Abstract
Nonhomologous end-joining (NHEJ) repairs DNA double-strand breaks (DSBs) during V(D)J recombination in developing lymphocytes and during immunoglobulin (Ig) heavy chain (IgH) class switch recombination (CSR) in peripheral B lymphocytes. We now show that CD21-cre–mediated deletion of the Xrcc4 NHEJ gene in p53-deficient peripheral B cells leads to recurrent surface Ig-negative B lymphomas (“CXP lymphomas”). Remarkably, CXP lymphomas arise from peripheral B cells that had attempted both receptor editing (secondary V[D]J recombination of Igκ and Igλ light chain genes) and IgH CSR subsequent to Xrcc4 deletion. Correspondingly, CXP tumors frequently harbored a CSR-based reciprocal chromosomal translocation that fused IgH to c-myc, as well as large chromosomal deletions or translocations involving Igκ or Igλ, with the latter fusing Igλ to oncogenes or to IgH. Our findings reveal peripheral B cells that have undergone both editing and CSR and show them to be common progenitors of CXP tumors. Our studies also reveal developmental stage-specific mechanisms of c-myc activation via IgH locus translocations. Thus, Xrcc4/p53-deficient pro–B lymphomas routinely activate c-myc by gene amplification, whereas Xrcc4/p53-deficient peripheral B cell lymphomas routinely ectopically activate a single c-myc copy.
Ig heavy (IgH) and light (IgL) chain variable region exons are assembled from component V, D, and J segments in developing B lymphocytes. V(D)J recombination is initiated by the RAG1/2 endonuclease, which introduces DNA double-strand breaks (DSBs) between V, D, and J segments and flanking recombination signal sequences (RSs) (1). Subsequently, cleaved coding segments are joined to form V(D)J exons and RSs are joined to form RS joins (2). Both coding and RS joining are performed by classical nonhomologous end-joining (C-NHEJ), which is a major general DSB repair pathway in mammalian cells (3). Xrcc4 and DNA Ligase IV (Lig4) form a complex that is required for V(D)J recombination (4, 5). In their absence, coding or RS ends are joined at low frequency, usually with substantial sequence deletion from one or both partners (6, 7). In mice, Xrcc4 inactivation results in severe combined immune deficiency owing to inability to complete V(D)J recombination (6).
In progenitor B (pro–B) cells in the mouse BM, productive assembly of variable region exons within the IgH locus (Igh) on chromosome 12 leads to production of IgH μ chains that signal differentiation to the precursor B (pre–B) cell stage in which IgL variable region exons are assembled (8). Mice, like humans, have two IgL families, termed Igκ and Igλ, which are encoded by Igκ and Igλ loci that, respectively, lie on chromosomes 6 and 16. Igκ and Igλ expression is “isotype” excluded, such that a given B cell usually expresses either Igκ or Igλ, but not both (9). In mice, ∼95% of mature B lymphocytes are Igκ+, with the remainder being Igλ+. In that context, Igκ assembly usually precedes that of Igλ (9). Thus, most Igκ+ B cells contain Igλ in germline configuration, with Igλ rearrangements occurring in cells in which both Igκ alleles are rearranged out-of-frame or that harbor deletions of the Jκ segments, κ enhancer, and/or Cκ exons (9). Such deletions usually occur via rearrangement of Vκs or an RS heptamer in the Jκ-Cκ intron (IRS) to a bona fide RS 25 kb downstream of Cκ (3′RS) (10). Recent analyses suggest that Igκ deletions via 3′RS rearrangements may play a role in progression to Igλ rearrangement (11).
Expression of complete Ig (IgH/IgL) leads to IgM+ B lymphocytes, which ultimately down-regulate RAG expression to enforce allelic exclusion (1). However, newly generated BM IgM+ B lymphocytes that express autoreactive B cell receptors can maintain RAG expression and continue to rearrange IgL loci to generate new IgL chains in a tolerance process termed “receptor editing” (12–14). Receptor editing can replace rearranged Igκ loci with secondary productive Igκ rearrangements, as well as with nonfunctional Igκ rearrangements or Igκ deletions that may lead to Igλ rearrangement (12–14). Thus, Igλ+ B cells can be generated developmentally from pre–B cells with two nonproductive Igκ rearrangements or via receptor editing from immature Igκ+ B cells. Receptor editing is initiated in immature BM B cells (15, 16). Yet, several studies suggested IgL gene rearrangement, sometimes called “revision,” in mouse and human peripheral B cells, including germinal center B cells (17–21). However, many peripheral mouse RAG+ B lineage cells are pro– or pre–B cells that migrate to the periphery after immunization (22, 23), and knock-in reporter studies suggested that although RAG genes are expressed in B cells that have just migrated from the BM (24, 25), they are not reinduced in peripheral B cells once expression is terminated (25, 26).
After antigen stimulation, mature IgM+ peripheral B cells can undergo IgH class switch recombination (CSR), a recombination/deletion process in which the IgH μ constant region exons (Cμ) are deleted and replaced by one of several sets of downstream CH exons (e.g., Cγ, Cε, and Cα; referred to as CH genes) (27), leading to switching from IgM to another Ig class (e.g., IgG, IgE, or IgA). The activation-induced cytidine deaminase (AID) initiates CSR (28) by deaminating cytidines in switch (S) regions (29), which are 1–10-kb repetitive sequences located 5′ of each CH gene. AID-generated lesions within the donor Sμ and a downstream acceptor S region are processed into DSBs, which are end-joined to complete CSR (27). In contrast to V(D)J recombination, substantial CSR occurs in the absence of Xrcc4 or Lig4 (C-NHEJ) via an alternative end-joining (A-EJ) pathway strongly biased to use microhomology (30). However, CSR is significantly impaired in Xrcc4-deficient B cells owing to failure to join broken S regions because up to 20% of Xrcc4-deficient B cells activated for CSR in vitro have IgH chromosomal breaks, with a substantial portion participating in chromosomal translocations (30).
Inactivation of Xrcc4 in mice results in impaired cellular proliferation and ionizing radiation sensitivity. Xrcc4 deficiency also results in extensive apoptosis of newly generated neurons and late embryonic death (6), both of which can be rescued by deficiency for the p53 tumor suppressor (31). In this context, p53 monitors the G1 cell cycle checkpoint, signaling apoptosis of certain cell types, such as neurons and progenitor lymphocytes, which harbor persistent DSBs (32). However, as p53 deficiency does not rescue defective NHEJ associated with Xrcc4 deficiency, Xrcc4/p53–double-deficient mice are still immunodeficient and inevitably succumb to pro–B cell lymphomas that harbor RAG-dependent complex translocations (33). These translocations usually join IgH on chromosome 12 to a region downstream of c-myc on chromosome 15, resulting in dicentric 12;15 translocations and c-myc amplification via breakage–fusion–bridge cycles (34). Such complex translocations are rare in human peripheral B cell lymphomas, which more frequently harbor reciprocal translocations that fuse IgH, or less frequently IgL loci, just upstream of c-myc, leading to ectopic c-myc activation (35).
In the current study, we have asked whether inactivation of C-NHEJ in WT or p53-deficient peripheral B cells leads to peripheral B cell lymphoma with CSR or V(D)J recombination-associated IgH or IgL locus translocations.
RESULTS
Inactivation of Xrcc4 in p53-deficient peripheral B cells leads to novel B cell lymphomas
We previously inactivated Xrcc4 specifically in transitional stage peripheral B cells by generating mice that harbored one copy of a loxP-flanked (floxed) Xrcc4 allele (Xc) and one copy of an Xrcc4-null allele (X−) plus a transgene that drives Cre recombinase expression via a CD21 promoter (termed CXc/– mice) (30). CD21 proteins are expressed during B cell development at the time when immature transitional B cells differentiate into mature long-lived peripheral B cells, including marginal zone B cells, follicular B cells, B1a (CD5+), and B1b cells (36). CD21cre mediates efficient Cre recombination in mature, but not immature, transitional B cells (37). Thus, CXc/– mice have normal IgM+ splenic B cell numbers as Xrcc4 is not deleted during early B cell developmental stages where V(D)J recombination occurs (30). Remarkably, CXc/– mice have a normal life span and are not cancer-prone (Fig. 1 A), despite high levels of general and IgH-specific genomic instability in ex vivo αCD40/IL-4–stimulated peripheral CXc/– B cells (30). Analogous to what we previously found for B lymphoid and neuronal progenitors (31), lack of CXc/– B cell transformation may reflect p53-dependent elimination of cells harboring DSBs and other types of genomic instability. To investigate this possibility, we bred CXc/– mice into a p53-deficient background to generate CXc/−P−/− mice.
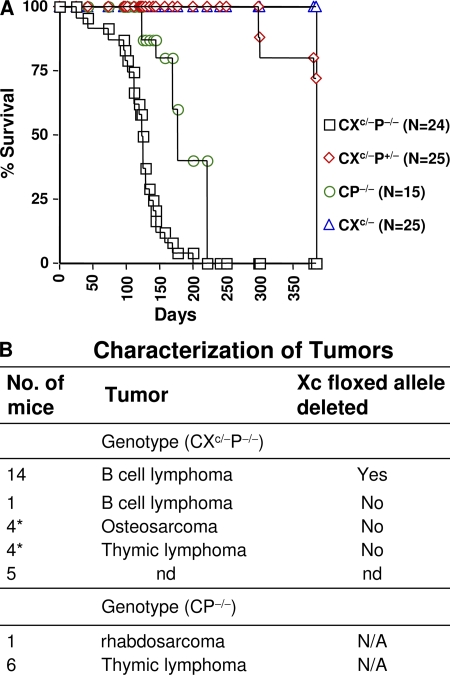
Figure 1.
Establishment of the CXP B cell lymphoma model via conditional inactivation of Xrcc4 in CD21Cre-expressing, p53-deficient mice. (A) Kaplan-Meier survival curve: percent survival of CXc/−P−/− (n = 24); CXc/−P+/− (n = 25); CP−/− (n = 15); and CXc/− (n = 25) mice versus age in days is shown. (B) Tumor types that developed in the indicated cohorts of CXc/−P−/− and CP−/− mice. The number of mice that developed a given type of tumor is indicated at the left. Deletion of the floxed Xrcc4 allele in given tumors is indicated at the right. nd, not determined; N/A, not applicable. The asterisk indicates CXc/−P−/− mice that developed osteosarcoma or thymic lymphomas, in addition to B cell lymphomas.
The majority of CXc/−P−/− mice become moribund by 3.5–4.5 mo of age, with >50% succumbing to Xrcc4/p53-deficient B lineage lymphomas that present predominantly in the mesenteric lymph nodes and that had deleted Xrcc4 (Fig. 1, A and B). The remaining CXc/−P−/− mice succumbed to thymic lymphomas and sarcomas characteristic of p53 deficiency alone and which had not deleted Xrcc4 (Fig. 1 B). In contrast, none of the CP−/−, CXc/–, or CXc/−P+/− littermates developed B lymphomas (Fig. 1, A and B). Of eight CXc/−P−/− B lineage lymphomas (termed CXP lymphomas) characterized in detail, all were surface Ig negative (sIg−) and all were B220+, CD43low, CD138/Syndecan-1+, and CD19+ (Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20082271/DC1). In addition, all had clonal IgH and Igκ rearrangements and many had clonal Igλ rearrangements, even though they usually lacked expression of IgH and IgL proteins (Table S1; see below). RNA expression analyses indicated that CXP lymphomas lacked expression of λ5 and Vpre–B, two diagnostic early B cell markers, but did show very low-level RAG expression (Fig. S1, B and C, and not depicted). We also analyzed six additional Xrcc4/p53-deficient B lymphomas derived from CXc/− mice in which the p53 gene was also floxed (CXc/−Pc/c) and found that the majority had a phenotype similar to CXc/−P−/− lymphomas (Fig. S2 and Table S2). Likewise, eight additional CXc/−P−/− B lymphomas that were also heterozygous for a mutation in the 3′ IgH regulatory region (CXPR lymphomas) had a similar phenotype, including lack of surface Ig expression (unpublished data). As expected, we found little or no evidence of Xrcc4 deletion in sorted BM B220+IgM− cells or in sorted HSAhighCD21low peripheral immature B cells, but essentially complete Xrcc4 deletion in purified HSAlowCD21high mature splenic B cells (Fig. S3). By PCR, we occasionally observed very low levels of a product corresponding to Xrcc4 deletion in BM and immature B cell samples, which theoretically may reflect very low-level deletion in earlier B cell stages, but which also may simply reflect recirculating peripheral B cells in the BM and/or low-level contamination of the sorted populations (unpublished data). Along with the appearance of these B lineage tumors being dependent on Xrcc4 deletion via CD21cre expression at transitional B cell stage (Fig. 1 B and Fig. S3) (30), these phenotypic characterizations indicate that CXP lymphomas, despite lack of surface Ig expression, derive from peripheral B cells.
IgH, Igκ, and Igλ rearrangements in CXP tumors
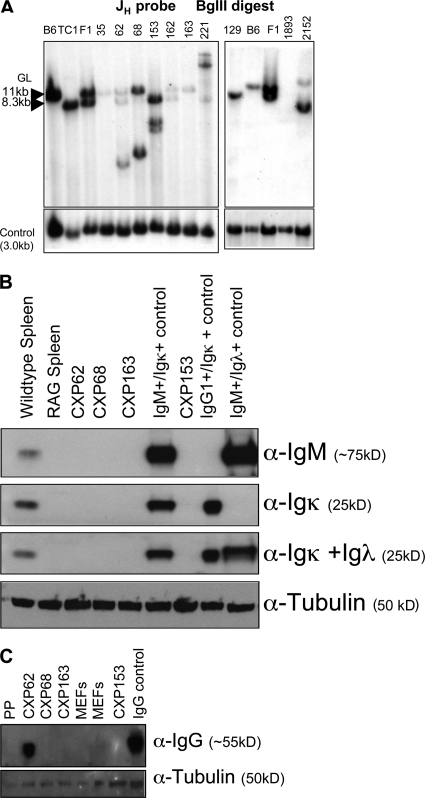
Recurrent sIg− peripheral lymphomas have not been observed previously. To elucidate the basis for this striking phenotype, we first assayed for expression of IgH chains via Western blotting of tumor extracts. Most analyzed CXP B lymphomas lacked readily detectable IgH chain expression, although CXP62 expressed cytoplasmic γ chains (Fig. 2, B and C). Both normal and C-NHEJ–deficient pro–B lines display rearrangements of both JH alleles (31). Assays of CXP tumor DNA for JH rearrangements via Southern blotting revealed that all primary CXP tumors showed JH rearrangements, along with varying degrees of a germline JH band (Fig. 2 A). However, the germline band usually occurred in low levels and was often further reduced in passaged tumors (unpublished data), indicating derivation from non–B lineage cells within the tumor. Despite lack of detectable IgH expression, molecular cloning demonstrated that CXP tumors contained structurally normal in-frame or out-of-frame V(D)J rearrangements, indicating that they occurred before Xrcc4 inactivation (see the following section). Finally, most CXP tumors had rearrangements in or around IgH S regions. At least five out of eight CXP tumors had clonal Sμ/Cμ rearrangements; three had one or more clonal Sα rearrangements of which one was molecularly cloned (from CXP68) and found to be an Sγ2a to Sα rearrangement; one CXP tumor had an Sγ2b rearrangement; and another had a clonal Sγ3 rearrangement in addition to the Sγ2a to Sα rearrangement (Table S1 and Fig. S4, A and B, available at http://www.jem.org/cgi/content/full/jem.20082271/DC1). Thus, most CXP tumors derived from peripheral B cells that had attempted IgH CSR.
Figure 2.
CXP lymphomas harbor clonal JH rearrangements, but do not express IgM or IgL chains. (A) Southern blot analyses of CXP lymphoma genomic DNA samples for rearrangement or deletion of JH loci (representative of at least two separate analyses for each sample). (B) Western blot analyses of CXP lymphoma cellular extracts for expression of μ heavy chain and κ or λ light chains using HRP-conjugated anti-IgM (μ heavy chain specific), anti-κ, or anti-λ antibodies, respectively. (C) Western blot analyses of CXP lymphoma cellular extracts for expression of γ heavy chains using HRP-conjugated anti–mouse IgG antibody. An α-tubulin antibody was used to generate a loading control in both panels. PP indicates extracts from Peyer's patches. Data presented in this figure shows results representative of two or three independent sets of analyses.
No analyzed CXP tumor expressed readily detectable Igκ and Igλ chains (Fig. 2 B and not depicted). Approximately 60% of normal B cells have one productively rearranged Igκ allele and one germline allele, and ∼40% have one productive and one nonproductively rearranged or deleted Jκ allele (38). We assayed for Jκ rearrangements by Southern blotting and found that all eight analyzed CXP tumors lacked both copies of the Jκ locus (Fig. 3 A and Fig. S5 A, available at http://www.jem.org/cgi/content/full/jem.20082271/DC1). Although there was a variable level of a germline-sized Jκ band, the level was far below that expected for a single germline allele and roughly correlated with that of contaminating nonlymphoid DNA as judged by the JH Southern blot (Fig. 2 A). Southern blotting revealed that CXP tumors also lacked both copies of the Cκ exons (Fig. 3 A). We tested for downstream RS rearrangement in CXP tumors via Southern blotting and found rearrangement and/or deletion of the 3′RS sequences in all analyzed tumors (Fig. 3 A and Fig. S5 A). As normal 3′RS rearrangements do not result in deletions larger than a few nucleotides (39), we tested the hypothesis that 3′RS rearrangements in CXP lymphomas had occurred after Xrcc4 inactivation by cloning and sequencing rearrangements. Of three RS rearrangements isolated, two had apparent Vκ and 3′RS region junctions accompanied by large deletions (30–80 bp into Vκ and 26–43 bp beyond 3′RS sequence), whereas the other had a normal Vκ to 3′RS join (Fig. S5 B). In addition, Southern blotting with a 3′RS probe showed CXP163 had an aberrant RS rearrangement with a large deletion (>4 kb) downstream of the 3′RS sequence (Fig. S5 C). Southern blotting also showed that a significant proportion of CXc/−Pc/c B lymphomas (four out of six; Fig. S2) and CXPR B lymphomas (five out of eight; not depicted) had deleted one or both Igκ alleles. We conclude that deletional Igκ 3′RS rearrangements in CXP tumors are reminiscent of those associated with attempted V(D)J joining in the absence of C-NHEJ, suggesting they occurred in progenitors of CXP tumors subsequent to Xrcc4 inactivation via CD21cre expression.
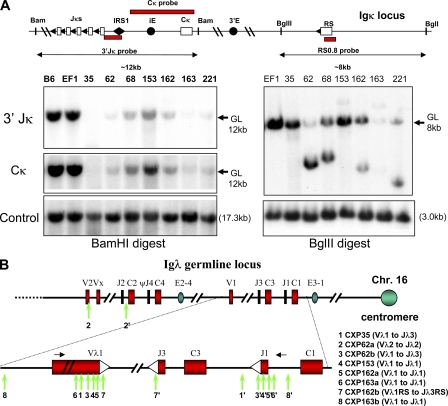
Figure 3.
Aberrant IgL rearrangements in CXP lymphomas. (A, right) Southern blot analyses for Jκ rearrangements in CXP lymphoma genomic DNA using BamHI digestion and the 3′ Jκ and Cκ probes indicated in schematic at top of panel. (left) Southern blot analysis for RS rearrangements in CXP lymphoma DNA using BglII digestion and the indicated RS0.8 probe. Germline (GL) bands and their known sizes are indicated (arrows). Blots were performed at least two times. (B) Assays for rearrangements occurring in or near Vλ and Jλ segments in DNA samples from CXP lymphomas. Rearrangements were isolated either by PCR or by cloning from λ-phage genomic DNA libraries generated from individual tumor DNA samples. Black arrows denote the position of the PCR primers used for the studies, and primer sequences are indicated in the Materials and methods section. The two breakpoints for a given rearrangement in the vicinity of Vλ and Jλ in a particular tumor are indicated by the same number with the breakpoint in the Jλ region indicated by a (′) symbol. PCR analyses were performed at least three times from one or multiple independent DNA samples from a given tumor. PCR fragments were subcloned into the pGEM vector, and 10–20 subclones were sequenced. Identical rearrangements that were isolated from >80% of the subclones were scored as clonal rearrangements. At least two independent phage clones were isolated from each genomic library and the junctions found in phage clones were independently confirmed as clonal junctions in a given tumor by PCR analyses of tumor cell DNA. Sequences of cloned Vλ and Jλ rearrangement junctions are shown in Fig. S5E.
Complete Igκ deletion is generally observed in a subset of mouse peripheral Igλ+ B cells (39). Therefore, we assayed CXP lymphoma DNA by Southern blotting with probes to detect VλJλ rearrangements within both Igλ clusters (Fig. S5 D). These analyses demonstrated that at least six out of nine assayed CXP or CXc/−Pc/c B cell lymphomas had clear-cut Igλ rearrangements, with CXP62, 162, and 163 showing rearrangements of both alleles (Fig. S5, A and D). Southern blotting analyses also showed that several analyzed CXPR B lymphomas had Igλ rearrangements (unpublished data). To elucidate the nature of the Igλ rearrangements in CXP tumors, we performed PCR using Vλ1/Vλ2- and Jλ1/Jλ2-specific primers to isolate six different rearrangements involving one or the other Jλ cluster (Fig. 3 B and Fig. S5 E). Nearly all isolated Igλ rearrangements involved joining of sequences in or upstream of a Vλ to sequences in or downstream of a Jλ, with the majority of breakpoints leading to deletions larger than the few basepair deletions observed from one or both segments in normal VλJλ joins (Fig. 3 B and Fig. S5 E) (40). As PCR only allows isolation of junctions within the primer boundaries, we cloned several Igλ rearrangements from genomic phage libraries generated from CXP163 and CXP162 DNA and identified a deletional rearrangement that went ∼2.8-kb and ∼800-bp, respectively, beyond the Vλ1 and Jλ1 segments, as well as aberrantly deleted RS rearrangements (Fig. 3 B and Fig. S5 E). Overall, the observed pattern of Igλ rearrangements in CXP lymphomas, namely large deletions from the participating segments on one or both sides of the join, is fully consistent with the types of junctions previously characterized for attempted V(D)J recombination of endogenous loci in C-NHEJ–deficient progenitor B or T lymphocytes (3, 41). Therefore, CXP tumors appear to arise from B cells that attempted Igλ rearrangement subsequent to Xrcc4 deletion via CD21cre expression.
C-NHEJ suppresses oncogenic CSR-related translocations between IgH and c-myc
Spectral karyotyping (SKY) revealed that six out of eight characterized CXP tumors (CXP68, 153, 162, 163, 221, and 2152) harbored clonal translocations between chromosomes 12 and 15, possibly involving IgH (Chr12) and c-myc (Chr15) loci (Fig. 4 A, Table S3, and Fig. S6 A, available at http://www.jem.org/cgi/content/full/jem.20082271/DC1). To test this possibility, we performed fluorescence in situ hybridization (FISH) with 5′ and 3′ IgH BAC probes plus a BAC probe containing c-myc (Fig. 4 A). These analyses demonstrated that 5 out of 6 tumors (CXP68, 153, 162, 163, and 2152) harbored reciprocal t(12;15) and t(15;12) translocations in which the centromeric portion (CH) of IgH (3′ IgH probe) was fused proximal to the telomeric portion of Chr15 in or around c-myc (c-myc BAC probe) to yield the t(12;15) translocation, and the telomeric (VH) portion of IgH (5′ IgH probe) was fused proximal to the centromeric portion of Chr15 in or just upstream of c-myc to yield the reciprocal t(15;12) translocation (Fig. 4 A and Fig. S6 A). CXP221 harbored a clonal t(12;15) translocation involving IgH and c-myc loc, but lacked the reciprocal t(15;12) translocation, and instead contained a clonal t(15;7) translocation (Fig. S6 A). Based on FISH, neither c-myc nor IgH appeared amplified in CXP tumors. Similar to CXP tumors, three out of four analyzed CXc/−Pc/c B lymphomas and six out of six CXPR tumors analyzed had translocations involving chromosomes 12 and 15 that involved the IgH and c-myc loci (Fig. S6 A, Table S4, and not depicted).
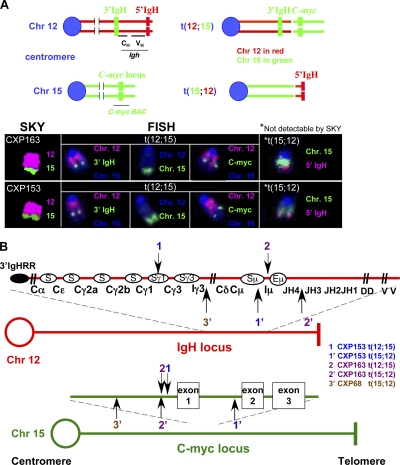
Figure 4.
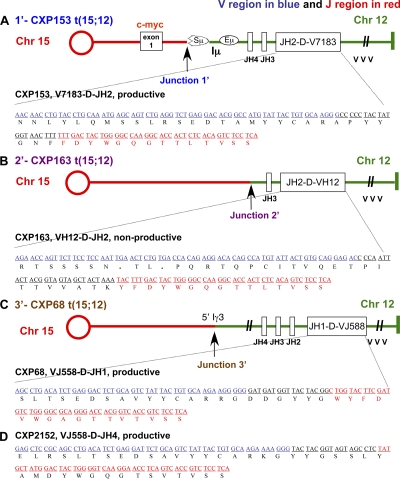
Reciprocal IgH/c-myc translocations in CXP lymphoma. (A) SKY, FISH, and chromosome paint analyses (Chr12 pink, Chr15 green) for IgH/c-myc translocations in CXP163 and CXP153. A 3′ IgH BAC (light green) and a c-myc BAC (light green) were used in combination with chromosome paints to detect t(12;15) translocations. The t(15;12) translocation, not detectable by SKY, was visualized using the 5′ IgH BAC (pink) and a Chr15 paint (green). At least 15 metaphase spreads were analyzed for SKY, FISH, and chromosomal paints, respectively, for each CXP tumor. Translocations shown were observed in the majority of metaphases, and were scored as clonal if observed in >80% of metaphases. (B) Schematic of IgH/c-myc translocation breakpoint junctions cloned and sequenced from λ-phage tumor genomic DNA libraries. At least two independent phage clones were isolated from each genomic DNA library, and breakpoint junctions identified within them were further independently confirmed by PCR and sequencing from the appropriate tumor DNA samples. The numbers on the top indicate t(12;15) translocation breakpoints and the same numbers containing a (′) on the bottom indicate the “reciprocal” t(15;12) translocation breakpoints in the corresponding tumor: CXP153 (junction 1 and 1′), 163 (junction 2 and 2′), and 68 (junction 3′).
To further analyze CXP B lymphoma t(12;15) and t(15;12) translocation junctions, we performed Southern blotting with probes within or just upstream of c-myc (Fig. S6 B). These analyses demonstrated that the six CXP lymphomas with t(12;15) translocations had clonal rearrangements in or around c-myc (Fig. S6 B and not depicted), with one additional tumor that lacks a t(12;15) translocation (CXP62; detailed in the following section), also showing a clonal c-myc rearrangement (Fig. S6 B). Southern blotting confirmed that none of the CXP tumors had amplified c-myc (Fig. S6 B). Northern blotting with a c-myc probe further demonstrated that all CXP tumors with t(12;15) translocations had greatly increased c-myc expression, indicating ectopic overexpression from a single copy of the translocated c-myc gene (Fig. S6 C). Of note, only two CXP tumors lacked t(12;15) translocations; however, one (CXP62) activated c-myc via an Igλ/c-myc translocation (see below) and the other (CXP35) had a complex Chr12 translocation that resulted in N-myc amplification and overexpression (Fig. S6 C and Fig. S7 A, available at http://www.jem.org/cgi/content/full/jem.20082271/DC1) reminiscent of that observed in Artemis/p53-deficient pro–B cell tumors (42). Thus, all CXP tumors overexpressed a myc family gene.
To further elucidate IgH/c-myc translocations in CXP tumors, we isolated translocation junctions by screening genomic DNA libraries from individual tumors. Nucleotide sequence analyses showed that all five characterized junctions corresponded to translocations between IgH and c-myc (Fig. 4 B and Fig. S6 D). All junctions on Chr12 (Fig. 4 B, red), including those associated with both t(12;15) and t(15;12) occurred within IgH, most in the CH region within or around S regions consistent with derivation via aberrant CSR. The CXP153 t(12;15) junction linked Sγ1 to sequences just 5′ of c-myc (junction 1, CXP153), whereas the t(15;12) linked Sμ to sequences in the first intron of c-myc (Fig. 4 B and Fig. S6 D). The CXP163 t(12;15) junction linked Iμ to a sequence 288 bp upstream of c-myc exon 1, and the t(15;12) linked sequences between JH3 and JH4 to sequences 289 bp upstream of c-myc exon 1 (Fig. 4 B and Fig. S6 D); notably, the CXP163 t(15;12) junction occurred downstream of a nonproductive VH(D)JH rearrangement (Fig. 5 B), indicating that it did not happen during V(D)J recombination and, rather, may have resulted in association with a large deletion from a breakpoint downstream in Iμ-Sμ region. Finally, the t(15;12) CXP68 junction linked sequences near Iγ3 to sequences 1.4-kb upstream of c-myc exon 1 (Fig. 4 B), again, likely resulting from attempted CSR to Sγ3. Four of these five cloned junctions contained junctional microhomology suggestive of A-EJ (Fig. S6 D). Overall, CXP IgH/c-myc translocations are remarkably similar to those of human sporadic Burkitt's lymphoma that fuse IgH S regions upstream of c-myc.
Figure 5.
CXP lymphomas harbor normal V(D)J rearrangements in IgH locus. Nucleotide sequence analysis of primary VH(D)JH rearrangements cloned from the following: CXP153 by both λ-phage library and PCR (A); CXP163 by λ-phage library (B); CXP68 by PCR (C); and CXP2152 by PCR (D). Additional sequence analyses revealed that CXP153 had a productive V7183-D-JH2 rearrangement juxtaposed into to the first intron of c-myc and CXP163 has a nonproductive VH12-D-JH2 rearrangement juxtaposed to the sequence upstream of c-myc exon1. V regions are shown in blue and J regions in red. The open reading frame is indicated under the DNA sequence. PCR analyses were performed at least three times from one or multiple independent tumor DNA samples. PCR fragments were subcloned into pGEM vector, 10–20 subclones were sequenced, and identical rearrangements observed in >80% clones were scored as clonal rearrangements.
Because of the location of the characterized translocation junctions downstream of the IgH JH region, we considered the possibility that many CXP tumors were sIg− because they separated a productive VH(D)JH exon from the CH region. In this context, two out of three analyzed CXP tumors with IgH/c-myc translocations (CXP68 and CXP153) contained an in-frame (productive) VH(D)JH rearrangement fused to the c-myc locus on Chr15; whereas the other (CXP163) involved a translocated nonproductive VH(D)JH rearrangement (Fig. 5, A–C). In CXP163, which has the nonproductive VH(D)JH translocated to c-myc, the other Chr12 was involved in a t(12;16) translocation that may have resulted in loss of the presumably productive VH(D)JH exon, as the breakpoint is centromeric to IgH (Fig. 7 A and Fig. S7 A). Of the four VH(D)JH junctions and downstream flanks sequenced (3,884 bp), one carried two somatic point mutations and the other three lacked mutations (unpublished data). Finally, cloned VH(D)JH rearrangements appeared normal in respect to junctional deletions (Fig. 5, A–D), indicating they occurred in C-NHEJ–proficient cells. Therefore, primary V(D)J recombination at the IgH locus was normal in B lineage cells that gave rise to CXP lymphomas.
CXP lymphomas frequently harbor Igλ translocations
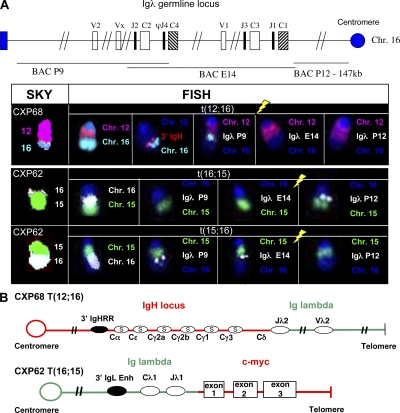
SKY analyses demonstrated that five out of eight characterized CXP B lymphomas (CXP35, 62, 68, 162, and 163), many of the same ones that harbored translocations between chromosomes 12 and 15, harbored Chr16 translocations to various other chromosomes with two others (CXP153 and 2152) harboring nonclonal Chr16 translocations (Fig. 6 A, Table S3, and Fig. S7 A). In contrast to the Chr12 translocations, all but one Chr16 translocation (i.e., CXP62) appeared nonreciprocal (Fig. 6 A and Fig. S7 A), similar to RAG-initiated oncogenic IgH translocations in Xrcc4/p53–deficient pro–B lymphomas (34). As the Igλ locus is on Chr16, we used FISH with three different BAC probes covering the Igλ locus to determine whether the Chr16 breakpoints involved Igλ (Fig. 6 A; P12 is centromeric to Igλ, E14 spans both Igλ clusters, P9 is telomeric to Igλ). These analyses revealed that all five Chr16 breakpoints occurred directly within various regions of Igλ (Fig. 6 A and Fig. S7 A). We also observed clonal translocations of Chr6 (which contains Igκ) in one of eight CXP tumors and in two of four characterized CXc/−Pc/c B lymphomas (Tables S3 and S4). Notably two tumors (CXP162 and CXc/−Pc/c 1893) harbored clonal t(6;16) translocations that fused Igλ to Chr6. However, cytogenetic analyses indicated that these t(6;16) translocations did not harbor Igκ (Fig. S7 A), although Igκ may still have been involved and deleted during joining (see the following section).
Figure 6.
Recurrent chromosome 16 translocations. (A) SKY/FISH analyses of Igλ translocations in CXP lymphomas. (A, top) schematic map of Igλ locus in germline configuration and the location of the three BACs used for FISH (P9, E14, and P12).(bottom) SKY revealed a t(12;16) translocation in CXP68, and a reciprocal t(16;15) and t(15;16) translocation in CXP62. FISH and chromosome paints (Chr12 pink, Chr16 light blue or white, Chr15 green) showed the breakpoints of Chr16 translocations occurred in different regions of the Igλ locus, indicated by yellow lightning symbols over metaphases hybridized with different Igλ BACs. 3′ IgH (pink) and P9 (light blue) BACs were colocalized at the junction of t(12;16) in CXP68. CXP62 has a reciprocal translocation with P12 BAC (white) on the der(16)t(16;15) and E14 and P9 (white) BACs on the der(15)t(15;16). At least 15 metaphase spreads were analyzed for each of the cytogenetic analyses, and representative clonal translocations observed in >80% of the metaphases are shown. (B, top) Cloning of t(12;16) breakpoints from a genomic library made from CXP68 DNA. Nucleotide sequence analyses of cloned junction (shown in Fig. S7 B) indicated breakpoints occurred between Cδ in IgH and Jλ2 in Igλ locus. (bottom) Cloning of t(16; 15) breakpoints from a genomic library made from CXP62 DNA. Nucleotide sequence analyses indicated breakpoints occurred between sequences upstream of c-myc exon 1 and Jλ1 with 3′ Igλ enhancer in close proximity. At least two independent phage clones were isolated from each genomic library and the junctions found in phage clones were independently confirmed as clonal junctions in a given tumor by PCR analyses of tumor DNA sample. Fig. S7 is available at http://www.jem.org/cgi/content/full/jem.20082271/DC1.
Characterization of several Igλ breakpoints by FISH and/or molecular cloning revealed partner loci and showed that sequences around translocation junctions lacked somatic hypermutation (SHM; Fig. S7 B and not depicted). By SKY, the CXP62 tumor, which was the only one that did not activate a myc-family gene by an IgH translocation, contained reciprocal t(16;15) and t(15;16) translocations, which juxtaposed the Igλ P12 BAC probe to the telomeric portion of Chr15 in the t(16;15) translocation, and the Igλ P9 and E14 BAC probes to the centromeric portion of Chr15 in the t(15;16) translocation (Fig. 6 A). Based on analyses of cloned junctions, the t(16;15) translocation fused sequences upstream of Jλ1 to sequences 5′ of c-myc exon 1 (Fig. 6 B and Fig. S7 B), which led to high level expression of c-myc transcripts (Fig. S6 C), whereas the t(15;16) translocation contained the reciprocal product (Fig. S7 B). FISH analyses of the complex t(12;12;16) translocation that led to N-myc amplification in CXP35 revealed that it involved both Igλ and IgH loci (Fig. S7 A). Likewise, FISH analyses of the nonreciprocal t(12;16) CXP68 translocation revealed colocalization of 3′ IgH and Igλ P9 BAC probes at the junction (Fig. 6 A). By phage cloning, we mapped the CXP68 breakpoint to Cδ in IgH and just 3′ of Jλ2 in Igλ (Fig. 6 B and Fig. S7 B).
Comparative genomic hybridization analyses of CXP tumors
We used comparative genomic hybridization (CGH) to assay for sequence copy number differences among tumor and control tissues samples. Among the four samples derived from the CXP or CXc/−Pc/c cohort, three had clearly lost one copy of either the entire Chr6 or a large region of Chr6, with the boundary of the loss occurring near or spanning Igκ (Fig. S8, available at http://www.jem.org/cgi/content/full/jem.20082271/DC1, and not depicted). Out of nine CXPR B lymphoma DNAs analyzed, seven again had a clear loss of one copy of the entire Chr6 or a large region that in two extended directly from Igκ (Fig. S8 and not depicted). CGH also showed focal deletion of Igκ in most analyzed CXP, CXc/−Pc/c, or CXPR tumors (Fig. S8 and not depicted), which is consistent with results of Southern blotting analyses. In contrast, three thymic lymphomas and two forebrain tumors, most likely of glial origin, analyzed did not show loss of Chr6 or a large region proximal to Igκ (Fig. S8 and not depicted).
DISCUSSION
Xrcc4 deficiency in peripheral B cells does not predispose to B cell lymphoma, despite dramatic IgH instability in Xrcc4-deficient B cells activated for CSR by treatment with αCD40 plus IL-4 (30). In this context, αCD40 plus IL-4–activated splenic CXP B cells had at most a very modest increase in IgH breaks compared with activated Xrcc4-deficient peripheral B cells (28 ± 13% versus 17 ± 7% of metaphases containing IgH breaks). Yet, CD21cre deletion of Xrcc4 in p53-deficient peripheral B cells, or deletion of both genes in peripheral B cells, recurrently led to sIg− B lymphomas that harbor activated myc, likely reflecting the critical role of p53 in eliminating B cells with activated c-myc expression and/or eliminating B cells with high genomic instability (33, 43, 44). CXP B lymphomas are remarkable in several ways. First, most CXP lymphomas arise from B cells that have attempted IgH CSR and a majority appear to arise from B cells that have undergone receptor editing, with both processes occurring after Xrcc4 inactivation by CD21Cre. Second, many CXP tumors harbored two separate, clonal translocations that, respectively, involved IgH and IgL loci. Occurrence of clonal IgH and IgL translocations in the same tumor may reflect dual CSR and editing activities of B cells from which they arose. In this context, some CXP lymphomas had clonal translocations that fused IgH and Igλ, clearly indicating simultaneous presence of breaks in both loci in tumor progenitors. As outlined below, these unusual features of CXP tumors have novel implications for B cell developmental processes and for oncogenic translocation mechanisms.
Germline Xrcc4 plus p53 deficiency in mice leads to pro–B lymphomas with RAG-dependent translocations that fuse JH sequences to sequences far downstream of c-myc, resulting in dicentric chromosomes and c-myc activation via breakage-fusion-bridge amplification (34). Thus, it is remarkable that inactivation of same two genes in mature B cells recurrently leads to ectopic activation of a single copy c-myc gene by fusion of IgH into its upstream region, analogous to similar IgH/c-myc translocations in human Burkitt's lymphoma and diffuse large B cell lymphomas (35). In pristane- or IL-6–induced mouse plasmacytomas, similar IgH/c-myc translocations occur and are initiated by AID (45, 46). In this context, CXP lymphoma IgH/c-myc translocations occur in and around IgH S regions and downstream of normal VH(D)JH exons, indicating their likely selection from the abundant pool of IgH translocations that result from aberrant CSR in activated Xrcc4-deficient peripheral B cells (30). As the intronic IgH enhancer is generally absent from CXP IgH/c-myc translocations, long-range 3′ IgH regulatory region activity (47), or that of unknown IgH regulatory elements, may ectopically activate c-myc. The recurrent differences in IgH/c-myc translocations in Xrcc4/p53-deficient pro–B versus CXP peripheral B cell tumors might reflect RAG versus AID-dependent DSB initiation in IgH and/or c-myc, other differential stage-specific mechanisms that lead to DSBs in and around c-myc, stage-specific differences in nuclear localization of IgH and c-myc loci, or mechanisms that differentially activate c-myc after translocation, including stage-specific IgH enhancer activities. As both Xrcc4/p53-deficient pro–B and CXP peripheral B cell lymphomas frequently have IgH/c-myc junctions that appear to be mediated by A-EJ, repair pathway choice is not likely to be a major factor in generating a particular form of translocation. Given the uniform and recurrent IgH/c-myc translocations, the CXP B lymphoma system should allow putative roles of cis elements and other factors to be tested.
CXP lymphomas also frequently harbor chromosome 16 translocations that involve aberrant V(D)J recombination of Igλ after Xrcc4 deletion. At least one CXP Igλ translocation was oncogenic. CXP62 had reciprocal t(15;16) and t(16;15) translocations, the latter of which fused the downstream portion of Igλ to sequences just upstream of c-myc, resulting in ectopic c-myc activation, reminiscent of Igλ/c-myc translocations observed in certain human B lymphomas (35). Characterization of additional CXP Igλ translocations may lead to identification of additional oncogenes. Notably, we also observe loss of all or part of one copy of chromosome 6 with some large deletions extending from the vicinity of Igκ. The latter findings suggest that rearrangements of Igκ, likely in association with aberrant editing, may also lead to translocations or chromosome loss. In the latter context, unrepaired DSBs can lead to chromosome loss in yeast (48). The frequent loss of all or a large portion of chromosome 6 in CXP tumors is striking and suggests a potential role in etiology of these tumors, perhaps by deletion of a tumor suppressor sequence.
More than 95% of mouse peripheral B cells express Igκ and, correspondingly, have VκJκ joins on one or both Igκ alleles and Igλ in germline configuration (9). A small percentage of mouse B cells express Igλ, and these have Igκ alleles that are nonproductively rearranged or deleted by 3′RS or related rearrangements (39, 49). Notably, Igλ-expressing mouse B cells frequently arise via receptor editing, often in the context of Igκ deletion via 3′RS rearrangement (39). Thus, it is striking that CXP lymphomas frequently had clonal Igκ deletions via aberrant Vκ to 3′RS rearrangements and aberrant Vλ to Jλ rearrangements, with both types of rearrangements occurring subsequent to CD21cre expression. Based on these findings, we speculate that a common CXP tumor progenitor derives from an Igκ-expressing B cell that attempted receptor editing subsequent to Xrcc4 deletion. Attempted editing in the absence of Xrcc4 would lead to aberrant rearrangement/deletion of Igκ and progression to Igλ rearrangement, which also would be aberrant. Thus, editing Xrcc4-deficient B cells would lose IgL expression caused by inability to undergo productive IgL editing.
As a working model, we suggest that aberrant receptor editing in CXP tumor progenitors generates an initial transforming event and/or activates aberrant CSR through loss of parts of chromosome 6 or translocations of Igλ. Aberrant CSR in the absence of Xrcc4 would lead to a high level of IgH translocations (30), including IgH/c-myc translocations that would complete the transformation process. In the case of the CXP tumor that harbors an Igλ/c-myc instead of an IgH/c-myc translocation, the former may have been sufficient to fully transform the cells. This scenario provides a potential role for chromosome 6 losses, and also could explain why all CXP Igλ translocations are not clearly oncogenic, why some CXP Igλ translocations have IgH as a partner, and why CXP IgH translocations can involve either productive or nonproductive IgH alleles.
Receptor editing is generally thought to occur in immature B cells in the BM (16); yet a substantial fraction of CXP tumor progenitors appear to have both breaks associated with attempted editing of the Igλ locus and breaks associated with attempted IgH CSR. In this regard, BM immature B cells in quasimonoclonal (QM) mice can express both AID and RAG1/2 (50), although other studies of developing B cells in WT mice concluded that AID is not expressed in BM B cells (51). QM mice harbor a productive IgH knock-in V(D)J allele and a homozygous gene-targeted deletion of both Jκ alleles and, thus, only have Igλ-producing B cells (52). The striking similarities between immature QM B cells and proposed CXP tumor progenitors suggest a potential relationship. In this regard, CXP tumor progenitors might derive from an undetected, low-level population of CD21cre-expressing BM immature B cells that delete Xrcc4 and undergo aberrant editing and CSR before migration to the periphery. However, we note that CXP tumors arise in the periphery, have features of peripheral B cells, and harbor clonal Igκ and Igλ rearrangements, as well as IgH CSR-associated rearrangements that occurred subsequent to CD21cre expression. Conceivably, the Igλ breaks that lead to translocations found in CXP tumor progenitors might reflect RAG-initiated breaks in BM developing B cells that persist because of p53 deficiency, as observed for RAG-initiated IgH breaks in ATM-deficient or 53BP1/p53–double-deficient pro–B cells (53, 54). However, we also observed dual IgH and Igλ translocations in a CXc/−Pc/c tumor in which both p53 and Xrcc4 were deleted peripherally. Therefore, we must consider the alternative possibility that CXP tumors arise from peripheral B cells driven into editing. Such cells might also participate in extrafollicular responses in which CSR occurs with limited SHM (55), a possibility consistent with most CXP tumor IgH variable region exons lacking SHM.
Transitional B cell populations express very low levels of RAG upon arrival in the periphery, with RAG expression being essentially undetectable in more mature splenic B cell populations (24, 25). Thus, if the RAG breaks in CXP tumor progenitors, indeed, are generated in the periphery, it remains to be determined in what cell type they occur and how apparently low-level RAG expression could generate them. Given that CXP progenitors nearly all arise from cells that harbor IgH/c-myc translocations, it is quite striking that CXP tumors do not arise from sIg+ B cells simply in the context of aberrant CSR. Indeed, the recurrent occurrence of sIg− lymphomas with aberrant Igκ and Igλ rearrangements and translocations indicates that inability to productively complete receptor editing confers a major predisposition to transformation of peripheral CXP B cells. Perhaps editing CXP B cells represent a larger fraction of the CD21-expressing B cell pool than anticipated or are enriched in certain locations. For example, receptor editing might especially contribute to antibody diversification in B cells of the gut-associated lymphoid tissue (e.g., mesenteric lymph nodes), where the CXP tumors appear to arise most frequently and in which lymphocytes are chronically activated and driven into immune responses by products of the gut microflora.
MATERIALS AND METHODS
Mice.
CD21cre/Xrcc4c/− mice were generated as previously described (30) and crossed into p53 germline-deficient mice (56) or mice carrying conditional p53 alleles (57). Animal work was approved by the Institutional Animal Care and Use Committee of Children's Hospital Boston.
Southern, Northern, and Western blot.
Genomic DNA was isolated from tumor masses or normal tissues from control mice, and Southern blotting was performed as previously described (58). 3′Jκ probe was a 1-kb XbaI–HindIII fragment from plasmid pSPIg8 containing the Jκ and Cκ region (59), or generated by PCR using the following primers: 3′ Jκ5, 5′-CATCCAAGAGATTGGATCGGAGAATAAGCA-3′; MA35, 5′-AACACTGGATAAAGCAGTTTATGCCCTTTC-3′; and plasmid pSPIg8 as template. Cκ probe was a 1.7-kb HindIII–BglII fragment covering the Cκ region. RS probes were previously described (60). RNA samples were extracted from tumor masses or normal tissues from control mice using TriPure Isolation Reagent (Roche). Northern blotting was performed following a standard protocol. Western blotting was performed using HRP-conjugated antibodies at 1:1,000 dilution (HRP goat anti–mouse IgM [μ chain–specific antibody], HRP goat anti–mouse λ, HRP goat anti–mouse κ, and HRP goat anti–mouse IgG [γ chain–specific antibody]; SouthernBiotech). Additional primers used for generating probes are detailed in the Supplemental materials and methods (available at http://www.jem.org/cgi/content/full/jem.20082271/DC1).
Phage and PCR cloning.
Tumor DNA samples were digested to completion with EcoRI, and fragments were cloned into either λZAP II vector (Stratagene) or λDASH II vector (Stratagene) according to the size of the fragments. Libraries were screened according to standard protocols (Stratagene) using Cλ1 or Jλ2 probe for λ rearrangements or translocations; c-myc probe A or c-myc probe D for c-myc translocations; Cα, Cγ1, and Sγ1 probes for S region rearrangement and translocations (see details for probes in the Supplemental materials and methods). Single plaques were purified, subcloned, and sequenced. In the case of λZAPII clones, the inserts were excised according to the protocol provided (Stratagene). Positive clones were verified by restriction analysis and hybridization. Sequencing of subcloned inserts was performed using T7 or T3 primers (DF-HCC DNA Resource Core). To obtain junctional sequences, internal primers were required for some tumors. For PCR cloning of Igλ rearrangements: Vlambda1int:5′-ttgtgactcaggaatctgca-3′ and Lamin 5′-ggagcagtctgaaatgagacaaagcat-3′ were used. Additional primers used for PCR cloning of junctions are detailed in Online supplemental material.
SKY and FISH analyses.
Preparation of metaphase chromosomes, SKY analyses, FISH, and whole-chromosome painting using single-chromosome–specific paints were performed as previously described (61). FISH probes were as follows: a BAC that covered the 3′ region of the IgH locus encompassing 3′ IgH enhancer and 100 kb downstream (3′ IgH BAC), a BAC just upstream of the IgH VH region (5′ IgH BAC) as described previously (62), and a BAC that contained the c-myc gene (RP23-307D14, c-myc BAC). BACs for Igλ regions are RP23-382P9, RP23-60E14, RP23-374P12, and BACs for Igκ regions are RP23-84F6 and RP23-64I9. All the BACs were obtained from BACPAC CHORI database, except for the IgH BACs.
Online supplemental material.
Fig. S1 shows the FACS, Northern blot, and RT-PCR data for assays used to characterize indicated surface protein and transcript expression by CXP lymphomas. Fig. S2 shows a Kaplan-Meier curve and summary of tumors that develop in CXc/−Pc/c lymphomas. Fig. S3 shows the evidence for specific Xrcc4 deletion via CD21cre. Fig. S4 shows assays for IgH S region rearrangements in CXP lymphomas. Fig. S5 shows Southern blot data for Igκ and Igλ rearrangements and sequences of aberrant VλJλ joins in CXP lymphomas. Fig. S6 shows various characterizations of translocations in CXP tumors, including representative cytogenetic data (SKY, FISH, and chromosome paints) for analyses that were used to characterize Igh/c-myc translocations, Southern blots for characterization of c-myc rearrangements, Northern blot assays for c-myc expression, and sequences of translocation breakpoints. Fig. S7 displays cytogenetic data (SKY, FISH, and chromosome paints) for Igλ translocations, and sequence data for translocation breakpoints from CXP lymphomas. Fig. S8 displays CGH array data for analysis of genetic amplifications and deletions in CXP lymphomas. Table S1 summarizes the phenotypes of CXP lymphomas. Table S2 summarizes the phenotypes of CXc/−Pc/c lymphomas. Table S3 summarizes the characterized clonal translocations in CXP lympomas. Table S4 summarizes the characterized clonal translocations in CXc/−Pc/c lymphomas. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20082271/DC1.
Acknowledgments
We thank Drs. L. Chin and Y. Xiao (The Belfer Cancer Genomics Center at Dana Farber Cancer Institute, Boston MA) for analysis of CGH data; Dr. S. Casola, P. Goff, and T. Hickernell for technical assistance and suggestions; Drs. J. Kutok and J. Aster (Brigham and Women's Hospital, Boston, MA) for histology analysis; and members of the Alt laboratory for discussions.
This work was supported by National Institutes of Health (NIH) grants 5P01 CA92625 (to F.W. Alt and K. Rajewsky), an NIH postdoctoral training grant NRSA T32 CA7008 (to C.T. Yan), Leukemia and Lymphoma Society (LLS) senior fellowships (to J.H. Wang and M. Gostissa), a LLS fellowship (to A. Datta). F.W. Alt is an investigator for the Howard Hughes Medical Institute.
The authors have no conflicting financial interests.
Abbreviations used: A-EJ, alternative end-joining; AID, activation-induced cytidine deaminase; C-NHEJ, classical NHEJ; CSR, class switch recombination; DSB, double-strand break; FISH, fluorescence in situ hybridization; NHEJ, nonhomologous end-joining; QM, quasimonoclonal; RS, recombination signal sequence; SHM, somatic hypermutation; SKY, spectral karyotyping.
J.H. Wang and C.T. Yan contributed equally to this paper.
References
- 1.Jung, D., C. Giallourakis, R. Mostoslavsky, and F.W. Alt. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24:541–570. [DOI] [PubMed] [Google Scholar]
- 2.Jung, D., and F.W. Alt. 2004. Unraveling V(D)J recombination; insights into gene regulation. Cell. 116:299–311. [DOI] [PubMed] [Google Scholar]
- 3.Rooney, S., J. Chaudhuri, and F.W. Alt. 2004. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 200:115–131. [DOI] [PubMed] [Google Scholar]
- 4.Li, Z., T. Otevrel, Y. Gao, H.L. Cheng, B. Seed, T.D. Stamato, G.E. Taccioli, and F.W. Alt. 1995. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 83:1079–1089. [DOI] [PubMed] [Google Scholar]
- 5.Grawunder, U., D. Zimmer, P. Kulesza, and M.R. Lieber. 1998. Requirement for an interaction of XRCC4 with DNA ligase IV for wild-type V(D)J recombination and DNA double-strand break repair in vivo. J. Biol. Chem. 273:24708–24714. [DOI] [PubMed] [Google Scholar]
- 6.Gao, Y., Y. Sun, K.M. Frank, P. Dikkes, Y. Fujiwara, K.J. Seidl, J.M. Sekiguchi, G.A. Rathbun, W. Swat, J. Wang, et al. 1998. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 95:891–902. [DOI] [PubMed] [Google Scholar]
- 7.Frank, K.M., J.M. Sekiguchi, K.J. Seidl, W. Swat, G.A. Rathbun, H.L. Cheng, L. Davidson, L. Kangaloo, and F.W. Alt. 1998. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 396:173–177. [DOI] [PubMed] [Google Scholar]
- 8.Bassing, C.H., W. Swat, and F.W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 109(Suppl):S45–S55. [DOI] [PubMed] [Google Scholar]
- 9.Gorman, J.R., and F.W. Alt. 1998. Regulation of immunoglobulin light chain isotype expression. Adv. Immunol. 69:113–181. [DOI] [PubMed] [Google Scholar]
- 10.Schlissel, M.S. 2004. Regulation of activation and recombination of the murine Igkappa locus. Immunol. Rev. 200:215–223. [DOI] [PubMed] [Google Scholar]
- 11.Vela, J.L., D. Ait-Azzouzene, B.H. Duong, T. Ota, and D. Nemazee. 2008. Rearrangement of mouse immunoglobulin kappa deleting element recombining sequence promotes immune tolerance and lambda B cell production. Immunity. 28:161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gay, D., T. Saunders, S. Camper, and M. Weigert. 1993. Receptor editing: an approach by autoreactive B cells to escape tolerance. J. Exp. Med. 177:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radic, M.Z., J. Erikson, S. Litwin, and M. Weigert. 1993. B lymphocytes may escape tolerance by revising their antigen receptors. J. Exp. Med. 177:1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiegs, S.L., D.M. Russell, and D. Nemazee. 1993. Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 177:1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelanda, R., S. Schwers, E. Sonoda, R.M. Torres, D. Nemazee, and K. Rajewsky. 1997. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 7:765–775. [DOI] [PubMed] [Google Scholar]
- 16.Nemazee, D. 2006. Receptor editing in lymphocyte development and central tolerance. Nat. Rev. Immunol. 6:728–740. [DOI] [PubMed] [Google Scholar]
- 17.Han, S., B. Zheng, D.G. Schatz, E. Spanopoulou, and G. Kelsoe. 1996. Neoteny in lymphocytes: Rag1 and Rag2 expression in germinal center B cells. Science. 274:2094–2097. [DOI] [PubMed] [Google Scholar]
- 18.Han, S., S.R. Dillon, B. Zheng, M. Shimoda, M.S. Schlissel, and G. Kelsoe. 1997. V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science. 278:301–305. [DOI] [PubMed] [Google Scholar]
- 19.Hikida, M., M. Mori, T. Takai, K. Tomochika, K. Hamatani, and H. Ohmori. 1996. Reexpression of RAG-1 and RAG-2 genes in activated mature mouse B cells. Science. 274:2092–2094. [DOI] [PubMed] [Google Scholar]
- 20.Papavasiliou, F., R. Casellas, H. Suh, X.F. Qin, E. Besmer, R. Pelanda, D. Nemazee, K. Rajewsky, and M.C. Nussenzweig. 1997. V(D)J recombination in mature B cells: a mechanism for altering antibody responses. Science. 278:298–301. [DOI] [PubMed] [Google Scholar]
- 21.Hertz, M., V. Kouskoff, T. Nakamura, and D. Nemazee. 1998. V(D)J recombinase induction in splenic B lymphocytes is inhibited by antigen-receptor signalling. Nature. 394:292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gartner, F., F.W. Alt, R.J. Monroe, and K.J. Seidl. 2000. Antigen-independent appearance of recombination activating gene (RAG)–positive bone marrow B cells in the spleens of immunized mice. J. Exp. Med. 192:1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagaoka, H., G. Gonzalez-Aseguinolaza, M. Tsuji, and M.C. Nussenzweig. 2000. Immunization and infection change the number of recombination activating gene (RAG)–expressing B cells in the periphery by altering immature lymphocyte production. J. Exp. Med. 191:2113–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monroe, R.J., K.J. Seidl, F. Gaertner, S. Han, F. Chen, J. Sekiguchi, J. Wang, R. Ferrini, L. Davidson, G. Kelsoe, and F.W. Alt. 1999. RAG2:GFP knockin mice reveal novel aspects of RAG2 expression in primary and peripheral lymphoid tissues. Immunity. 11:201–212. [DOI] [PubMed] [Google Scholar]
- 25.Yu, W., H. Nagaoka, M. Jankovic, Z. Misulovin, H. Suh, A. Rolink, F. Melchers, E. Meffre, and M.C. Nussenzweig. 1999. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature. 400:682–687. [DOI] [PubMed] [Google Scholar]
- 26.Igarashi, H., N. Kuwata, K. Kiyota, K. Sumita, T. Suda, S. Ono, S.R. Bauer, and N. Sakaguchi. 2001. Localization of recombination activating gene 1/green fluorescent protein (RAG1/GFP) expression in secondary lymphoid organs after immunization with T-dependent antigens in rag1/gfp knockin mice. Blood. 97:2680–2687. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhuri, J., U. Basu, A. Zarrin, C. Yan, S. Franco, T. Perlot, B. Vuong, J. Wang, R.T. Phan, A. Datta, et al. 2007. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 94:157–214. [DOI] [PubMed] [Google Scholar]
- 28.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 29.Di Noia, J.M., and M.S. Neuberger. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22. [DOI] [PubMed] [Google Scholar]
- 30.Yan, C.T., C. Boboila, E.K. Souza, S. Franco, T.R. Hickernell, M. Murphy, S. Gumaste, M. Geyer, A.A. Zarrin, J.P. Manis, et al. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 449:478–482. [DOI] [PubMed] [Google Scholar]
- 31.Gao, Y., D.O. Ferguson, W. Xie, J.P. Manis, J. Sekiguchi, K.M. Frank, J. Chaudhuri, J. Horner, R.A. DePinho, and F.W. Alt. 2000. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 404:897–900. [DOI] [PubMed] [Google Scholar]
- 32.Vogelstein, B., D. Lane, and A.J. Levine. 2000. Surfing the p53 network. Nature. 408:307–310. [DOI] [PubMed] [Google Scholar]
- 33.Mills, K.D., D.O. Ferguson, and F.W. Alt. 2003. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 194:77–95. [DOI] [PubMed] [Google Scholar]
- 34.Zhu, C., K.D. Mills, D.O. Ferguson, C. Lee, J. Manis, J. Fleming, Y. Gao, C.C. Morton, and F.W. Alt. 2002. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 109:811–821. [DOI] [PubMed] [Google Scholar]
- 35.Kuppers, R., and R. Dalla-Favera. 2001. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 20:5580–5594. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi, K., Y. Kozono, T.J. Waldschmidt, D. Berthiaume, R.J. Quigg, A. Baron, and V.M. Holers. 1997. Mouse complement receptors type 1 (CR1;CD35) and type 2 (CR2;CD21): expression on normal B cell subpopulations and decreased levels during the development of autoimmunity in MRL/lpr mice. J. Immunol. 159:1557–1569. [PubMed] [Google Scholar]
- 37.Kraus, M., M.B. Alimzhanov, N. Rajewsky, and K. Rajewsky. 2004. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 117:787–800. [DOI] [PubMed] [Google Scholar]
- 38.Mostoslavsky, R., F.W. Alt, and K. Rajewsky. 2004. The lingering enigma of the allelic exclusion mechanism. Cell. 118:539–544. [DOI] [PubMed] [Google Scholar]
- 39.Retter, M.W., and D. Nemazee. 1998. Receptor editing occurs frequently during normal B cell development. J. Exp. Med. 188:1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boudinot, P., A.M. Drapier, P.A. Cazenave, and P. Sanchez. 1994. Mechanistic and selective constraints act on the establishment of V lambda J lambda junctions in the B cell repertoire. J. Immunol. 152:2248–2255. [PubMed] [Google Scholar]
- 41.Malynn, B.A., T.K. Blackwell, G.M. Fulop, G.A. Rathbun, A.J. Furley, P. Ferrier, L.B. Heinke, R.A. Phillips, G.D. Yancopoulos, and F.W. Alt. 1988. The scid defect affects the final step of the immunoglobulin VDJ recombinase mechanism. Cell. 54:453–460. [DOI] [PubMed] [Google Scholar]
- 42.Rooney, S., J. Sekiguchi, S. Whitlow, M. Eckersdorff, J.P. Manis, C. Lee, D.O. Ferguson, and F.W. Alt. 2004. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc. Natl. Acad. Sci. USA. 101:2410–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsson, J.A., and J.L. Cleveland. 2003. Myc pathways provoking cell suicide and cancer. Oncogene. 22:9007–9021. [DOI] [PubMed] [Google Scholar]
- 44.Hemann, M.T., A. Bric, J. Teruya-Feldstein, A. Herbst, J.A. Nilsson, C. Cordon-Cardo, J.L. Cleveland, W.P. Tansey, and S.W. Lowe. 2005. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 436:807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramiro, A.R., M. Jankovic, T. Eisenreich, S. Difilippantonio, S. Chen-Kiang, M. Muramatsu, T. Honjo, A. Nussenzweig, and M.C. Nussenzweig. 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 118:431–438. [DOI] [PubMed] [Google Scholar]
- 46.Kovalchuk, A.L., W. duBois, E. Mushinski, N.E. McNeil, C. Hirt, C.F. Qi, Z. Li, S. Janz, T. Honjo, M. Muramatsu, et al. 2007. AID-deficient Bcl-xL transgenic mice develop delayed atypical plasma cell tumors with unusual Ig/Myc chromosomal rearrangements. J. Exp. Med. 204:2989–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boxer, L.M., and C.V. Dang. 2001. Translocations involving c-myc and c-myc function. Oncogene. 20:5595–5610. [DOI] [PubMed] [Google Scholar]
- 48.Kaye, J.A., J.A. Melo, S.K. Cheung, M.B. Vaze, J.E. Haber, and D.P. Toczyski. 2004. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 14:2096–2106. [DOI] [PubMed] [Google Scholar]
- 49.Nadel, B., P.A. Cazenave, and P. Sanchez. 1990. Murine lambda gene rearrangements: the stochastic model prevails over the ordered model. EMBO J. 9:435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mao, C., L. Jiang, M. Melo-Jorge, M. Puthenveetil, X. Zhang, M.C. Carroll, and T. Imanishi-Kari. 2004. T cell-independent somatic hypermutation in murine B cells with an immature phenotype. Immunity. 20:133–144. [DOI] [PubMed] [Google Scholar]
- 51.Crouch, E.E., Z. Li, M. Takizawa, S. Fichtner-Feigl, P. Gourzi, C. Montano, L. Feigenbaum, P. Wilson, S. Janz, F.N. Papavasiliou, and R. Casellas. 2007. Regulation of AID expression in the immune response. J. Exp. Med. 204:1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cascalho, M., A. Ma, S. Lee, L. Masat, and M. Wabl. 1996. A quasi-monoclonal mouse. Science. 272:1649–1652. [DOI] [PubMed] [Google Scholar]
- 53.Callen, E., M. Jankovic, S. Difilippantonio, J.A. Daniel, H.T. Chen, A. Celeste, M. Pellegrini, K. McBride, D. Wangsa, A.L. Bredemeyer, et al. 2007. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 130:63–75. [DOI] [PubMed] [Google Scholar]
- 54.Difilippantonio, S., E. Gapud, N. Wong, C.Y. Huang, G. Mahowald, H.T. Chen, M.J. Kruhlak, E. Callen, F. Livak, M.C. Nussenzweig, B.P. Sleckman, and A. Nussenzweig. 2008. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. [DOI] [PMC free article] [PubMed]
- 55.MacLennan, I.C., K.M. Toellner, A.F. Cunningham, K. Serre, D.M. Sze, E. Zuniga, M.C. Cook, and C.G. Vinuesa. 2003. Extrafollicular antibody responses. Immunol. Rev. 194:8–18. [DOI] [PubMed] [Google Scholar]
- 56.Donehower, L.A., M. Harvey, B.L. Slagle, M.J. McArthur, C.A. Montgomery Jr., J.S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 356:215–221. [DOI] [PubMed] [Google Scholar]
- 57.Jonkers, J., R. Meuwissen, H. van der Gulden, H. Peterse, M. van der Valk, and A. Berns. 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29:418–425. [DOI] [PubMed] [Google Scholar]
- 58.Frank, K.M., N.E. Sharpless, Y. Gao, J.M. Sekiguchi, D.O. Ferguson, C. Zhu, J.P. Manis, J. Horner, R.A. DePinho, and F.W. Alt. 2000. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell. 5:993–1002. [DOI] [PubMed] [Google Scholar]
- 59.Van Ness, B.G., M. Weigert, C. Coleclough, E.L. Mather, D.E. Kelley, and R.P. Perry. 1981. Transcription of the unrearranged mouse C kappa locus: sequence of the initiation region and comparison of activity with a rearranged V kappa-C kappa gene. Cell. 27:593–602. [DOI] [PubMed] [Google Scholar]
- 60.Moore, M.W., J. Durdik, D.M. Persiani, and E. Selsing. 1985. Deletions of kappa chain constant region genes in mouse lambda chain-producing B cells involve intrachromosomal DNA recombinations similar to V-J joining. Proc. Natl. Acad. Sci. USA. 82:6211–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharpless, N.E., D.O. Ferguson, R.C. O'Hagan, D.H. Castrillon, C. Lee, P.A. Farazi, S. Alson, J. Fleming, C.C. Morton, K. Frank, et al. 2001. Impaired nonhomologous end-joining provokes soft tissue sarcomas harboring chromosomal translocations, amplifications, and deletions. Mol. Cell. 8:1187–1196. [DOI] [PubMed] [Google Scholar]
- 62.Franco, S., M. Gostissa, S. Zha, D.B. Lombard, M.M. Murphy, A.A. Zarrin, C. Yan, S. Tepsuporn, J.C. Morales, M.M. Adams, et al. 2006. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 21:201–214. [DOI] [PubMed] [Google Scholar]