Abstract
Natural killer (NK) cells and CD8 T cells require adhesion molecules for migration, activation, expansion, differentiation, and effector functions. DNAX accessory molecule 1 (DNAM-1), an adhesion molecule belonging to the immunoglobulin superfamily, promotes many of these functions in vitro. However, because NK cells and CD8 T cells express multiple adhesion molecules, it is unclear whether DNAM-1 has a unique function or is effectively redundant in vivo. To address this question, we generated mice lacking DNAM-1 and evaluated DNAM-1–deficient CD8 T cell and NK cell function in vitro and in vivo. Our results demonstrate that CD8 T cells require DNAM-1 for co-stimulation when recognizing antigen presented by nonprofessional antigen-presenting cells; in contrast, DNAM-1 is dispensable when dendritic cells present the antigen. Similarly, NK cells require DNAM-1 for the elimination of tumor cells that are comparatively resistant to NK cell–mediated cytotoxicity caused by the paucity of other NK cell–activating ligands. We conclude that DNAM-1 serves to extend the range of target cells that can activate CD8 T cell and NK cells and, hence, may be essential for immunosurveillance against tumors and/or viruses that evade recognition by other activating or accessory molecules.
Cytotoxic lymphocytes, such as NK cells and CD8 T cells, require adhesion molecules to migrate into sites of infection and the tumor microenvironment and establish tight contact with virally infected and tumor cells (1). Moreover, in secondary lymphoid organs, adhesion molecules facilitate CD8 T cell and NK cell interactions with professional APCs, which induce their activation, expansion, and differentiation. Adhesion molecules act directly as signaling molecules, initiating intracellular pathways that trigger activation and adhesiveness of cytotoxic lymphocytes. They also act as accessory molecules, sustaining cellular contacts that are necessary for the TCRs and NK cell receptors to engage their cognate ligands and deliver intracellular signals.
CD8 T cells and NK cells express multiple adhesion molecules. Although several studies have addressed the importance of integrins (1), attention has recently focused on a novel Ig superfamily adhesion molecule called DNAX accessory molecule 1 (DNAM-1; also known as CD226). In humans, DNAM-1 is normally expressed on NK cells and CD8 T cells, as well as CD4 T cells and monocytes (2). In mice, ∼40–50% of NK cells and all CD8 T cells constitutively express DNAM-1, whereas CD4 T cells express DNAM-1 preferentially upon activation (3). DNAM-1 binds to CD155, also known as Necl-5 or poliovirus receptor (4, 5). CD155 is expressed on epithelial cells, endothelial cells, and APCs. DNAM-1 also binds CD112 (Nectin-2), which is found on epithelial cells (4, 5). In vitro studies have shown that DNAM-1 triggers NK cell–mediated killing of tumor cells expressing CD155 and CD112 (4, 6–10). DNAM-1 also facilitates NK cell interaction with subsets of DCs that express CD155 (11). Moreover, DNAM-1 promotes co-stimulation of CD4 and CD8 T cells (3, 12, 13) and mediates adhesion of monocytes to endothelial cells facilitating transendothelial migration (14). DNAM-1 physically associates with the integrin lymphocyte function-associated antigen 1 (αLβ2) and modulates its activation and capacity to bind intercellular adhesion molecule 1 (2, 15). Mechanistically, DNAM-1 recruits actin-binding proteins such as Discs-large and 4.1G, which rearrange actin cytoskeleton, promoting clustering of lymphocyte function-associated antigen 1 and formation of an adhesive complex that polarizes lymphocytes toward other cells (16).
In summary, DNAM-1 appears to contribute to multiple innate and adaptive responses, which include (a) leukocyte migration; (b) activation, expansion, and differentiation of CD8 T cells and CD4 T cells; and (c) effector responses of T cells and NK cells. However, because cytotoxic lymphocytes express multiple adhesion molecules, it is difficult to determine whether DNAM-1 plays a unique role in these functions or is redundant. To address this question, we generated mice lacking expression of DNAM-1 and evaluated DNAM-1–deficient CD8 T cell and NK cell function in vitro and in vivo. Our results demonstrate that CD8 T cells require co-stimulation by DNAM-1 when the antigen is presented by nonprofessional APCs, whereas DNAM-1 is dispensable in the presence of DCs. Thus, DNAM-1 can promote CD8 T cell responses to tumors expressing CD155 and/or CD112. In an analogous fashion, NK cells require DNAM-1 for elimination of tumor cells that are relatively resistant to NK cell–mediated cytotoxicity caused by the absence of other NK cell–activating ligands. In brief, DNAM-1 broadens the range of target cells that can activate CD8 T cells and NK cells and, hence, it may be critical for immunosurveillance of tumors and/or viruses that escape recognition by other activating or accessory molecules.
RESULTS AND DISCUSSION
Normal development of DNAM-1–deficient mice
To generate DNAM-1−/− mice, we designed a targeting vector to replace exons two and three, encoding the start site and the first 129 amino acids of DNAM-1 with an MC1-neor gene flanked by loxP sites (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20081752/DC1). One targeted E14.1 (129P2/OlaHsd) embryonic stem cell was injected into C57BL/6 blastocysts, and resulting chimeras were bred to C57BL/6 mice expressing a Cre transgene under the CMV promoter to delete the MC1-neor gene. The DNAM-1 deletion was then backcrossed onto a C57BL/6 background. Homozygote DNAM-1−/− mice were obtained at expected frequencies from intercrosses of heterozygote mice. DNAM-1−/− mice were phenotypically normal and healthy, and developed no gross abnormalities up to 1 yr of age. Flow cytometric analysis of DNAM-1−/− mice did not reveal major differences in the lymphoid cell populations (NK cells, CD8 T cells, CD4 T cells, DCs, plasmacytoid DCs, and B cells) in the spleen, thymus, peripheral blood, and lymph nodes or in bone marrow precursor populations (Figs. S1–S3; and not depicted). Therefore, DNAM-1 appears to be dispensable for development, including that of the immune system. Additionally, lack of DNAM-1 did not result in any compensatory change in the expression of CD96, the alternative lymphocyte receptor for CD155 (Fig. S1) (7, 17).
DNAM-1 is essential for CD8 T cell responses to nonprofessional APCs
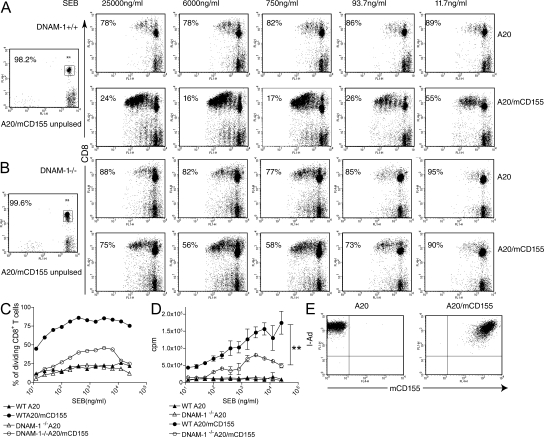
To assess the co-stimulatory function of DNAM-1 on CD8 T cells, we isolated T cells from WT and DNAM-1−/− mice, labeled them with CFSE, and stimulated them in vitro with APCs pulsed with graded doses of staphylococcal enterotoxin B (SEB) superantigen. As APCs, we used the B cell line A20, which expresses MHC class II, or A20 cells transfected with murine CD155 (mCD155; Fig. 1 E). After 72 h, we harvested cells and evaluated CD8 T cell division. The percentage of dividing WT CD8 T cells was significantly higher when T cells were stimulated with A20-mCD155 rather than A20 parental cells (Fig. 1, A and C). In contrast, CD155 had a relatively minor effect on the proliferation of DNAM-1−/− CD8 T cells; the percentage of dividing DNAM-1−/− CD8 T cells was only slightly higher when SEB was presented by A20-mCD155 cells (Fig. 1, B and C). Similar results were obtained when the CD8 T cell proliferative response to SEB was measured by thymidine incorporation (Fig. 1 D). These data demonstrate a clear role of DNAM-1 in priming and expansion of CD8 T cells in response to a superantigen presented by B cells.
Figure 1.
DNAM-1 augments CD8 T cell division in response to superantigen. (A and B) FACS analysis of CFSE dilution 72 h after (A) WT or (B) DNAM-1−/− T cells were co-cultured with A20 cells or A20 cells expressing mCD155 in the presence of the indicated concentrations of SEB. The percentage of undivided CD8 T cells is indicated. (left) Two control panels show the CFSE content of T cells incubated with unpulsed APCs. (C) Percentage of dividing CD8 T cells stimulated by A20 or A20-mCD155 at graded concentrations of SEB. (D) Proliferation of WT or DNAM-1−/− T cells stimulated by A20 or A20-mCD155 in the presence of graded concentrations of SEB measured by [3H]thymidine incorporation. Results are means ± SEM. **, P < 0.01. (E) Expression of MHC class II and mCD155 on control and CD155-transfected A20 B cell lymphoma.
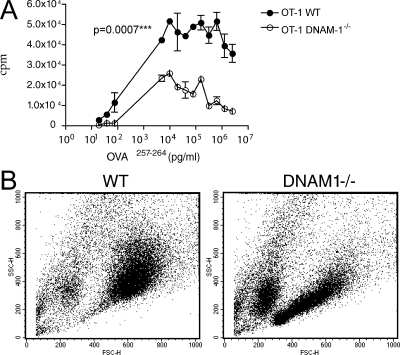
To evaluate the impact of DNAM-1 on CD8 T cell responses to antigens, we crossed OT-1 transgenic mice to DNAM-1−/− mice. We then evaluated the proliferation of purified WT and DNAM-1−/− CD8 transgenic T cells stimulated with the T cell lymphoma cell line EL-4 transfected with mCD155 and pulsed with OVA peptide 257–264 (Fig. 2 A). In this setting, OT-1 CD8 T cells lacking DNAM-1 proliferated less efficiently than WT OT-1 cells and also failed to become activated, as demonstrated by a lack of increase in size and granule content detected in forward scatter (FSC)/side scatter (SSC) profiles (Fig. 2 B). Collectively, these data firmly establish that DNAM-1 provides an important co-stimulatory signal for CD8 T cell proliferation when T cells are stimulated by nonprofessional APCs.
Figure 2.
DNAM-1 promotes CD8 T cell proliferation in response to antigen presented by nonprofessional APCs. DNAM-1−/− OT-1 transgenic CD8 T cells proliferate less efficiently than WT OT-1 cells in response to SIINFEKL peptide presented by EL-4 T lymphoma cells expressing mCD155. Proliferation was measured (A) by [3H]thymidine incorporation or (B) by evaluating the increase in SSC and FSC by flow cytometry in blasting cells. Dot plot analysis shows stimulation induced by 10 ng/ml SIINFEKL. One representative experiment out of three is shown. Results in A are means ± SEM.
DNAM-1 co-stimulation is not essential for CD8 T cell stimulation by professional APCs
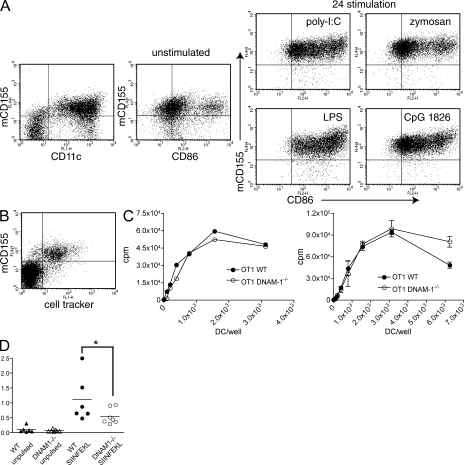
We then asked whether DNAM-1–CD155 interaction contributes to the priming and expansion of CD8 T cells by professional APCs. To evaluate this possibility, we first determined whether CD155 is expressed on mouse DCs, as has been shown for human monocyte-derived DCs (11). Consequently, we established a mCD155-specific antibody (Fig. S1) and stained immature and activated bone marrow–derived DCs (BMDCs). BMDCs constitutively expressed mCD155 and the expression level was further augmented by stimulation with Toll-like receptor agonists, including LPS, poly I:C, zymosan, and CpG oligonucleotides (Fig. 3 A). To determine whether mCD155 is expressed on primary DCs involved in antigen presentation, we applied a mixture of a green fluorescent cell tracker and a mild irritant to the ears of mice. We extracted draining cervical lymph nodes (CLNs) 48 h later and examined DC by flow cytometry. mCD155 was selectively expressed on cell tracker+ DCs that had migrated from the skin (Fig. 3 B). DCs that migrate to the CLNs at this time point are mainly dermal DCs, whereas Langerhans cells reach the CLNs at later time points (18). Thus, mCD155 is expressed on dermal DCs that migrate into the lymph nodes. In contrast, we were unable to detect mCD155 expression on primary spleen mouse DCs (unpublished data). We conclude that mCD155 is expressed on primary DCs that migrate from the periphery into the lymph nodes and, therefore, may potentially contribute to DC-mediated priming of T cell responses.
Figure 3.
DC-induced CD8 T cell stimulation is DNAM-1 independent. (A) BMDCs express of CD155, which is up-regulated by Toll-like receptor–induced DC maturation. (B) Expression of CD155 on CD11c+ DCs migrating from the periphery to CLNs 48 h after ear painting. A gate was applied on SSC/FSC to exclude small lymphocytes. (C) Proliferation of WT and DNAM-1−/− OT-1 in response to immature BMDCs pulsed with OVA for 18 h (left) or with OVA for 3 h followed by stimulation with CpG ODN1826 for 7 h (right). Proliferation was evaluated by [3H]thymidine incorporation. Results are means ± SEM. (D) Percentages of CD8 T cells producing IFN-γ after in vitro restimulation with EL-4–CD155 cells either pulsed or not pulsed with OVA peptide 257–264. Mice (six per group) were tested 8 d after footpad immunization. Horizontal bars represent means. *, P < 0.05.
To evaluate this possibility, we stimulated WT and DNAM-1−/− CD8 OT-1 cells with BMDCs that had been pulsed with OVA peptide 257–264. We used immature BMDCs and mature BMDCs activated with a CpG oligonucleotide. In contrast to our results obtained using A20 and EL-4 as APCs, lack of DNAM-1 had no impact on the capacity of OT-1 T cells to proliferate in response to DCs (Fig. 3 C). We conclude that co-stimulation by DNAM-1 is not required when T cells are stimulated by professional APCs, at least in vitro. To assess the impact of DNAM-1 deficiency on the generation of antigen-specific T cells in vivo, we immunized WT and DNAM-1−/− mice in the footpad with OVA peptide 257–264. After 1 wk, we removed draining lymph nodes and restimulated all cells in vitro with either BMDCs or EL-4–mCD155 pulsed with OVA peptide 257–264. The frequency of OVA-specific CD8 T cells was determined by intracellular staining for IFN-γ. No significant differences in the frequency of IFN-γ–secreting T cells was observed after restimulation of DNAM-1−/− and WT lymph nodes with DCs (unpublished data), indicating that DNAM-1 deficiency had no marked impact on CD8 T cell priming. In contrast, fewer IFN-γ–secreting T cells were evident in lymph nodes from the DNAM-1−/− mice after restimulation with EL-4–mCD155 (Fig. 3 D), corroborating a role for DNAM-1 in promoting CD8 T cell activation and function in response to stimulation by nonprofessional APCs.
NK cells require DNAM-1 to eliminate targets resistant to cytotoxicity
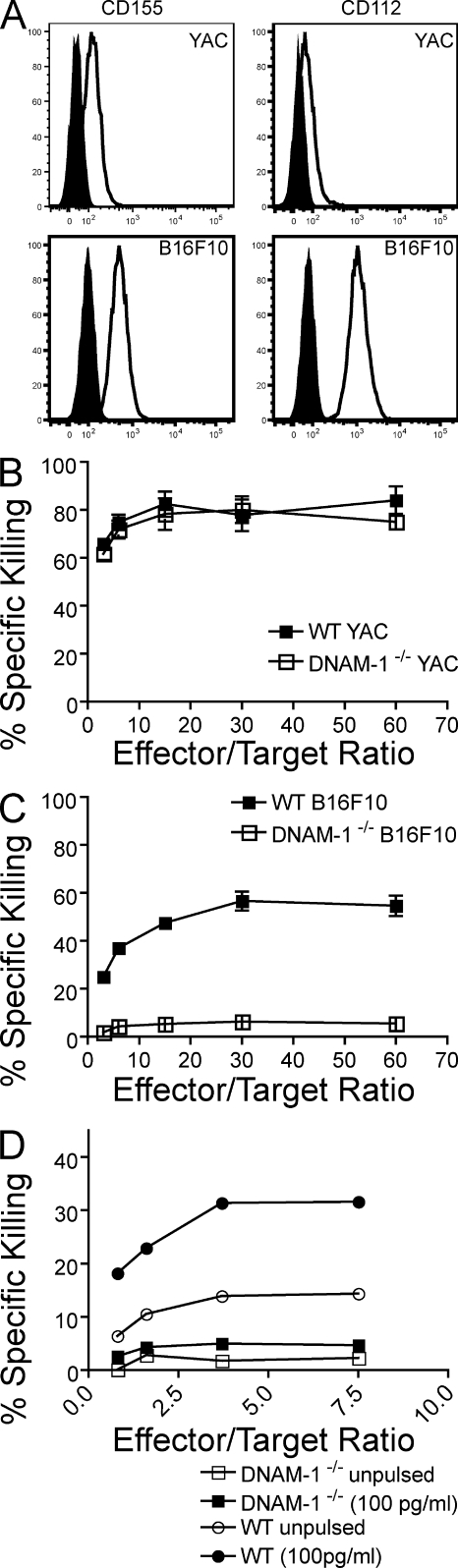
Because CD155 is expressed on many tumor cells, DNAM-1 has been implicated as a major trigger for NK cell antitumor activity (4, 6–10). However, NK cells express additional receptors that mediate antitumor activity. NKG2D triggers NK cell surveillance against tumor cells expressing stress-induced MHC class I–related ligands such as MIC-A, MIC-B, and UL16-binding protein in humans, and Rae1, H60, and Mult1 in mice (19–21). Natural cytotoxicity receptors, such as NKp46, NKp44, and NKp30, have also been shown to mediate an important role in antitumor activity (22). The relative contribution of DNAM-1 versus other receptors has not been addressed in vivo. We first examined the cytotoxic ability of purified DNAM-1−/− and WT NK cells cultured with IL-2 to kill target cells in vitro. As target cells, we chose YAC-1 and B16F10. YAC-1 expresses the ligands for NKG2D, whereas B16F10 does not (23, 24); both targets express ligands for DNAM-1 (Fig. 4 A). We found that IL-2–cultured NK cells efficiently killed YAC-1 cells whether or not they expressed DNAM-1 (Fig. 4 B). In contrast, although WT NK cells were capable of killing B16F10 tumors, DNAM-1−/− NK cells did not appreciably kill this tumor target (Fig. 4 C). Interestingly, DNAM-1−/− OT-1 CD8 T cells killed B16 cells pulsed with OVA peptide 257–264 less effectively than WT OT-1 CD8 T cells (Fig. 4 D), indicating that DNAM-1 may contribute to the cytotoxic function of both innate and adaptive lymphocytes.
Figure 4.
Lack of DNAM-1 impairs NK cell and CD8 T cell cytotoxicity. (A) Differential expression of DNAM-1 ligands on B16F10 and YAC cells. B16F10 and YAC-1 cells were analyzed for the expression of CD155 and CD112 by flow cytometry. Open histograms represent staining with the respective antibody, whereas shaded histograms show isotype staining. These data represent three independent experiments. (B and C) Lack of DNAM-1 affects NK cell–mediated death of tumor cells lacking NKG2D ligands. IL-2–activated purified NK cells were tested for cytotoxic activity against (B) YAC and (C) B16F10 targets in a standard 4-h 51Cr release assay. Each data point is the mean ± SEM of two independent experiments done in triplicate for each effector/target ratio. (D) Reduced killing of B16 cells pulsed with OVA peptide by DNAM-1−/− CD8 T cells. Splenocytes from OT-1 transgenic mice were stimulated with OVA peptide 257–264 and cultured in vitro for 15 d in the presence of 20 U/ml IL-2. Antigen-specific cytotoxicity was assessed using B16 cells, either pulsed or not pulsed with 100 pg/ml OVA peptide 257–264. Naive WT OT-1 CD8+ T cells did not kill peptide-pulsed target cells. The data represent two independent experiments.
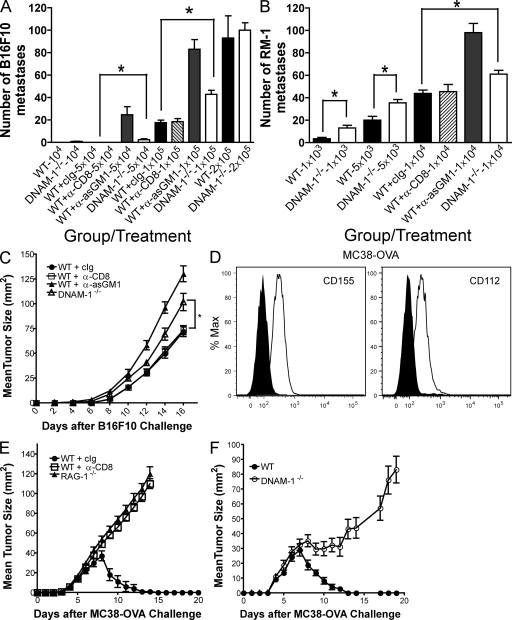
To determine whether DNAM-1 contributes to NK cell–mediated surveillance against tumors in vivo, we initially chose to examine the B16F10 experimental metastases model. Tumor cells were injected i.v. and lung metastases were counted 2 wk after inoculation. At lower tumor doses (5 × 104 and 105 cells), DNAM-1−/− mice had significantly higher numbers of B16F10 lung metastases than did WT mice (Fig. 5 A). At the highest dose of B16F10 (2 × 105 cells), DNAM-1 deficiency had no effect, suggesting that the host response was completely overwhelmed at this dose. The rejection of B16F10 was entirely NK cell dependent, as illustrated by the lack of rejection of metastases after depletion of NK cells by the anti-asGM1 antibody (Fig. 5 A). Interestingly, the augmented tumor growth caused by DNAM-1 deficiency was not as severe as that caused by NK cell depletion, indicating that DNAM-1–CD155 interaction is not the only mechanism by which NK cells mediate rejection of B16F10. Accordingly, we observed that only a subset of ∼50% of NK cells expresses DNAM-1 (Fig. S1). Similar results were obtained in another experimental metastases model based on the injection of the mouse prostate carcinoma, RM-1, that also expresses mCD155 (unpublished data). At all tumor doses inoculated (103, 5 × 103, and 104 cells), DNAM-1−/− mice had significantly higher numbers of RM-1 lung metastases than WT mice (Fig. 5 B), although, again, mice depleted of NK cells had the highest numbers of metastases. Consistent with the metastases data, accelerated tumor growth was also observed after s.c. injection of B16F10 in DNAM-1−/− mice as compared with WT mice (Fig. 5 C). These results corroborate the increased susceptibility of DNAM-1–deficient mice to tumors because of the reduced function of NK cells.
Figure 5.
Reduced NK cell– and CD8 T cell–mediated suppression of tumors in DNAM-1−/− mice. (A and B) B6 WT and DNAM-1−/− mice were challenged i.v. with various doses (as indicated) of either (A) B16F10 or (B) RM-1, as described in Materials and methods. Lungs were harvested on day 14 and fixed in Bouin's solution, and tumor colonies were counted under a dissecting microscope. Data are depicted as mean tumor colonies ± SEM (n = 5–14 mice for each dose/group). Statistical differences between WT and DNAM-1−/− mice are depicted by asterisks (P < 0.05 using the Mann-Whitney test). (C) WT and DNAM-1−/− mice were injected with 5 × 104 B16F10 tumor cells s.c., and tumor growth was monitored at least every second day using a caliper square to determine the product of two perpendicular tumor diameters. Some groups of WT mice were depleted of CD8 T cells or NK cells, as described in Materials and methods. Results were recorded as the means ± SEM of five mice and are representative of two to four independent experiments. Statistical differences between WT + cIg and DNAM-1−/− mice are depicted by an asterisk (P < 0.05 using the Mann-Whitney test). (D) Expression of CD155 and CD112 on MC38-OVA cells as assessed by flow cytometry. Open histograms represent staining with the respective antibody, whereas shaded histograms show isotype staining. (E) WT or RAG1−/− mice were injected with 106 MC38-OVA tumor cells s.c., and tumor growth was monitored as described in C. Some groups of WT mice were depleted of CD8 T cells. (F) WT and DNAM-1−/− mice were injected with 106 MC38-OVA tumor cells s.c., and tumor growth was monitored as described in C. Results were recorded as the means ± SEM of five (E) or nine (F) mice.
DNAM-1 promotes CD8 T cell–mediated tumor rejection
To determine whether DNAM-1 contributes to CD8 T cell–mediated tumor immunosurveillance, we used a tumor model based on inoculation of a colon carcinoma cell line, MC38, that has been transfected with OVA (MC38-OVA). We chose MC38-OVA because this tumor cell line expresses ligands for DNAM-1, CD112, and CD155 (Fig. 5 D). In addition, our initial studies demonstrated that s.c. inoculation of MC38-OVA induces marked tumor growth in WT mice depleted of CD8 T cells and RAG1−/− mice in comparison to control Ig–treated WT mice (Fig. 5 E), which indicates that CD8 T cells are critical for MC38-OVA tumor rejection. MC38-OVA grew similarly for the first 5 d after injection into DNAM-1−/− and WT mice. Subsequently, however, DNAM-1−/− mice failed to reject the tumor as compared with WT control (Fig. 5 F). We conclude that DNAM-1 contributes to CD8 T cell–mediated tumor immunosurveillance, most likely by co-stimulating CD8 T cells.
Concluding remarks
This report is the first describing mice specifically lacking DNAM-1 and definitively establishes an in vivo role for DNAM-1 during CD8 T cell and NK cell responses. Our results demonstrate that CD8 T cells require DNAM-1 for efficient stimulation by APCs that are weakly immunogenic, whereas a lack of DNAM-1 has no detectable impact on CD8 T cell stimulation by DCs. Thus, although DNAM-1 accessory function may be redundant in secondary lymphoid organs, it may be essential in peripheral tissues to promote optimal activation and effector functions of CD8 T cells that recognize their cognate MHC-peptide antigen on cells with limited expression of co-stimulatory ligands, such as tumor cells. Intraepithelial lymphocytes, γδ T cells, and lamina propria lymphocytes may also depend on DNAM-1 for co-stimulatory signals from epithelial cells expressing CD155. Intraepithelial and γδ T cells have been shown to rely on NKG2D-mediated co-stimulation and are in some cases directly activated by NKG2D ligands on tumor cells (25–27). Given this, DNAM-1 may either augment or, in some cases, compensate for NKG2D.
As indicated by our results with the B16F10 melanoma, expression of DNAM-1 enables NK cells to more efficiently kill tumor cells that are comparatively resistant to NK cell–mediated death because they lack NKG2D ligands. In contrast, DNAM-1 is dispensable for NK cell–mediated killing of highly susceptible targets that express NKG2D ligands, like YAC-1. Overall, our data suggest that DNAM-1 may play an important role in immunosurveillance against tumors and perhaps viral infections that are weakly immunogenic and refractory to NK cell–mediated cytotoxicity. Moreover, we have shown that DNAM-1 significantly contributes to CD8 T cell–mediated surveillance against tumor cells expressing CD112 and/or CD155. We and others have shown that T cells express at least one additional receptor for CD155, CD96, which has distinct functional characteristics (7, 17). Although it is possible that the lack of DNAM-1 is at least in part compensated by CD96, future studies will be necessary to determine the relative contributions of these two receptors to T cell and NK cell stimulation.
Genome-wide analysis of type I diabetes has recently revealed robust association of type I diabetes with a nonsynonymous single nucleotide polymorphism within the DNAM-1 gene (28). This observation, together with the observed expression of both DNAM-1 and CD155 in thymocytes (Fig. S3), suggests that DNAM-1–CD155 interactions may regulate the generation of autoreactive T cells, further emphasizing the importance of DNAM-1 in the immune response. We have shown that DNAM-1 considerably expands the range of cells that can activate CD8 T cells and NK cells. DNAM-1 is also up-regulated on CD4 T cells after Th1 differentiation but is down-regulated on Th0 and Th2 cells (3). How DNAM-1 influences CD4 T cell and perhaps T reg cell responses remains an open question.
In conclusion, this report provides the first demonstration that DNAM-1 promotes CD8 T cell co-stimulation and NK cell responses in vivo, broadening the spectrum of target cells that are effectively recognized. It has been shown that human CMV reduces expression of CD155 in infected cells through UL141 (29). Moreover, some tumors overexpress CD155 and release it as a soluble molecule (30), possibly to block DNAM-1 and prevent recognition of tumor cells. These mechanisms of evasion underscore the potential importance of DNAM-1–CD155 interactions in immunosurveillance against tumors and/or viral infections, particularly those that are weakly immunogenic because they do not engage major activating or accessory molecules.
MATERIALS AND METHODS
DNAM-1−/− mice.
A targeting vector was designed to replace exons two and three (encoding the start site and first 129 amino acids) of DNAM-1 with an MC1-neor gene flanked by loxP sites (Fig. S1). The construct was linearized and electroporated into E14.1 (129P2/OlaHsd) embryonic stem cells; 747 clones from two independent experiments were expanded and screened by Southern blotting using an external probe. One targeted clone was identified and confirmed using multiple restriction enzyme digests and external and internal probes. The clone was injected into C57BL/6 blastocysts, and resulting chimeras were bred to C57BL/6 mice expressing a Cre transgene under the CMV promoter to delete the MC1-neor gene. Germline transmission was assessed by coat color and the presence of the DNAM-1 mutation by PCR. The DNAM-1 deletion was backcrossed onto a C57BL/6 background, facilitated by a genome-wide screening of polymorphic microsatellite markers at 10-centiMorgan intervals at each generation (performed by the Rheumatic Disease Core Center's Speed Congenics Laboratory, Washington University School of Medicine). A 96% C57BL/6 male (B6N4) was identified for further backcrossing; B6N6 heterozygotes were intercrossed to generate the DNAM-1−/− mice used for this study. C57BL/6 WT, and RAG1−/− and OT-1 transgenic mice (obtained from the Jackson Laboratory) were bred and maintained at an Association for Assessment and Accreditation of Laboratory Animal Care–approved facility at Washington University School of Medicine and/or the Peter MacCallum Cancer Centre. All experiments were approved by the Washington University Animal Care Committee and the Peter MacCallum Animal Experimentation Ethics Committee.
Cell lines.
RM-1, B16F10, YAC-1, MC38-OVA, RMA-S, A20, and EL-4 cells were maintained in RPMI 1640 supplemented with 10% FCS. Stable transfectants of mCD155 were generated by electroporation of mCD155 cDNA. Positive cells were purified by FACS sorting. The MC38-OVA line was generated by retrovirally infecting the MC38 parental line with a pMIG/MSCV-IRES-eGFP plasmid-encoding membrane-bound OVA (provided by J. Villadangos, Walter and Eliza Hall Institute for Medical Research, Parkville, Australia).
Antibodies and reagents.
We generated the mAb 480.1 anti–mouse DNAM-1 (mDNAM-1) or mAb 3.1 anti–mouse CD96 (mCD96) by immunizing rats with RBL cells transfected with mDNAM-1 cDNA (RBL–mDNAM-1) or mouse CD96 cDNA (RBL-mCD96) and selecting hybridomas that reacted with 293–mDNAM-1 or 293-mCD96 transfectants but not with untransfected 293 cells. To generate the mAb 4.24 to mCD155, we immunized rats with a recombinant protein consisting of the mCD155 extracellular region fused to human Fc and selected hybridomas that reacted with RMA-S–mCD155 transfectants but not with untransfected RMA-S cells. Anti-asGM1 for depletion of NK cells was obtained from Wako Chemicals USA, Inc., and anti-CD8 mAb (53.6.7) was produced in our laboratory. Some groups of B6 mice were depleted of CD8+ T lymphocytes or NK cells in vivo by treatment with 100 μg anti-CD8 mAb or anti-asGM1 on days −1, 0 (day of i.v. tumor inoculation), and 7. These schedules effectively deplete T cell or NK cell subsets, as assessed by analysis using FITC-labeled mAbs. IL-2 was a gift from Novartis.
Cell preparations and FACS analysis.
Single-cell suspensions were prepared from lymph nodes and spleens and depleted of erythrocytes by ammonium chloride lysis. Lung lymphocytes were enriched in a Percoll (GE Healthcare) density gradient (45 and 67.5%) at 800 g for 15 min followed by ammonium chloride lysis. Cells were stained with anti-NK1.1–PE (PK136), -TCRαβ–PECy5.5 (H57-597), -CD27–allophycocyanin (Lg.7F9), and -CD11b–FITC (M1/70). All antibodies were from eBioscience. Dead cells were excluded with 1 μg/ml fluorogold (Sigma-Aldrich) in the final wash. Cells were acquired on an LSR-II (BD). Analysis was performed using FlowJo software (Tree Star, Inc.). YAC-1, B16F10, RM-1, and MC38-OVA tumor cells were stained with biotinylated anti-mCD155 (clone 4.24) and purified anti-CD112 (clone 502-57; Abcam), followed by streptavidin-PE (eBioscience) and anti–rat–PE (Invitrogen), respectively. Cell acquisition and analyses were performed as described above. In some experiments, splenocytes or thymocytes were treated with Fc block (HB-197; American Type Culture Collection) and were stained with mAb 480.1 and anti-CD4, -CD8, -CD3, -NK1.1, -CD25, and -CD44 (all from BD). Samples were processed on a FACSCalibur and analyzed with CellQuest software (both from BD).
Stimulation with SEB.
Total T cells were purified from spleens of WT and DNAM-1−/− mice by positive selection with CD5 microbeads (Miltenyi Biotec). Contamination with CD5+ B cells was always <3% of the total cells recovered. Percentages of TCRVβ3, Vβ7, and Vβ8 within the CD4 and CD8 T cell compartments were always comparable in WT and DNAM-1−/− mice, as assessed by staining for CD4, CD8, and TCRVβ3, Vβ7, and Vβ8. A20 and mCD155-A20 cells were irradiated (11,000 rads) and pulsed for 4 h with serial dilutions of SEB (Toxin Technology Inc.). Cells were extensively washed and incubated with T cells labeled with CFSE (Invitrogen). After 72 h, cells were stained for CD4 and CD8 and analyzed by flow cytometry. In some experiments, cell proliferation was measured by [3H]thymidine incorporation.
Antigen stimulation.
OT-1 transgenic CD8 T cells were enriched from spleens of WT and DNAM-1−/− mice by negative selection with the CD8 T cell isolation kit (Miltenyi Biotec). Purity was monitored by staining with OVA257-264 tetramers (Beckman Coulter) and was always found to be >90%. EL-4 (American Type Culture Collection) and EL-4–mCD155 were irradiated (6,000 rads) and pulsed with SIINFEKL peptide (Bachem) for 3 h. Cells were extensively washed and incubated with purified OT-1 transgenic CD8 T cells for 72 h. Cell proliferation was measured by standard [3H]thymidine incorporation. Cell blasting was monitored by flow cytometry by recording changes in FSC/SSC.
In vitro and in vivo analyses of DCs.
Bone marrow cells were cultured with GM-CSF and analyzed at day 7 for expression of CD11c, mCD155, and CD86. Cells were challenged with 20 μg/ml poly I:C (InvivoGen), 1 μg/ml zymosan (Sigma-Aldrich), 6 μM CpG 1826 (QIAGEN), and 100 ng/ml LPS (Ultrapure LPS from Escherichia coli; List Biological Laboratories). Primary migrating DCs were visualized by painting ears of mice with CellTracker Green CMFDA (Invitrogen) dissolved in a mixture of acetone and dibutyl phthalate, as a source of irritation. CLNs were analyzed 48 h later by flow cytometry.
Detection of OVA-specific CD8 T cells in vivo.
WT and DNAM-1–deficient mice were immunized s.c. in the footpad with CFA and 100 nmol OVA peptide 257–264 (SIINFEKL). On day 7 or 8, mice were killed and popliteal lymph nodes were extracted. Lymph nodes were disrupted by scraping and were passed over 70-μm nylon cell strainers to make single-cell suspensions. 1.5 million lymph node cells were co-cultured in 24-well plates with 5 × 105 APCs either pulsed for 4 h with 1 μM SIINFEKL or left unpulsed. Two types of APCs were used: EL-4 cells transfected with CD155 or BMDCs (day 6 or 7 of GM-CSF culture). Co-cultures were incubated for 4 h at 37°C. 2 μM monensin was added to block cytokine export, and co-cultures were incubated overnight. Cells were surface stained with Fc receptor–blocking mAb 2.4G2, followed by anti-CD8α–FITC (BD). To prepare for intracellular staining, cells were fixed in 2% paraformaldehyde (Sigma-Aldrich) and permeablilized in 0.5% saponin buffer. Cells were then stained with anti–IFN-γ–allophycocyanin (BD) and analyzed by flow cytometry.
NK cell in vitro assays.
Splenocyte suspensions were prepared as described above. For purification of NK cells by MACS, splenocytes were incubated with the NK cell isolation kit (Miltenyi Biotec), and cells were sorted by autoMACS (Miltenyi Biotec). NK cell media consisted of RPMI 1640 containing 10% FCS, β-2ME, nonessential amino acids, sodium pyruvate, l-glutamine, and penicillin/streptomycin. NK cells were expanded in NK cell media containing 1,000 U/ml IL-2 for 5 d. Cytotoxicity was assessed by standard 4-h 51Cr release assays.
In vivo tumor assays.
For lung metastasis assays, single-cell suspensions of B16F10 or RM-1 were prepared in PBS, and from 103 to 2 × 105 cells were injected i.v. in the tail vein of recipient mice. Lungs were harvested on day 14 and fixed in Bouin's solution, and tumor nodules were counted with the aid of a dissection microscope (model SZX7; Olympus). For s.c. tumor assays, mice were injected with various doses of B16F10 or MC38-OVA s.c., and tumor growth was monitored at least every second day using a caliper square to determine the product of two perpendicular tumor diameters.
Statistical analysis.
Statistical significance was assessed through the use of the Mann-Whitney rank sum test, as appropriate.
Online supplemental material.
Fig. S1 shows the generation of DNAM1−/− mice and of anti-mCD155 and -mDNAM1 antibodies. Fig. S2 shows NK cell percentages and maturation in WT and DNAM1−/− mice. Fig. S3 shows thymic T cell subsets in WT and DNAM1−/− mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081752/DC1.
Supplementary Material
Acknowledgments
We thank Mike White for blastocyst injections and the Rheumatic Disease Core Center's Speed Congenics Laboratory at Washington University School of Medicine for high density microsatellite mapping.
This work was supported by National Institutes of Health grant R01AI056139-05 and Juvenile Diabetes Research Foundation grant 24-2008-938. We thank the National Health and Medical Research Council of Australia for the support of program grant 454569, a Senior Principal Research Fellowship (to M.J. Smyth), a Doherty Fellowship (to D.M. Andrews), and a CJ Martin Fellowship (to N.M. Haynes), as well as the Leukaemia Foundation of Australia for a research scholarship (to C.J. Chan).
The authors have no conflicting financial interests.
S. Gilfillan, C.J. Chan, M.J. Smyth, and M. Colonna contributed equally to this paper.
References
- 1.Bryceson, Y.T., M.E. March, H.G. Ljunggren, and E.O. Long. 2006. Activation, coactivation, and costimulation of resting human natural killer cells. Immunol. Rev. 214:73–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shibuya, A., D. Campbell, C. Hannum, H. Yssel, K. Franz-Bacon, T. McClanahan, T. Kitamura, J. Nicholl, G.R. Sutherland, L.L. Lanier, and J.H. Phillips. 1996. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity. 4:573–581. [DOI] [PubMed] [Google Scholar]
- 3.Dardalhon, V., A.S. Schubart, J. Reddy, J.H. Meyers, L. Monney, C.A. Sabatos, R. Ahuja, K. Nguyen, G.J. Freeman, E.A. Greenfield, et al. 2005. CD226 is specifically expressed on the surface of Th1 cells and regulates their expansion and effector functions. J. Immunol. 175:1558–1565. [DOI] [PubMed] [Google Scholar]
- 4.Bottino, C., R. Castriconi, D. Pende, P. Rivera, M. Nanni, B. Carnemolla, C. Cantoni, J. Grassi, S. Marcenaro, N. Reymond, et al. 2003. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J. Exp. Med. 198:557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tahara-Hanaoka, S., K. Shibuya, Y. Onoda, H. Zhang, S. Yamazaki, A. Miyamoto, S. Honda, L.L. Lanier, and A. Shibuya. 2004. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int. Immunol. 16:533–538. [DOI] [PubMed] [Google Scholar]
- 6.Castriconi, R., A. Dondero, M.V. Corrias, E. Lanino, D. Pende, L. Moretta, C. Bottino, and A. Moretta. 2004. Natural killer cell-mediated killing of freshly isolated neuroblastoma cells: critical role of DNAX accessory molecule-1-poliovirus receptor interaction. Cancer Res. 64:9180–9184. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs, A., M. Cella, E. Giurisato, A.S. Shaw, and M. Colonna. 2004. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J. Immunol. 172:3994–3998. [DOI] [PubMed] [Google Scholar]
- 8.Pende, D., C. Bottino, R. Castriconi, C. Cantoni, S. Marcenaro, P. Rivera, G.M. Spaggiari, A. Dondero, B. Carnemolla, N. Reymond, et al. 2005. PVR (CD155) and Nectin-2 (CD112) as ligands of the human DNAM-1 (CD226) activating receptor: involvement in tumor cell lysis. Mol. Immunol. 42:463–469. [DOI] [PubMed] [Google Scholar]
- 9.Tahara-Hanaoka, S., K. Shibuya, H. Kai, A. Miyamoto, Y. Morikawa, N. Ohkochi, S. Honda, and A. Shibuya. 2006. Tumor rejection by the poliovirus receptor family ligands of the DNAM-1 (CD226) receptor. Blood. 107:1491–1496. [DOI] [PubMed] [Google Scholar]
- 10.El-Sherbiny, Y.M., J.L. Meade, T.D. Holmes, D. McGonagle, S.L. Mackie, A.W. Morgan, G. Cook, S. Feyler, S.J. Richards, F.E. Davies, et al. 2007. The requirement for DNAM-1, NKG2D, and NKp46 in the natural killer cell-mediated killing of myeloma cells. Cancer Res. 67:8444–8449. [DOI] [PubMed] [Google Scholar]
- 11.Pende, D., R. Castriconi, P. Romagnani, G.M. Spaggiari, S. Marcenaro, A. Dondero, E. Lazzeri, L. Lasagni, S. Martini, P. Rivera, et al. 2006. Expression of the DNAM-1 ligands, Nectin-2 (CD112) and poliovirus receptor (CD155), on dendritic cells: relevance for natural killer-dendritic cell interaction. Blood. 107:2030–2036. [DOI] [PubMed] [Google Scholar]
- 12.Shibuya, K., J. Shirakawa, T. Kameyama, S. Honda, S. Tahara-Hanaoka, A. Miyamoto, M. Onodera, T. Sumida, H. Nakauchi, H. Miyoshi, and A. Shibuya. 2003. CD226 (DNAM-1) is involved in lymphocyte function–associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J. Exp. Med. 198:1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Della Chiesa, M., C. Romagnani, A. Thiel, L. Moretta, and A. Moretta. 2006. Multidirectional interactions are bridging human NK cells with plasmacytoid and monocyte-derived dendritic cells during innate immune responses. Blood. 108:3851–3858. [DOI] [PubMed] [Google Scholar]
- 14.Reymond, N., A.M. Imbert, E. Devilard, S. Fabre, C. Chabannon, L. Xerri, C. Farnarier, C. Cantoni, C. Bottino, A. Moretta, et al. 2004. DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J. Exp. Med. 199:1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibuya, K., L.L. Lanier, J.H. Phillips, H.D. Ochs, K. Shimizu, E. Nakayama, H. Nakauchi, and A. Shibuya. 1999. Physical and functional association of LFA-1 with DNAM-1 adhesion molecule. Immunity. 11:615–623. [DOI] [PubMed] [Google Scholar]
- 16.Ralston, K.J., S.L. Hird, X. Zhang, J.L. Scott, B. Jin, R.F. Thorne, M.C. Berndt, A.W. Boyd, and G.F. Burns. 2004. The LFA-1-associated molecule PTA-1 (CD226) on T cells forms a dynamic molecular complex with protein 4.1G and human discs large. J. Biol. Chem. 279:33816–33828. [DOI] [PubMed] [Google Scholar]
- 17.Seth, S., M.K. Maier, Q. Qiu, I. Ravens, E. Kremmer, R. Forster, and G. Bernhardt. 2007. The murine pan T cell marker CD96 is an adhesion receptor for CD155 and nectin-1. Biochem. Biophys. Res. Commun. 364:959–965. [DOI] [PubMed] [Google Scholar]
- 18.Kissenpfennig, A., S. Henri, B. Dubois, C. Laplace-Builhe, P. Perrin, N. Romani, C.H. Tripp, P. Douillard, L. Leserman, D. Kaiserlian, et al. 2005. Dynamics and function of Langerhans cells in vivo: dermal dendritic cells colonize lymph node areas distinct from slower migrating Langerhans cells. Immunity. 22:643–654. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez, S., V. Groh, and T. Spies. 2006. Immunobiology of human NKG2D and its ligands. Curr. Top. Microbiol. Immunol. 298:121–138. [DOI] [PubMed] [Google Scholar]
- 20.Raulet, D.H. 2003. Roles of the NKG2D immunoreceptor and its ligands. Nat. Rev. Immunol. 3:781–790. [DOI] [PubMed] [Google Scholar]
- 21.Lanier, L.L. 2008. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 9:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moretta, A., C. Bottino, M. Vitale, D. Pende, C. Cantoni, M.C. Mingari, R. Biassoni, and L. Moretta. 2001. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu. Rev. Immunol. 19:197–223. [DOI] [PubMed] [Google Scholar]
- 23.Diefenbach, A., A.M. Jamieson, S.D. Liu, N. Shastri, and D.H. Raulet. 2000. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat. Immunol. 1:119–126. [DOI] [PubMed] [Google Scholar]
- 24.Cerwenka, A., A.B. Bakker, T. McClanahan, J. Wagner, J. Wu, J.H. Phillips, and L.L. Lanier. 2000. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 12:721–727. [DOI] [PubMed] [Google Scholar]
- 25.Roberts, A.I., L. Lee, E. Schwarz, V. Groh, T. Spies, E.C. Ebert, and B. Jabri. 2001. NKG2D receptors induced by IL-15 costimulate CD28-negative effector CTL in the tissue microenvironment. J. Immunol. 167:5527–5530. [DOI] [PubMed] [Google Scholar]
- 26.Das, H., V. Groh, C. Kuijl, M. Sugita, C.T. Morita, T. Spies, and J.F. Bukowski. 2001. MICA engagement by human Vgamma2Vdelta2 T cells enhances their antigen-dependent effector function. Immunity. 15:83–93. [DOI] [PubMed] [Google Scholar]
- 27.Girardi, M., D.E. Oppenheim, C.R. Steele, J.M. Lewis, E. Glusac, R. Filler, P. Hobby, B. Sutton, R.E. Tigelaar, and A.C. Hayday. 2001. Regulation of cutaneous malignancy by gammadelta T cells. Science. 294:605–609. [DOI] [PubMed] [Google Scholar]
- 28.Todd, J.A., N.M. Walker, J.D. Cooper, D.J. Smyth, K. Downes, V. Plagnol, R. Bailey, S. Nejentsev, S.F. Field, F. Payne, et al. 2007. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 39:857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomasec, P., E.C. Wang, A.J. Davison, B. Vojtesek, M. Armstrong, C. Griffin, B.P. McSharry, R.J. Morris, S. Llewellyn-Lacey, C. Rickards, et al. 2005. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat. Immunol. 6:181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baury, B., D. Masson, B.M. McDermott Jr., A. Jarry, H.M. Blottiere, P. Blanchardie, C.L. Laboisse, P. Lustenberger, V.R. Racaniello, and M.G. Denis. 2003. Identification of secreted CD155 isoforms. Biochem. Biophys. Res. Commun. 309:175–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.