Abstract
Since the identification of ligands for human and mouse DNAM-1, emerging evidence has suggested that DNAM-1 plays an important role in the T cell– and natural killer (NK) cell–mediated recognition and lysis of tumor cells. However, it remains undetermined whether DNAM-1 is involved in tumor immune surveillance in vivo. We addressed this question by using DNAM-1–deficient mice. DNAM-1–deficient cytotoxic T lymphocyte (CTL) and NK cells showed significantly less cytotoxic activity against DNAM-1 ligand-expressing tumors in vitro than wild-type (WT) cells. The methylcholanthrene (MCA)-induced fibrosarcoma cell line Meth A expressed the DNAM-1 ligand CD155, and DNAM-1–deficient mice showed increased tumor development and mortality after transplantation of Meth A cells. Moreover, the DNAM-1–deficient mice developed significantly more DNAM-1 ligand-expressing fibrosarcoma and papilloma cells in response to the chemical carcinogens MCA and 7,12-dimethylbenz[a]anthracene (DMBA), respectively, than did WT mice. These results indicate that DNAM-1 plays an important role in immune surveillance of tumor development.
Several lines of evidence demonstrate that the immune system protects the host against tumor development. CTL and NK cells are major players in cell-mediated immunity to tumors (1, 2). Tumor recognition by and activation of CTL and NK cells are mediated by antigen receptors and a variety of adhesion and costimulatory molecules including CD2, LFA-1 (leukocyte function-associated antigen-1; CD11a/CD18), CD27, 2B4, CD28, ICOS (inducible costimulator), NKG2D, TRAIL (TNF-related apoptosis-inducing ligand), and others (2, 3). Interactions of these cell surface receptors with their respective ligands expressed on tumors induce cytotoxic activity of CTL and NK cells against tumors (4).
DNAM-1 (CD226) is a member of the immunoglobulin superfamily and is expressed on the majority of NK cells, T cells, monocytes, and platelets (5–7). DNAM-1 constitutively associates with LFA-1 on NK cells (8). Ligation of LFA-1 with antibodies causes Fyn-dependent phosphorylation of tyrosine residues in the cytoplasmic domain of DNAM-1, which are necessary for DNAM-1–dependent NK cell functions (8). DNAM-1 also associates with LFA-1 in activated T cells, for which the protein kinase C–induced serine phosphorylation in the cytoplasmic domain of DNAM-1 is responsible (8), and is involved in an LFA-1–mediated costimulatory signal for naive T cell differentiation and proliferation (9). The poliovirus receptor (PVR) CD155 and its family member CD112 (PRR-2 [PVR-related family 2], also called nectin-2) are ligands for human and mouse DNAM-1 (10–12). Interactions between DNAM-1 on NK cells and T cells and its ligands CD112 and CD155 on tumor cells augment cell-mediated cytotoxicity and cytokine production (10, 11). Although human CD112 and CD155 are broadly distributed on epithelial and endothelial cells in many tissues (13, 14), they are overexpressed on certain tumors, both nonhematopoietic and hematopoietic, including colorectal carcinomas (15, 16), gastric cancers (16), ovarian cancers (17), neuroblastomas (18), myeloid leukemias (19), and multiple myeloma (20), suggesting that DNAM-1 ligand expression might be induced by tumorigenesis and, thus, stimulates CTL and NK cells. However, it remains uncertain whether DNAM-1 is involved in immune surveillance of tumors in vivo. We therefore generated KO mice lacking the Cd226 (DNAM-1) gene and investigated the in vivo role of DNAM-1 in tumor immunity.
RESULTS AND DISCUSSION
Mouse DNAM-1 mediates cytotoxic activity of CTL and NK cells
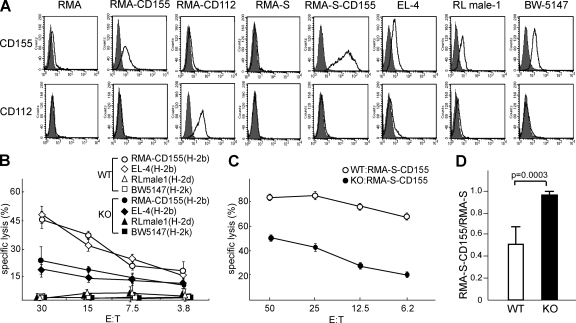
Previous studies demonstrated that the addition of human DNAM-1–neutralizing antibodies to in vitro cytotoxicity assays inhibited CTL or NK cell–mediated lysis of several tumor cell lines expressing DNAM-1 ligands (5, 10, 11), suggesting that human DNAM-1 is involved in CTL and NK cell–mediated cytotoxicity. However, the role of DNAM-1 in cytotoxicity of mouse CTL and NK cells has not been investigated. To address this issue, we generated mice lacking the Cd226 (DNAM-1) gene in the BALB/c background (DNAM-1 KO mice; Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20081611/DC1). Naive DNAM-1–deficient mice showed a normal composition of lymphocyte populations in the spleen, BM, and LN (Fig. S2 and not depicted), suggesting that the disruption of Cd226 did not affect lymphocyte differentiation. We then examined the expression of DNAM-1 ligands CD155 and CD112 in mouse tumor cell lines EL-4 (H-2b), RL male-1 (H-2d), and BW5147 (H-2k). These cell lines expressed comparable amounts of CD155 (Fig. 1 A). In contrast, CD112 was expressed on RMA transfectant expressing CD112 (RMA-CD112) but was barely detected on the other tumor cells (Fig. 1 A). We next generated CTL by coculture of spleen cells from WT and DNAM-1–deficient BALB/c mice (H-2d) with the RMA cell line (H-2b) and then purified CD8+ T cells by sorting. Because RMA cells did not express DNAM-1 ligands (Fig. 1 A) and the population of CD8+ T cells after the culture was comparable between WT and KO spleen cells (not depicted), DNAM-1 deficiency did not affect the induction of DNAM-1–deficient CTL cells. The purified WT and DNAM-1–deficient CD8+ T cells specifically killed EL-4 tumor cells as well as RMA transfectant expressing CD115 (RMA-CD155), but not RL male-1 or BW5147 (H-2k) target cells, suggesting that both CTL were H-2b specific (Fig. 1 B). However, DNAM-1–deficient CTL were significantly less cytotoxic to RMA-CD155 and EL-4 than were the WT CTL cells (Fig. 1 B).
Figure 1.
DNAM-1 is involved in NK cell– and CTL-mediated cytotoxicity against DNAM-1 ligand–expressing tumor cell lines. (A) Tumor cell lines or transfectants indicated were stained with anti-CD155 or anti-CD112 mAbs and analyzed by flow cytometry. (B and C) H-2b–specific CD8+ CTL (B) or DX5+ NK (C) cells from WT or DNAM-1–deficient BALB/c mice (H-2d) were cocultured with 51Cr-labeled target cells indicated for 3 h. Culture supernatants were harvested and 51Cr releases were counted. (D) WT (n = 5) or DNAM-1–deficient (n = 5) mice were injected i.p. with a 1:1 mixture of CFSE-labeled RMA-S and RMA-S–CD155. 12 h after injection, cells were harvested from the peritoneal cavities and analyzed for the ratio of RMA-S–CD155 to RMA-S cells by flow cytometry. Data are representative of three independent experiments. Error bars show SD.
We also examined the cytotoxicity of DNAM-1–deficient NK cells. These cells showed no defect in cytolysis of Yac-1 tumor cells (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20081611/DC1), which substantially express NKG2D ligands (21) but not DNAM-1 ligands. However, they showed less cytotoxicity to RMA-S–CD155 cells than did WT NK cells (Fig. 1, A and C). These results together formally demonstrate that DNAM-1 is involved in both CTL and NK cell–mediated cytotoxicity.
DNAM-1–deficient mice showed impaired clearance of CD155-expressing tumor cells
We next investigated the role of DNAM-1 in tumor clearance in vivo. WT or DNAM-1–deficient mice were injected i.p. with a 1:1 mixture of CFSE-labeled RMA-S and RMA-S–CD155 cells. At 12 h, tumor cells were harvested from the peritoneal cavity and analyzed for the ratio of cell numbers by flow cytometry. We recovered significantly fewer RMA-S–CD155 cells than RMA-S cells from WT mice than from DNAM-1–deficient mice (Fig. 1 D). Considering the short time interval, NK cells, rather than CTL, might be involved in the clearance of RMA-S–CD155 cells. These results suggest that DNAM-1 plays an important role in the clearance of tumors expressing CD155 in vivo.
DNAM-1–deficient mice showed higher mortality after transplantation of Meth A cells
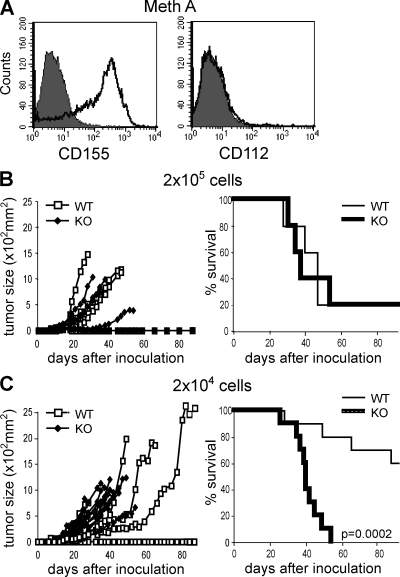
We previously demonstrated that although RMA tumor cells grew in syngeneic mice, RMA-CD155 or RMA-CD112 cells were rejected by CD8+ T cells and NK cells, suggesting that DNAM-1 was involved in the rejection (16). To provide formal evidence for the role of DNAM-1 in protecting against DNAM-1 ligand-expressing tumors in vivo, we transplanted Meth A tumors that expressed CD155, but not CD112, into WT and DNAM-1–deficient mice (Fig. 2 A). When inoculated with 2 × 105 cells, both groups of mice developed tumors by 3–4 wk after inoculation, and their survival and mortality did not differ (Fig. 2 B). However, when inoculated with 2 × 104 cells, all DNAM-1–deficient mice developed tumors by 20 d after inoculation, but 6 out of 10 WT mice showed no evidence of tumor development and the mortality of WT mice was significantly lower (Fig. 2 C). These results indicate that DNAM-1 was involved in the rejection of the Meth A tumors.
Figure 2.
DNAM-1 is involved in tumor rejection in vivo. (A) Meth A cells were stained with anti-CD155 or anti-CD112 mAbs and analyzed by flow cytometry. (B and C) WT (B, n = 5; C, n = 10) or DNAM-1–deficient (B, n = 5; C, n = 10) mice were inoculated s.c. with 2 × 104 (B) or 2 × 105 (C) Meth A on day 0. Tumor size in each mouse was measured three times a week. Survival data per group are shown. The experiments were performed twice with similar results. One experiment is shown.
DNAM-1 deficiency enhanced the development of 3-methylcholanthrene (MCA)–induced fibrosarcoma
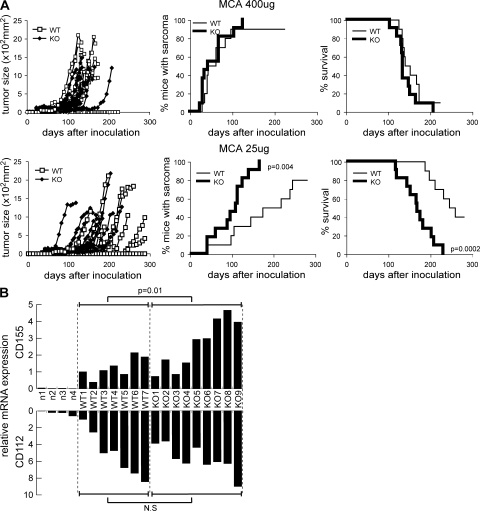
Because Meth A is an MCA-induced fibrosarcoma cell line, we examined whether DNAM-1 might also be involved in immune surveillance of fibrosarcoma after MCA injection. WT and DNAM-1–deficient mice were injected s.c. with 25 or 400 μg MCA. At 400 μg MCA, both groups of mice similarly developed fibrosarcoma by 30 d after injection (Fig. 3 A) and showed no difference in mortality. In contrast, at 25 μg MCA, WT mice developed tumors more slowly and survived significantly longer than DNAM-1–deficient mice (Fig. 3 A). These results suggest that DNAM-1 played an important role in immune surveillance of MCA-induced fibrosarcoma.
Figure 3.
DNAM-1 is involved in immune surveillance against MCA-induced fibrosarcoma. (A) WT (n = 10) or DNAM-1–deficient (n = 10) mice were injected s.c. with 400 μg or 25 μg MCA on day 0. Tumor size in each mouse was measured once a week. Tumor incidence and survival data for 10 mice per group are shown. (B) When 25 μg MCA-induced fibrosarcomas in seven WT or nine DNAM-1–deficient mice reached a size of 1 cm in diameter, fibrosarcoma was resected from each mouse and subjected to quantitative RT-PCR for CD155 and CD112. The skins resected from four naive mice were also subjected to quantitative RT-PCR. The data are the mean of triplicates. The experiments were performed twice with similar results. One experiment is shown.
To further assess the role of DNAM-1 in immune surveillance of MCA-induced fibrosarcoma, we used quantitative RT-PCR to evaluate DNAM-1 ligand expression on fibrosarcoma cells that were developed after injection of 25 μg MCA into WT and DNAM-1–deficient mice. Although CD155 and CD112 messenger RNAs (mRNAs) were barely detected in naive derma, MCA-induced fibrosarcoma cells expressed significant quantities of both in both groups of mice (Fig. 3 B). Notably, although the fibrosarcoma cells expressed comparable amounts of CD112 mRNA in both groups, CD155 mRNA expression was significantly higher in the DNAM-1–deficient mice (Fig. 3 B). These results suggest that DNAM-1 recognition of CD155 on fibrosarcoma cells was involved in tumor immune surveillance, which is consistent with the “immunoediting” theory in which immune responses select for variant tumor cells which have lost or decreased expression of specific molecules that are targeted by immune effector cells (22). In contrast to CD155, CD112 seemed not to be involved in immunoediting. This may be explained by our previous observations that the affinities of homotypic binding of CD112 were likely to be higher than those of CD112 binding to DNAM-1 (12).
Recent studies using genetically engineered animals have demonstrated that a variety of immune cells in both the innate and adaptive immune systems are involved in tumor immunoediting of MCA-induced fibrosarcoma (4). Mice lacking cellular components such as αβ T cells (23), γδ T cells (23, 24), NK cells (25, 26), or NK T cells (25–27) developed higher rates of MCA-induced fibrosarcoma. The development of fibrosarcoma induced by MCA was also enhanced in mice deficient in TRAIL (28) or injected with a neutralizing antibody against TRAIL (29), which is constitutively expressed on liver NK cells and might be involved in NK cell–mediated immunity to fibrosarcoma development. However, activating receptors constitutively expressed on T cells and NK cells in the spleen have not yet been identified. In the present and previous studies, we showed that DNAM-1 was involved in CTL and NK cell–mediated tumor cytolysis in vitro (Fig. 1) and in the rejection of transplanted tumor cell lines (29), suggesting that DNAM-1 was responsible for CTL and/or NK cell–mediated immune surveillance of fibrosarcoma development induced by MCA.
DNAM-1 deficiency enhanced the development of 7,12-dimethylbenz[a]anthracene (DMBA)–induced papilloma
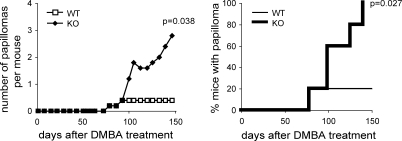
To examine whether DNAM-1 is involved in immune surveillance of tumors other than MCA-induced fibrosarcoma, we treated WT and DNAM-1–deficient mice with DMBA, a chemical carcinogen that induces papillomas on the skin. One out of five mice in each group developed papillomas by 80 d after treatment with 400 μg DMBA alone, without 12-O-tetradecanoylphorbol 13-acetate (Fig. 4). The number of DNAM-1–deficient mice with papillomas increased by 100 d and all DNAM-1–deficient mice developed papillomas by 150 d, yet only the one WT mouse developed papillomas (Fig. 4). The mean number of papillomas in DNAM-1–deficient mice was significantly higher than in WT mice (Fig. 4). To further assess the role of DNAM-1 in immune surveillance of DMBA-induced papillomas, we examined the expression of CD155 and CD112 mRNAs in the papillomas by quantitative RT-PCR. We detected both in significant amounts in all eight papillomas developed in DNAM-1–deficient mice (unpublished data). We also detected both in two papillomas developed in the WT mouse, but we could not statistically compare the expressions between groups. Nonetheless, these results indicate that DNAM-1 plays an important role in immune surveillance of DMBA-induced papilloma as well as that of MCA-induced fibrosarcoma.
Figure 4.
DNAM-1 is involved in immune surveillance against DMBA-induced papilloma. WT (n = 5) or DNAM-1–deficient (n = 5) mice were treated with 400 μg DMBA on day 0. The number of papillomas developed in each mouse was observed twice a week. The mean number of papillomas and tumor incidence per group are shown. The experiments were performed twice with similar results. One experiment is shown.
Emerging evidence suggests that DNAM-1 plays an important role in T cell– and NK cell–mediated immunity to a variety of tumors expressing DNAM-1 ligands (15–20). This is the first study using DNAM-1–deficient mice to demonstrate direct genetic evidence for a role of the DNAM-1 receptor in immune surveillance of chemical carcinogen-induced tumors. Future studies aimed at the clinical application of DNAM-1 as a potential molecular target for the treatment of cancer should be interesting.
MATERIALS AND METHODS
Generation of DNAM-1 KO mice.
To construct the DNAM-1 targeting vector, we replaced exon 2–3 of the Cd226 (DNAM-1) gene with a neomycin-resistant gene cassette using the bacterial artificial chromosome system (30). BALB/c embryonic stem cells were electroporated with the linearized DNAM-1 targeting vector, and cells were selected for G418 antibiotic resistance. Correctly targeted embryonic stem cell clones with normal karyotypes were injected into 3.5-d-old BALB/c blastocysts to create chimeric mice (Cd226+/−). Cd226+/− mice were intercrossed to generate cd226−/− mice. Homozygous mice were obtained at the expected Mendelian frequencies and developed normally. Cd226−/− mice and control WT mice were bred under specific pathogen-free conditions in the same room of our animal facility. All experiments were performed according to the guidelines of the animal ethics committee of the University of Tsukuba Animal Research Center.
Cell lines and antibodies.
RMA and EL-4 (31) were H-2b+ T lymphoma cell lines. Meth A, RL male-1, A20, and SP2/0 were H-2d+ fibroblast, T cell, B cell, and myeloma cell lines, respectively. BW5147 was an H-2k+ T lymphoma cell line. Anti-CD8 and -DX5 monoclonal antibodies were purchased from BD. Anti-DNAM-1 (TX42) (16), anti-CD155 (TX56), and anti-CD112 (TX78) antibodies were generated in our laboratory.
Generation of CTL and NK cells.
We cocultured 5 × 106 spleen cells with 106 RMA cells, pretreated with 100 μg/ml mitomycin C, in a 24-well plate for 5 d and then with purified CD8+ T cells by using the IMAG sorting system (BD). DX5+ cells were purified from the spleen by using magnetic activated cell sorting (MACS; Miltenyi Biotec) and cultured in the presence of 200 ng/ml IL-2 for 5 d. Purities of CD8+ and DX5+ cells were >95%, as determined by flow cytometry.
Cytotoxicity assay.
Cytotoxicity assays were performed as described previously (11).
Tumor clearance assay.
WT or DNAM-1–deficient mice were injected i.p. with a 1:1 mixture of CFSE-labeled RMA-S and RMA-S–CD155 cells (106 each). 12 h after injection, tumor cells were harvested from the peritoneal cavities and analyzed by flow cytometry.
Tumor growth assay and survival of mice.
Groups of 5 or 10 WT or DNAM-1–deficient male mice were injected s.c. in the back with transplantable tumor cell lines, with 25 or 400 μg MCA (Sigma-Aldrich) dissolved in 0.1 ml maize oil, or were painted with 400 μg DMBA (Sigma-Aldrich) dissolved in 4 mM of acetone. Mice were examined once or three times a week for tumor size with a caliper square, as previously described (16).
Quantitative RT-PCR.
Total RNA was extracted from fibrosarcomas and naive derma with Isogen reagent (Nippon Gene). For RT, we used 2–5 μg of total RNA in a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) at a final volume of 20 μl. Real-time PCR analysis of CD155 and CD112d was performed with an ABI 7700 sequence detector (Applied Biosystems) with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). The primers were as follows: CD155 forward, 5′-CAACTGGTATGTTGGCCTCA-3′ and reverse, 5′-ATTGGTGACTTCGCACACAA-3′; and CD112d forward, 5′-CTCTGTGGATCGAATGGTCA-3′ and reverse, 5′-GGCAGCGATAATACCTCCAA-3′. The GAPDH level was measured as an internal control (upper primer, 5′-CTTCACCACCATGGAGAAGGC-3′; and lower primer, 5′-GGCATGGACTGTGGTCATGAG-3′) to normalize the data. The thermal cycling conditions comprised an initial denaturation step at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Standard curves were generated by using serial dilutions of cDNA from Meth A cells for CD155 and BaF-CD112d cells for CD112d. All values were determined in triplicate.
Statistics.
Statistical analyses were performed by using the unpaired Student's t test.
Online supplemental material.
Fig. S1 shows the construct of DNAM-1 targeting vector, genomic Cd226 analyzed by RT-PCR, the expression of Cd226 mRNA in the tail, and DNAM-1 protein on the spleen cells by staining with FITC-conjugated anti-CD8 and biotin-conjugated anti–DNAM-1 (TX42), followed with PE-conjugated streptavidin in the indicated mice. Fig. S2 shows the lymphocytes population in the thymus and spleen cells derived from WT (n = 5) and DNAM-1–deficient (n = 5) mice. Fig. S3 show the cytotoxicities of Yac-1 by WT and DNM-1–deficient NK cells, as analyzed by 51Cr releases assay. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081611/DC1.
Supplementary Material
Acknowledgments
We thank Lewis Lanier (University of California, San Francisco) for critical reading of the manuscript, Eiichi Nakayama (Okayama University) for discussion, and Yurika Soeda for secretarial assistance.
This research was supported in part by grants provided by the Ministry of Education, Science and Culture of Japan and the Program for Promotion of Fundamental Studies in Health Science of the National Institute of Biomedical Innovation (NIBIO).
The authors have no conflicting financial interests.
A. Iguchi-Manaka and H. Kai contributed equally to this paper.
References
- 1.Dunn, G.P., L.J. Old, and R.D. Schreiber. 2004. The three Es of cancer immunoediting. Annu. Rev. Immunol. 22:329–360. [DOI] [PubMed] [Google Scholar]
- 2.Swann, J.B., and M.J. Smyth. 2007. Immune surveillance of tumors. J. Clin. Invest. 117:1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanier, L.L. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225–274. [DOI] [PubMed] [Google Scholar]
- 4.Dunn, G.P., C.M. Koebel, and R.D. Schreiber. 2006. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 6:836–848. [DOI] [PubMed] [Google Scholar]
- 5.Shibuya, A., D. Campbell, C. Hannum, H. Yssel, K. Franz-Bacon, T. McClanahan, T. Kitamura, J. Nicholl, G.R. Sutherland, L.L. Lanier, and J.H. Phillips. 1996. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity. 4:573–581. [DOI] [PubMed] [Google Scholar]
- 6.Kojima, H., H. Kanada, S. Shimizu, E. Kasama, K. Shibuya, H. Nakauchi, T. Nagasawa, and A. Shibuya. 2003. CD226 mediates platelet and megakaryocytic cell adhesion to vascular endothelial cells. J. Biol. Chem. 278:36748–36753. [DOI] [PubMed] [Google Scholar]
- 7.Shibuya, A., S. Tahara-Hanaoka, and K. Shibuya. 2005. DNAM-1 (CD226): a two-sword fencer for innate and adaptive immunity. Current Medical Chemistry - Anti-Inflammatory and Anti-Allergy Agents. 4:53–58. [Google Scholar]
- 8.Shibuya, K., L.L. Lanier, J.H. Phillips, H.D. Ochs, K. Shimizu, E. Nakayama, H. Nakauchi, and A. Shibuya. 1999. Physical and functional association of LFA-1 with DNAM-1 adhesion molecule. Immunity. 11:615–623. [DOI] [PubMed] [Google Scholar]
- 9.Shibuya, K., J. Shirakawa, T. Kameyama, S. Honda, S. Tahara-Hanaoka, A. Miyamoto, M. Onodera, T. Sumida, H. Nakauchi, H. Miyoshi, and A. Shibuya. 2003. CD226 (DNAM-1) is involved in lymphocyte function–associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J. Exp. Med. 198:1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottino, C., R. Castriconi, D. Pende, P. Rivera, M. Nanni, B. Carnemolla, C. Cantoni, J. Grassi, S. Marcenaro, N. Reymond, et al. 2003. Identification of PVR (CD155) and nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J. Exp. Med. 198:557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tahara-Hanaoka, S., K. Shibuya, Y. Onoda, H. Zhang, S. Yamazaki, A. Miyamoto, S. Honda, L.L. Lanier, and A. Shibuya. 2004. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int. Immunol. 16:533–538. [DOI] [PubMed] [Google Scholar]
- 12.Tahara-Hanaoka, S., A. Miyamoto, A. Hara, S. Honda, K. Shibuya, and A. Shibuya. 2005. Identification and characterization of murine DNAM-1 (CD226) and its poliovirus receptor family ligands. Biochem. Biophys. Res. Commun. 329:996–1000. [DOI] [PubMed] [Google Scholar]
- 13.Lopez, M., M. Aoubala, F. Jordier, D. Isnardon, S. Gomez, and P. Dubreuil. 1998. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood. 92:4602–4611. [PubMed] [Google Scholar]
- 14.Iwasaki, A., R. Welker, S. Mueller, M. Linehan, A. Nomoto, and E. Wimmer. 2002. Immunofluorescence analysis of poliovirus receptor expression in Peyer's patches of humans, primates, and CD155 transgenic mice: implications for poliovirus infection. J. Infect. Dis. 186:585–592. [DOI] [PubMed] [Google Scholar]
- 15.Masson, D., A. Jarry, B. Baury, P. Blanchardie, C. Laboisse, P. Lustenberger, and M.G. Denis. 2001. Overexpression of the CD155 gene in human colorectal carcinoma. Gut. 49:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahara-Hanaoka, S., K. Shibuya, H. Kai, A. Miyamoto, Y. Morikawa, N. Ohkochi, S. Honda, and A. Shibuya. 2006. Tumor rejection by the poliovirus receptor family ligands of the DNAM-1 (CD226) receptor. Blood. 107:1491–1496. [DOI] [PubMed] [Google Scholar]
- 17.Carlsten, M., N.K. Bjorkstrom, H. Norell, Y. Bryceson, T. van Hall, B.C. Baumann, M. Hanson, K. Schedvins, R. Kiessling, H.G. Ljunggren, and K.J. Malmberg. 2007. DNAX accessory molecule-1 mediated recognition of freshly isolated ovarian carcinoma by resting natural killer cells. Cancer Res. 67:1317–1325. [DOI] [PubMed] [Google Scholar]
- 18.Castriconi, R., A. Dondero, M.V. Corrias, E. Lanino, D. Pende, L. Moretta, C. Bottino, and A. Moretta. 2004. Natural killer cell-mediated killing of freshly isolated neuroblastoma cells: critical role of DNAX accessory molecule-1–poliovirus receptor interaction. Cancer Res. 64:9180–9184. [DOI] [PubMed] [Google Scholar]
- 19.Pende, D., G.M. Spaggiari, S. Marcenaro, S. Martini, P. Rivera, A. Capobianco, M. Falco, E. Lanino, I. Pierri, R. Zambello, et al. 2005. Analysis of the receptor–ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: evidence for the involvement of the poliovirus receptor (CD155) and nectin-2 (CD112). Blood. 105:2066–2073. [DOI] [PubMed] [Google Scholar]
- 20.El-Sherbiny, Y.M., J.L. Meade, T.D. Holmes, D. McGonagle, S.L. Mackie, A.W. Morgan, G. Cook, S. Feyler, S.J. Richards, F.E. Davies, et al. 2007. The requirement for DNAM-1, NKG2D, and NKp46 in the natural killer cell-mediated killing of myeloma cells. Cancer Res. 67:8444–8449. [DOI] [PubMed] [Google Scholar]
- 21.Guerra, N., Y.X. Tan, N.T. Joncker, A. Choy, F. Gallardo, N. Xiong, S. Knoblaugh, D. Cado, N.M. Greenberg, and D.H. Raulet. 2008. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 28:571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khong, H.T., and N.P. Restifo. 2002. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat. Immunol. 3:999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girardi, M., D.E. Oppenheim, C.R. Steele, J.M. Lewis, E. Glusac, R. Filler, P. Hobby, B. Sutton, R.E. Tigelaar, and A.C. Hayday. 2001. Regulation of cutaneous malignancy by gammadelta T cells. Science. 294:605–609. [DOI] [PubMed] [Google Scholar]
- 24.Gao, Y., W. Yang, M. Pan, E. Scully, M. Girardi, L.H. Augenlicht, J. Craft, and Z. Yin. 2003. γδ T cells provide an early source of interferon γ in tumor immunity. J. Exp. Med. 198:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smyth, M.J., K.Y. Thia, S.E. Street, E. Cretney, J.A. Trapani, M. Taniguchi, T. Kawano, S.B. Pelikan, N.Y. Crowe, and D.I. Godfrey. 2000. Differential tumor surveillance by natural killer (NK) and NKT cells. J. Exp. Med. 191:661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smyth, M.J., N.Y. Crowe, and D.I. Godfrey. 2001. NK cells and NKT cells collaborate in host protection from methylcholanthrene-induced fibrosarcoma. Int. Immunol. 13:459–463. [DOI] [PubMed] [Google Scholar]
- 27.Crowe, N.Y., J.M. Coquet, S.P. Berzins, K. Kyparissoudis, R. Keating, D.G. Pellicci, Y. Hayakawa, D.I. Godfrey, and M.J. Smyth. 2005. Differential antitumor immunity mediated by NKT cell subsets in vivo. J. Exp. Med. 202:1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cretney, E., K. Takeda, H. Yagita, M. Glaccum, J.J. Peschon, and M.J. Smyth. 2002. Increased susceptibility to tumor initiation and metastasis in TNF-related apoptosis-inducing ligand-deficient mice. J. Immunol. 168:1356–1361. [DOI] [PubMed] [Google Scholar]
- 29.Takeda, K., M.J. Smyth, E. Cretney, Y. Hayakawa, N. Kayagaki, H. Yagita, and K. Okumura. 2002. Critical role for tumor necrosis factor–related apoptosis-inducing ligand in immune surveillance against tumor development. J. Exp. Med. 195:161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, Y., and B. Seed. 2003. Site-specific gene targeting in mouse embryonic stem cells with intact bacterial artificial chromosomes. Nat. Biotechnol. 21:447–451. [DOI] [PubMed] [Google Scholar]
- 31.Gorer, P.A. 1950. Studies in antibody response of mice to tumor inoculation. Br. J. Cancer. 4:372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.