle. 2006. Global transcriptional coactivators CREB-binding protein and p300 are highly essential collectively but not individually in peripheral B cells. Blood. 107:4407–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahon, S.B., and J.G. Monroe. 1995. Activation of the p21ras pathway couples antigen receptor stimulation to induction of the primary response gene egr-1 in B lymphocytes. J. Exp. Med. 181:417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harizi, H., C. Grosset, and N. Gualde. 2003. Prostaglandin E2 modulates dendritic cell function via EP2 and EP4 receptor subtypes. J. Leukoc. Biol. 73:756–763. [DOI] [PubMed] [Google Scholar]
- 31.Kabashima, K., T. Saji, T. Murata, M. Nagamachi, T. Matsuoka, E. Segi, K. Tsuboi, Y. Sugimoto, T. Kobayashi, Y. Miyachi, et al. 2002. The prostaglandin receptor EP4 suppresses colitis, mucosal damage and CD4 cell activation in the gut. J. Clin. Invest. 109:883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minakuchi, R., M.C. Wacholtz, L.S. Davis, and P.E. Lipsky. 1990. Delineation of the mechanism of inhibition of human T cell activation by PGE2. J. Immunol. 145:2616–2625. [PubMed] [Google Scholar]
- 33.Luckey, C.J., D. Bhattacharya, A.W. Goldrath, I.L. Weissman, C. Benoist, and D. Mathis. 2006. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 103:3304–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basso, K., A.A. Margolin, G. Stolovitzky, U. Klein, R. Dalla-Favera, and A. Califano. 2005. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 37:382–390. [DOI] [PubMed] [Google Scholar]
- 35.Alizadeh, A.A., M.B. Eisen, R.E. Davis, C. Ma, I.S. Lossos, A. Rosenwald, J.C. Boldrick, H. Sabet, T. Tran, X. Yu, et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 403:503–511. [DOI] [PubMed] [Google Scholar]
- 36.Legler, D.F., P. Krause, E. Scandella, E. Singer, and M. Groettrup. 2006. Prostaglandin E2 is generally required for human dendritic cell migration and exerts its effect via EP2 and EP4 receptors. J. Immunol. 176:966–973. [DOI] [PubMed] [Google Scholar]
- 37.Rosenwald, A., A.A. Alizadeh, G. Widhopf, R. Simon, R.E. Davis, X. Yu, L. Yang, O.K. Pickeral, L.Z. Rassenti, J. Powell, et al. 2001. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J. Exp. Med. 194:1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hummel, M., S. Bentink, H. Berger, W. Klapper, S. Wessendorf, T.F. Barth, H.W. Bernd, S.B. Cogliatti, J. Dierlamm, A.C. Feller, et al. 2006. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N. Engl. J. Med. 354:2419–2430. [DOI] [PubMed] [Google Scholar]
- 39.Schuhmacher, M., M.S. Staege, A. Pajic, A. Polack, U.H. Weidle, G.W. Bornkamm, D. Eick, and F. Kohlhuber. 1999. Control of cell growth by c-Myc in the absence of cell division. Curr. Biol. 9:1255–1258. [DOI] [PubMed] [Google Scholar]
- 40.Ryan, E.P., S.J. Pollock, T.I. Murant, S.H. Bernstein, R.E. Felgar, and R.P. Phipps. 2005. Activated human B lymphocytes express cyclooxygenase-2 and cyclooxygenase inhibitors attenuate antibody production. J. Immunol. 174:2619–2626. [DOI] [PubMed] [Google Scholar]
- 41.Mongini, P.K., J.K. Inman, H. Han, R.J. Fattah, S.B. Abramson, and M. Attur. 2006. APRIL and BAFF promote increased viability of replicating human B2 cells via mechanism involving cyclooxygenase 2. J. Immunol. 176:6736–6751. [DOI] [PubMed] [Google Scholar]
- 42.Graf, B.A., D.A. Nazarenko, M.A. Borrello, L.J. Roberts, J.D. Morrow, J. Palis, and R.P. Phipps. 1999. Biphenotypic B/macrophage cells express COX-1 and up-regulate COX-2 expression and prostaglandin E(2) production in response to pro-inflammatory signals. Eur. J. Immunol. 29:3793–3803. [DOI] [PubMed] [Google Scholar]
- 43.Gantner, F., C. Gotz, V. Gekeler, C. Schudt, A. Wendel, and A. Hatzelmann. 1998. Phosphodiesterase profile of human B lymphocytes from normal and atopic donors and the effects of PDE inhibition on B cell proliferation. Br. J. Pharmacol. 123:1031–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garrone, P., and J. Banchereau. 1993. Agonistic and antagonistic effects of cholera toxin on human B lymphocyte proliferation. Mol. Immunol. 30:627–635. [DOI] [PubMed] [Google Scholar]
- 45.Garrone, P., L. Galibert, F. Rousset, S.M. Fu, and J. Banchereau. 1994. Regulatory effects of prostaglandin E2 on the growth and differentiation of human B lymphocytes activated through their CD40 antigen. J. Immunol. 152:4282–4290. [PubMed] [Google Scholar]
- 46.Borghesi, L.A., G. Smithson, and P.W. Kincade. 1997. Stromal cell modulation of negative regulatory signals that influence apoptosis and proliferation of B lineage lymphocytes. J. Immunol. 159:4171–4179. [PubMed] [Google Scholar]
- 47.Shimozato, T., and P.W. Kincade. 1999. Prostaglandin E(2) and stem cell factor can deliver opposing signals to B lymphocyte precursors. Cell. Immunol. 198:21–29. [DOI] [PubMed] [Google Scholar]
- 48.Davis, R.E., K.D. Brown, U. Siebenlist, and L.M. Staudt. 2001. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 194:1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boily, G., J. Larose, S. Langlois, and D. Sinnett. 2007. Identification of transcripts modulated by ETV6 expression. Br. J. Haematol. 136:48–62. [DOI] [PubMed] [Google Scholar]
- 50.Rolink, A.G., J. Andersson, and F. Melchers. 2004. Molecular mechanisms guiding late stages of B-cell development. Immunol. Rev. 197:41–50. [DOI] [PubMed] [Google Scholar]
- 51.Quah, B.J., H.S. Warren, and C.R. Parish. 2007. Monitoring lymphocyte proliferation in vitro and in vivo with the intracellular fluorescent dye carboxyfluorescein diacetate succinimidyl ester. Nat. Protocols. 2:2049–2056. [DOI] [PubMed] [Google Scholar]
- 52.Thai, T.H., D.P. Calado, S. Casola, K.M. Ansel, C. Xiao, Y. Xue, A. Murphy, D. Frendewey, D. Valenzuela, J.L. Kutok, et al. 2007. Regulation of the germinal center response by microRNA-155. Science. 316:604–608. [DOI] [PubMed] [Google Scholar]
- 53.Kargul, G.J., D.B. Dudekula, Y. Qian, M.K. Lim, S.A. Jaradat, T.S. Tanaka, M.G. Carter, and M.S. Ko. 2001. Verification and initial annotation of the NIA mouse 15K cDNA clone set. Nat. Genet. 28:17–18. [DOI] [PubMed] [Google Scholar]
- 54.Rochon, C., V. Frouin, S. Bortoli, K. Giraud-Triboult, V. Duverger, P. Vaigot, C. Petat, P. Fouchet, B. Lassalle, O. Alibert, et al. 2006. Comparison of gene expression pattern in SP cell populations from four tissues to define common “stemness functions.” Exp. Cell Res. 312:2074–2082. [DOI] [PubMed] [Google Scholar]
- 55.Le Brigand, K., R. Russell, C. Moreilhon, J.M. Rouillard, B. Jost, F. Amiot, V. Magnone, C. Bole-Feysot, P. Rostagno, V. Virolle, et al. 2006. An open-access long oligonucleotide microarray resource for analysis of the human and mouse transcriptomes. Nucleic Acids Res. 34:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiriyama, M., F. Ushikubi, T. Kobayashi, M. Hirata, Y. Sugimoto, and S. Narumiya. 1997. Ligand binding specificities of the eight types and subtypes of the mouse prostanoid receptors expressed in Chinese hamster ovary cells. Br. J. Pharmacol. 122:217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhodes, D.R., J. Yu, K. Shanker, N. Deshpande, R. Varambally, D. Ghosh, T. Barrette, A. Pandey, and A.M. Chinnaiyan. 2004. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]