Abstract
Background
Teleost fishes do not have a vomeronasal organ (VNO), and their vomeronasal receptors (V1Rs, V2Rs) are expressed in the main olfactory epithelium (MOE), as are odorant receptors (ORs) and trace amine-associated receptors (TAARs). In this study, to obtain insights into the functional distinction among the four chemosensory receptor families in teleost fishes, their evolutionary patterns were examined in zebrafish, medaka, stickleback, fugu, and spotted green pufferfish.
Methodology/Principal Findings
Phylogenetic analysis revealed that many lineage-specific gene gains and losses occurred in the teleost fish TAARs, whereas only a few gene gains and losses have taken place in the teleost fish vomeronasal receptors. In addition, synonymous and nonsynonymous nucleotide substitution rate ratios (KA/KS) in TAARs tended to be higher than those in ORs and V2Rs.
Conclusions/Significance
Frequent gene gains/losses and high KA/KS in teleost TAARs suggest that receptors in this family are used for detecting some species-specific chemicals such as pheromones. Conversely, conserved repertoires of V1R and V2R families in teleost fishes may imply that receptors in these families perceive common odorants for teleosts, such as amino acids. Teleost ORs showed intermediate evolutionary pattern between TAARs and vomeronasal receptors. Many teleost ORs seem to be used for common odorants, but some ORs may have evolved to recognize lineage-specific odors.
Introduction
Olfaction is a sense for recognizing chemicals in the external environment. In many animals, olfaction is essential for various activities such as foraging, migration, and reproduction. Most terrestrial vertebrates have two distinct chemosensory organs, the main olfactory epithelium (MOE) and the vomeronasal organ (VNO). Generally, the MOE is considered to recognize environmental chemicals, while the VNO recognizes pheromones, although recent studies have reported some exceptions [1], [2]. It has been known that a distinct set of chemosensory receptors is expressed in each organ. In the MOE, main odorant receptors (ORs) and trace amine-associated receptors (TAARs) are expressed, whereas in the VNO, vomeronasal receptors type 1 (V1Rs) and type 2 (V2Rs) are expressed [reviewed in 3]. This implies that in terrestrial vertebrates, ORs and TAARs are mainly used to recognize “ordinary” odorants, whereas V1Rs and V2Rs are used to recognize pheromones. Indeed, several mammalian V1Rs and V2Rs respond to pheromone candidates [4]–[6].
Unlike the terrestrial vertebrates, teleost fishes do not have a VNO and their V1Rs and V2Rs are expressed in the MOE, as are ORs and TAARs [7], [8]. In teleost fishes, functional studies of the chemosensory receptors have not been reported. Thus, in teleost fishes, it is not clear which family of chemosensory receptors is used for detecting environmental chemicals or pheromones. Also, it remains unknown whether the different types of teleost fish chemosensory receptors respond to different classes of odorants. However, recent electrophysiological studies have indirectly suggested putative ligands of each type of odorant/pheromone receptors. For instance, goldfish sex pheromones 17α, 20β-dihydroxy-4-pregnen-3-one (17, 20-P) and F-prostaglandins are suggested to be perceived by ORs [9], whereas amino acids (major environmental odorants for teleost fishes) are considered to be detected by both ORs and V2Rs [9], [10].
The copy number, pattern of diversification, and selective constraints of a chemosensory receptor gene family seem to reflect the biological functions of receptors encoded to the gene family. For example, a chemosensory receptor family that perceives species-specific pheromones may show large variation across species. In contrast, if receptors in a family are used mainly for environmental odorants, the repertoire of these receptors may be more similar between different species. In terrestrial vertebrates, it is suggested that chemosensory receptors expressed in MOE (ORs and TAARs) are broadly tuned generalists, whereas vomeronasal receptors (V1Rs and V2Rs) are narrowly tuned specialists. In mammals, chicken, and frogs, this “differential tuning hypothesis” was supported by careful comparison of the evolutionary patterns between the two types of chemosensory receptor gene families [11]. However, the evolutionary patterns of the four chemosensory receptor families have not been studied in teleost fishes.
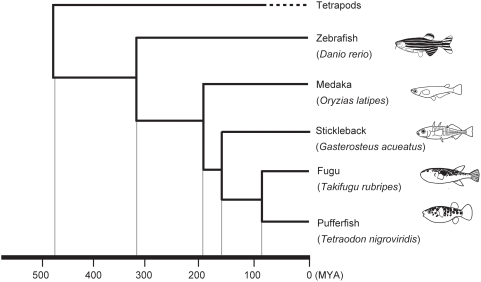
In this study, to obtain insights into the function and biological roles of the four chemosensory receptor families in teleost fishes, we analyzed the evolutionary patterns of the four gene families by comparing the gene repertoires from zebrafish Danio rerio, medaka Oryzias latipes, stickleback Gasterosteus aculeatus, fugu Takifugu rubripes, and spotted green pufferfish Tetraodon nigroviridis, for which draft genome sequences are publicly available. Their phylogenetic relationship and divergence times are well studied (Fig. 1), and these data enables us more detailed comparison of the evolutionary dynamics of chemosensory receptor gene families in teleost fishes. In this paper, we analyzed evolutionary patterns of the four chemosensory receptor gene families in teleost fishes. We also discussed the similarity and differences of the evolutionary modes of chemosensory receptors between teleost fishes and tetrapods.
Figure 1. Phylogenetic relationship and estimated divergence times of the five model fishes inferred from [12].
Results
Proportions of species-specific genes in the four chemosensory receptor gene families
From database searches and gene predictions, we identified 53 and 95 putatively functional (i.e. not disrupted by stop codon and/or frameshift) OR genes in medaka and stickleback, respectively. We also found 23 V2R genes in stickleback and 17 TAAR genes in pufferfish. Nucleotide sequences of these genes are available as supporting information (Texts S1, S2, S3, and S4; Data S1, S2, S3, and S4). In this study, phylogenetic trees of ORs, TAARs, V1Rs, and V2Rs in five model fishes were constructed using these sequences and published data (see Materials and methods).
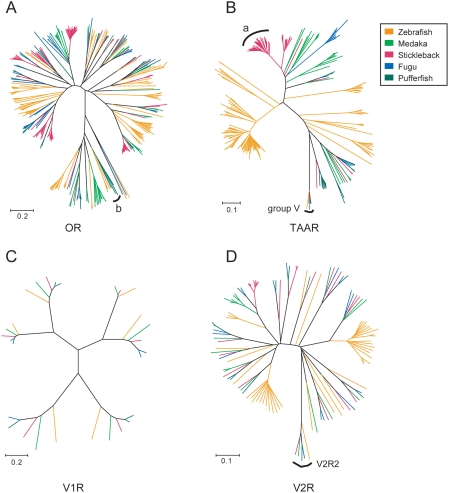
Fig. 2 shows the unrooted phylogenetic trees of ORs, TAARs, V1Rs, and V2Rs in the five fishes. Many species-specific clades were observed in teleost ORs and TAARs. In contrast, such species-specific clusters were rarely seen in teleost V2Rs. In teleost V1Rs, no species-specific gene duplications were found. To quantify the differences in phylogenetic patterns among the four evolutionary distinct chemosensory receptor families, the proportion of “species-specific” genes (i.e., genes that originated from species-specific gene gains and/or losses; see Materials and methods) was estimated for each chemosensory receptor family in each species. The numbers of chemosensory receptor genes and species-specific genes, as well as the proportion of species-specific genes in each chemosensory receptor family are shown in Table 1. For each of the five species, the proportion of species-specific genes in TAARs was higher than those of ORs and V2Rs (Table 1), although with the exception of one comparison between TAARs and V2Rs in fugu (p = 0.011, Fisher's exact test), the differences were not significant. With respect to the total numbers of genes in the five species, the proportion of species-specific TAAR genes was significantly higher than those of OR (p = 0.012) and V2R (p = 0.002) genes. The difference in the proportion of species-specific genes between ORs and V2Rs was not significant (p = 0.156). These trends might reflect the differences of the evolutionary patterns in the teleost fish chemosensory receptor families.
Figure 2. Unrooted phylogenetic trees of putatively functional (A) ORs, (B) TAARs, (C) V1Rs, and (D) V2Rs in zebrafish, medaka, stickleback, fugu, and pufferfish.
The trees were reconstructed using the neighbor-joining method with Poisson-corrected protein distances. The color of each branch indicates species.
Table 1. Numbers of putatively functional genes, species-specific genes, and the proportion of species-specific genes in OR, TAAR, V1R, and V2R gene families in five fishes.
| Gene family | Zebrafish | Medaka | Stickleback | Fugu | Pufferfish | Total | |
| OR | No. of genes | 102 | 53 | 95 | 44 | 42 | 336 |
| No. of species-specific genes | 71 | 27 | 70 | 10 | 15 | 193 | |
| Proportion of species-specific genes | 0.70 | 0.51 | 0.74 | 0.23 | 0.36 | 0.57 | |
| TAAR | No. of genes | 109 | 25 | 49 | 13 | 17 | 213 |
| No. of species-specific genes | 101 | 14 | 40 | 8 | 11 | 174 | |
| Proportion of species-specific genes | 0.93 | 0.56 | 0.82 | 0.62 | 0.65 | 0.82 | |
| V1R | No. of genes | 6 | 6 | 6 | 5 | 5 | 28 |
| No. of species-specific genes | 0 | 0 | 0 | 0 | 0 | 0 | |
| Proportion of species-specific genes | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| V2R | No. of genes | 46 | 17 | 23 | 15 | 11 | 112 |
| No. of species-specific genes | 31 | 6 | 10 | 0 | 1 | 48 | |
| Proportion of species-specific genes | 0.67 | 0.35 | 0.44 | 0.00 | 0.09 | 0.43 |
Selective forces operating on each teleost chemosensory receptor gene family
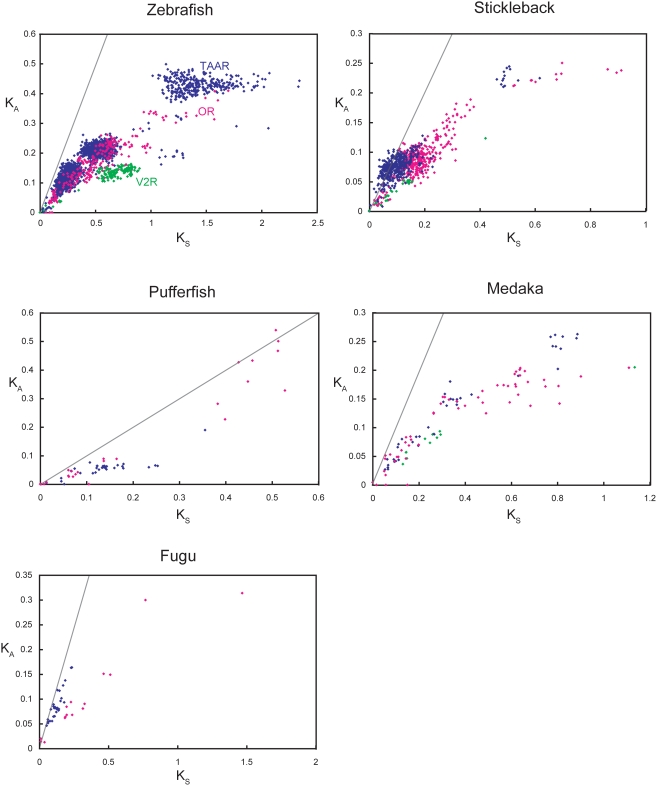
Fig. 3 shows scatterplots of pairwise non-synonymous (KA) and synonymous (KS) nucleotide substitution rates of the species-specific OR, TAAR, and V2R genes in the five fishes examined. In fugu and pufferfish, KA and KS values in V2Rs were not estimated because virtually no species-specific V2Rs were found. Selective constraints for each of three chemosensory receptor families were substantially different. In zebrafish, stickleback, and fugu, KA/KS ratios of TAARs were higher than those of ORs. In particular, most stickleback TAAR genes in clade a (Fig. 2) showed more than 1.0 (ca. 1.0–2.1) pairwise KA/KS ratios. This indicates that the clade a TAAR genes in stickleback have evolved under positive selection. Also in zebrafish and stickleback, V2R genes showed relatively lower KA/KS ratios than OR and TAAR genes (Fig. 3). This suggests that most V2R genes in these teleosts have evolved under strong functional constraints. However, these trends were not common in all teleost fish species examined. In pufferfish, KA/KS ratios of OR genes were higher than those of TAAR genes, which was opposite of the zebrafish, stickleback, and fugu results (Fig. 3). Interestingly, several distantly related (KS: 0.4–0.55) pairs of OR genes showed high KA/KS ratios (ca. 0.7–1.0) in pufferfish. All of these genes belonged to clade b (Fig. 2). In contrast to the other teleost fishes, the KA/KS ratios of species-specific genes in medaka did not clearly differ among OR, TAAR, and V2R gene families (Fig. 3).
Figure 3. Numbers of synonymous and nonsynonymous substitutions per site of the species-specific genes in five fishes.
The gray line indicates KS = KA.
Discussion
Evolutionary patterns of the four chemosensory receptor gene families in teleost fishes
In this study, we estimated the frequencies of species-specific gene gains/losses of the four evolutionary distinct chemosensory receptor gene families in zebrafish, medaka, stickleback, fugu, and pufferfish (Table 1), by calculating the proportions of species-specific genes from the phylogenetic trees (Fig. 2). We also estimated the selective constraints for each chemosensory receptor gene family (Fig. 3). In this section, we summarize the evolutionary patterns of the four chemosensory receptor gene families. Discussion in this section is somewhat speculative because there is no functional data of fish odorant/pheromone receptors.
The proportion of species-specific genes was lower in ORs than in TAARs, although some species-specific clades were observed in all species (Fig. 2). In addition, the KA/KS ratios of OR genes in zebrafish, stickleback, and fugu were clearly lower than those of TAAR genes (Fig. 3). These characteristics imply that most teleost ORs are conserved among lineages. Thus, teleost ORs are likely to perceive “common” odorants for fishes. However, a portion of teleost ORs may have evolved to recognize some species-specific chemicals. For instance, clade b ORs in pufferfish (Fig. 2) showed substantially high KA/KS ratios (Fig. 3). These ORs are considered to have evolved under positive selection. This implies that in the pufferfish lineage, the clade b ORs might adapt to odorants of some lineage-specific environments, such as freshwater habitats.
Phylogenetic analyses have revealed that many teleost TAARs formed species-specific clades (Fig. 2). In all species examined, the proportion of lineage-specific genes in the TAAR family was higher than those in the other families (Table 1). Exceptionally, group V TAARs [13] had strict orthologs in all teleost fish species (Fig. 2). It was suggested that these genes were not expressed in the olfactory organ [13]. Thus, the group V TAARs may have some other functions than odor detection. In zebrafish, stickleback, and fugu, KA/KS ratios of TAAR genes were substantially higher than those of OR and V2R genes (Fig. 3). In particular, the KA/KS ratios of many stickleback TAARs in clade a exceeded 1.0 (Fig. 3), indicating strong positive selection. Such a species-specific gene repertoire and high KA/KS ratios may imply that teleost TAARs are used for detecting species-specific chemicals, such as pheromones. In mice, TAARs are known to be the receptors for biogenic amines [14]. Physiological studies have shown that teleost fishes have olfactory sensitivity to several biogenic amines [15]–[17]. Interestingly, catecholamines or their metabolites are thought to be used for chemical communication in goldfish Carassius auratus [15]. Odor-based mate choice and conspecific recognition have been reported in stickleback [18], [19], suggesting that pheromonal cues are crucial for their reproduction and pre-mating isolation. The clade a TAAR genes might be involved in the reproductive behaviors in stickleback.
In contrast to ORs and TAARs, the repertoires of vomeronasal-type odorant receptors seem to be conserved among lineages. In teleost fishes, only five or six V1R genes have been identified in each species [20]. All V1R genes retain 1∶1 orthologs, no species-specific gene duplications being observed (Table 1). This may suggest that teleost V1Rs are used for detecting odor chemicals that are important for all teleost fishes.
Phylogenetic analysis revealed that most V2R genes including V2R2 genes had strict orthologs in five fishes examined (Fig. 2). The proportion of species-specific genes in teleost V2Rs is lower than those of ORs and TAARs (Table 1). The KA/KS ratios of V2R genes are also smaller than those of ORs and TAARs (Fig. 3). Teleost V2Rs are suggested to perceive amino acids [9], [10] that are very common environmental odors for teleost fishes [21]. In many teleost species, to recognize and to discriminate amino acids is very important for survival. Thus, the repertoire and functions of V2R genes may be maintained among teleost fish species.
Contrasting evolutionary modes between teleost fish and tetrapod chemosensory receptor genes
Our analysis revealed that, in teleost fishes, proportions of species-specific genes were not clearly different among different receptor families (Table 1), although slight differences were suggested. This is a very contrasting evolutionary pattern to the tetrapod chemoreceptors. In tetrapods, the proportions of species-specific OR and TAAR genes are much lower than those of V1Rs and V2Rs [11]. This pattern could be explained by the emergence of the vomeronasal organ in the tetrapod lineage. Mammalian V1Rs and V2Rs are expressed exclusively in the vomeronasal organ that is considered to detect pheromones [3]. Indeed, several V1Rs and V2Rs are shown to respond to pheromonal substances [6], [22]. In tetrapods, separation of the two chemosensory organs, the VNO and the MOE, may promote functional changes of vomeronasal receptors. On the other hand, teleost V1Rs and V2Rs might maintain their original functions as receptors for environmental odors, such as amino acids.
The most remarkable difference in the evolution of chemoreceptor gene families between tetrapods and teleosts is the difference of TAAR gene repertoires. Teleost fishes have relatively larger number of TAAR genes than that in tetrapods. In addition, the proportion of species-specifc TAAR genes was higher in teleost fishes, suggesting that fish TAARs have experienced frequent gene gains/losses. In tetrapods, pheromone recognition by vomeronasal receptors may reduce the functional significance of TAARs, or may cause the use of different type of pheromonal substances, instead of biogenic amines. This might explain the relatively small TAAR gene repertoires in frogs, chicken, and mammals [23].
In this study, we showed the contrasting evolutionary patterns among ORs, TAARs, V1Rs, and V2Rs in teleost fishes. We also revealed that several OR genes in pufferfish and TAAR genes in stickleback were under positive selection. These genes are possibly involved in some lineage-specific adaptive evolutions. Our results provide useful information for future functional studies in teleost fish olfaction.
Materials and Methods
Data mining
The sequence data of teleost fish chemoreceptor genes were collected from the following studies: zebrafish and fugu ORs [24]; pufferfish ORs [25]; teleost V1Rs [20]; V2Rs in zebrafish, medaka, fugu, and pufferfish [25]; TAARs in zebrafish, medaka, stickleback, and fugu [13]. Second, we newly identified medaka and stickleback ORs, stickleback V2Rs, and pufferfish TAARs from their draft genome sequences, by using TBLASTN searches and profile hidden Markov Model (HMM)-based gene prediction. The detailed method of gene identification is described in [25]. The nucleotide sequences of these genes and the list of their genomic positions are available as supplementary information.
Data analysis
For each of the four chemosensory receptor gene families, deduced amino acid sequences were aligned by the program MAFFT 5.861 [26]. Phylogenetic trees were constructed using the neighbor-joining method with Poisson-corrected protein distances implemented in the program MEGA 4 [27].
From the phylogenetic trees, we estimated the species-specific gene gains and losses by the method described in [11]. If a species-specific clade consists of n genes, at least n-1 events of gene gains/losses must have taken place in the clade since the species diverged from its closest relative in the five fishes. Thus, the total proportion of genes belonging to species-specific clades is defined as the sum of these n-1 genes for all species-specific clades divided by the total number of genes for that chemosensory receptor type in that species [11]. Species-specific clades were identified from the phylogenetic trees as clades supported with >50% bootstrap values.
To test selective constraints to the chemosensory receptors in teteost fishes, KS and KA values among all pairs of genes within each lineage-specific clade were calculated in OR, TAAR, and V2R gene families. The KA and KS values were estimated by Nei and Gojobori (1986) method [28] using DnaSP version 4.0 [29].
Supporting Information
Names and positions of medaka OR genes and pseudogenes.
(0.02 MB XLS)
Names and positions of stickleback OR genes and pseudogenes.
(0.04 MB XLS)
Names and positions of pufferfish TAAR genes and pseudogenes.
(0.03 MB XLS)
Names and positions of stickleback V2R genes and pseudogenes.
(0.04 MB XLS)
Nucleotide sequences of medaka OR genes and pseudogenes (FASTA format text file).
(0.06 MB TXT)
Nucleotide sequences of stickleback OR genes and pseudogenes (FASTA format text file).
(0.11 MB TXT)
Nucleotide sequences of pufferfish TAAR genes and pseudogenes (FASTA format text file).
(0.02 MB TXT)
Nucleotide sequences of stickleback V2R genes and pseudogenes (FASTA format text file).
(0.07 MB TXT)
Acknowledgments
The stickleback, medaka, and pufferfish genomic data were produced by the Broad Institute, National Institute of Genetics, and Genoscope, respectively. We thank Y. Yamanoue (University of Tokyo) for permission to use fish illustrations.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors are supported by Grant-in-Aid from the Japan Society for Promotion of Science (no. 19207707). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Baxi KN, Dorries KM, Eisthen HL. Is the vomeronasal system really specialized for detecting pheromones? Trends Neurosci. 2006;29:1–7. doi: 10.1016/j.tins.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Spehr M, Kelliher KR, Li X-H, Boehm T, Leinders-Zufall T, et al. Essential role of the main olfactory system in social recognition of major histocompatibility complex peptide ligands. J Neurosci. 2006;26:1961–1970. doi: 10.1523/JNEUROSCI.4939-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mombearts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- 4.Boschat C, Pelofi C, Randin O, Roppolo D, Luscher C, et al. Pheromone detection mediated by a V1r vomeronasal receptor. Nat Neurosci. 2002;5:1261–1262. doi: 10.1038/nn978. [DOI] [PubMed] [Google Scholar]
- 5.Leinders-Zufall T, Brennan P, Widmayer P, Chandramani SP, Maul-Pavicic A, et al. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science. 2004;306:1033–1037. doi: 10.1126/science.1102818. [DOI] [PubMed] [Google Scholar]
- 6.Kimoto H, Haga S, Sato K, Touhara K. Sex-specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature. 2005;437:898–901. doi: 10.1038/nature04033. [DOI] [PubMed] [Google Scholar]
- 7.Cao Y, Oh BC, Stryer L. Cloning and localization of two multigene receptor families in goldfish olfactory epithelium. Proc Natl Acad Sci U S A. 1998;95:11987–11992. doi: 10.1073/pnas.95.20.11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asano-Miyoshi M, Suda T, Yasuoka A, Osima S, Yamashita S, et al. Random expression of main and vomeronasal olfactory receptor genes in immature and mature olfactory epithelia of Fugu rubripes. J Biochem Tokyo. 2000;127:915–924. doi: 10.1093/oxfordjournals.jbchem.a022687. [DOI] [PubMed] [Google Scholar]
- 9.Sorensen PW, Sato K. Second messenger systems mediating sex pheromone and amino acid sensitivity in goldfish olfactory receptor neurons. Chem Senses. 2005;30(suppl. 1):i315–i316. doi: 10.1093/chemse/bjh241. [DOI] [PubMed] [Google Scholar]
- 10.Hansen A, Rolen SH, Anderson K, Morita Y, Caprio J, et al. Correlation between olfactory receptor cell type and function in the channel catfish. J Neurosci. 2003;15:9328–9339. doi: 10.1523/JNEUROSCI.23-28-09328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grus WE, Zhang J. Distinct evolutionary patterns between chemoreceptors of 2 vertebrate olfactory systems and the differential tuning hypothesis. Mol Biol Evol. 2008;25:1593–1601. doi: 10.1093/molbev/msn107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamanoue Y, Miya M, Inoue JG, Matsuura K, Nishida M. The mitochondrial genome of spotted green pufferfish Tetraodon nigroviridis (Teleostei: Tetraodontiformes) and divergence time estimation among model organisms in fishes. Genes Genet Syst. 2006;81:29–39. doi: 10.1266/ggs.81.29. [DOI] [PubMed] [Google Scholar]
- 13.Hashiguchi Y, Nishida M. Evolution of trace amine-associated receptor (TAAR) gene family in vertebrates: lineage-specific expansions and degradations of a second class of vertebrate chemosensory receptors expressed in the olfactory epithelium. Mol Biol Evol. 2007;24:2099–2107. doi: 10.1093/molbev/msm140. [DOI] [PubMed] [Google Scholar]
- 14.Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- 15.Hubbard PC, Barata EN, Canario AVM. Olfactory sensitivity to catecholamines and their metabolites in the goldfish. Chem Senses. 2003;28:207–218. doi: 10.1093/chemse/28.3.207. [DOI] [PubMed] [Google Scholar]
- 16.Michel WC, Sanderson MJ, Olson JK, Lipschitz DL. Evidence of a novel transduction pathway mediating detection of polyamines by the zebrafish olfactory system. J Exp Biol. 2003;206:1697–1706. doi: 10.1242/jeb.00339. [DOI] [PubMed] [Google Scholar]
- 17.Rolen SH, Sorensen PW, Mattson D, Caprio J. Polyamines as olfactory stimuli in goldfish Carassius auratus. J Exp Biol. 2003;206:1683–1696. doi: 10.1242/jeb.00338. [DOI] [PubMed] [Google Scholar]
- 18.Milinski M, Griffiths S, Wegner KM, Reusch TBH, Haas-Assenbaum A, et al. Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc Natl Acad Sci U S A. 2005;102:4414–4418. doi: 10.1073/pnas.0408264102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rafferty NE, Boughman JW. Olfactory mate recognition in a sympatric species pair of three-spined sticklebacks. Behav Ecol. 2006;17:965–970. [Google Scholar]
- 20.Saraiva LR, Korsching SI. A novel olfactory receptor gene family in teleost fish. Genome Res. 2007;17:1448–1457. doi: 10.1101/gr.6553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara TJ. Olfaction and gustation in fish: an overview. Acta Physiol Scand. 1994;152:207–217. doi: 10.1111/j.1748-1716.1994.tb09800.x. [DOI] [PubMed] [Google Scholar]
- 22.Del Punta K, Leinders-Zufall T, Rodriguez I, Jukam D, Wysocki CJ, et al. Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature. 2002;419:70–74. doi: 10.1038/nature00955. [DOI] [PubMed] [Google Scholar]
- 23.Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci U S A. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alioto TS, Ngai J. The odorant receptor gene repertoire of teleost fish. BMC Genomics. 2005;6:173. doi: 10.1186/1471-2164-6-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashiguchi Y, Nishida M. Evolution and origin of vomeronasal-type odorant receptor gene repertoire in fishes. BMC Evol Biol. 2006;6:76. doi: 10.1186/1471-2148-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 28.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 29.Rozas J, Sánchez-DelBarrio JC, Messegyer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Names and positions of medaka OR genes and pseudogenes.
(0.02 MB XLS)
Names and positions of stickleback OR genes and pseudogenes.
(0.04 MB XLS)
Names and positions of pufferfish TAAR genes and pseudogenes.
(0.03 MB XLS)
Names and positions of stickleback V2R genes and pseudogenes.
(0.04 MB XLS)
Nucleotide sequences of medaka OR genes and pseudogenes (FASTA format text file).
(0.06 MB TXT)
Nucleotide sequences of stickleback OR genes and pseudogenes (FASTA format text file).
(0.11 MB TXT)
Nucleotide sequences of pufferfish TAAR genes and pseudogenes (FASTA format text file).
(0.02 MB TXT)
Nucleotide sequences of stickleback V2R genes and pseudogenes (FASTA format text file).
(0.07 MB TXT)