Abstract
Conditional control of protein function in vivo offers tremendous potential for deconvoluting the roles of individual proteins in complex systems. We recently developed a method in which a small protein domain termed a destabilizing domain confers instability to fusion protein partners in cultured cells. Instability is reversed when a cell-permeable small molecule binds to this domain. Here we describe the use of this system to regulate protein function in living mammals. We demonstrate regulation of secreted proteins and their biological activity, with conditional secretion of an immunomodulatory cytokine resulting in tumor burden reduction in mouse models. Additionally, we use this approach to control the function of a specific protein following systemic delivery of its gene to a tumor, suggesting uses for enhancing the specificity and efficacy of targeted gene-based therapies. This method represents a novel strategy to regulate protein function in living organisms with an extraordinary level of control.
Introduction
Effective methods to regulate gene expression or protein function within mammalian hosts are essential for understanding basic biological mechanisms as well as directing gene-based therapies. Generating knockouts of a specific gene and subsequent monitoring of that perturbation in vivo is a powerful technique for understanding ways in which proteins contribute to complex biological processes (1-4). However, the gene of interest may be essential for development, or cellular or molecular compensation may occur during embryonic development, precluding or confounding the study of protein function in an adult organism (5, 6). Strategies to overcome this limitation have introduced elements of spatial and temporal control over gene expression (7). The Cre-lox system (8, 9) and tetracycline responsive transcriptional switch (10) have been used with great success; however, perturbations are either irreversible, or inherent reversibility is dependent upon stability of the protein product, the degradation of which can take days. Small-molecule activators or inhibitors of protein function target the protein product directly (11-15); however, the discovery process is arduous, and many small molecules lack either specificity (16) or optimal properties for use in vivo (17, 18).
We recently described a general method to achieve post-translational regulation of protein function in cultured cells (19). We engineered small, inherently unstable domains that confer instability to any fused partner proteins. When expressed in cells, these fusions are rapidly and constitutively degraded in a proteasome-dependent fashion. Addition of a stabilizing ligand that binds specifically to this destabilizing domain protects the fusion protein from degradation, allowing the protein of interest to perform its normal cellular function. Specifically, a point mutant (L106P) of the 107-amino acid protein FKBP confers instability to fusion partners, and this instability is reversed by a synthetic ligand named Shield-1. The combination of genetic manipulation coupled with small-molecule regulation endows this system with the specificity of gene targeting along with the speed, tunability, and reversibility inherent to small molecule control. Here, we describe the use of this system in living animals.
Results
Tunable regulation of protein stability in vivo
To assess the potential of this technology in mice, we attempted to conditionally regulate luciferase stability, using bioluminescence as an indicator of intracellular luciferase levels. The FKBP L106P destabilizing domain was cloned at the N-terminus of a thermostable luciferase (L106P-tsLuc) and stably integrated into the HCT116 colon cancer line. Cells expressing the fusion protein were tested for conditional regulation of luciferase activity and then xenografted into immunodeficient mice. Shield-1 was delivered intraperitoneally (ip) at a dose of 10 mg/kg and luciferase activity was measured in the mice using in vivo bioluminescence imaging (Fig. 1a). Maximum expression levels were observed 12 hours after treatment with Shield-1, and bioluminescent signals returned to background within 48 hours. These data suggest that Shield-1 is delivered systemically and maintained at sufficient levels within target cells to stabilize the fusion protein for a significant period of time before being cleared. Quantification of this data shows an approximate 10-fold increase in signal (Fig. 1b), in agreement with the dynamic range observed for luciferase activity in this cell line in culture (Supplementary Fig. 1). While this dynamic range is smaller than that observed for HCT116 cells expressing the L106P domain fused to the tomato fluorescent protein (which displayed an approximate 50-fold dynamic range with little detectable background signal; Supplementary Fig. 2), it is sufficient for studying the dynamics and kinetics of protein stabilization in vivo.
Figure 1.
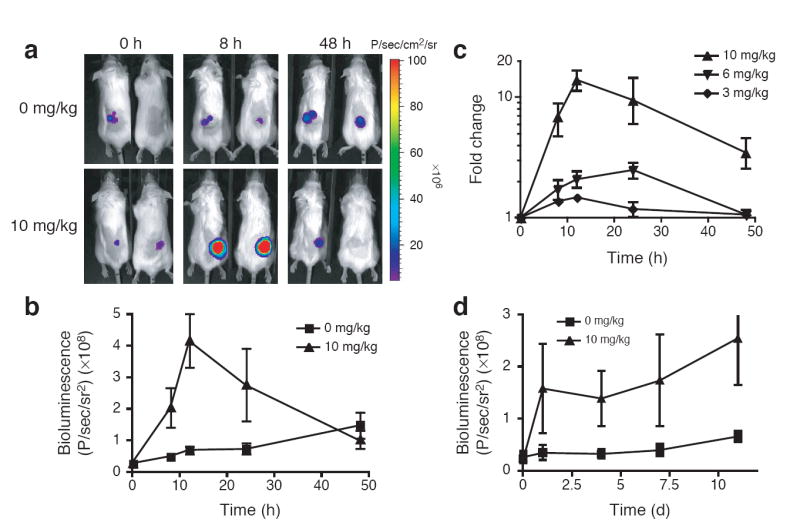
Conditional regulation of protein stability in vivo. (a) SCID mice bearing HCT116 L106P-tsLuc xenografts (50-100 mm3) were either untreated (top) or treated ip with Shield-1 (10 mg/kg, bottom) and bioluminescent signals were imaged over time. (b) Quantification of tumor signals from panel A. (c) Mice as in a were treated ip with Shield-1 at 3 mg/kg (diamonds), 6 mg/kg (inverted triangles), or 10 mg/kg (triangles) and imaged over time. (d) Mice bearing HCT116-tsLuc xenografts were either untreated (squares) or treated with Shield-1 (10 mg/kg, triangles) every 48 hr and imaged over time. Data for panels b-d are presented as the average bioluminescence detected within regions of interest drawn around the tumors ± SEM (n=4 to 10).
Small molecules are attractive as modulators of protein function due in part to their ability to regulate protein activity in a dose-dependent manner ((19) and Supplementary Figs. 1 and 2), so we tested for dose-dependent protein stability in the more complex environment of a living biological system. Mice bearing HCT116 L106P-tsLuc xenografts were treated with various doses of Shield-1 (3-10 mg/kg). As expected, bioluminescence levels increased with increasing doses of Shield-1 (Fig. 1c). The ability to tunably regulate protein levels in living systems should prove to be a powerful perturbation technique, especially when the function of a target protein is dependent upon its intracellular concentration (20, 21).
In cases where a gene is essential for development, levels of Shield-1 must be maintained until a time at which protein function may be abrogated. Therefore, we wanted to ascertain whether repeated Shield-1 delivery would allow for constant expression levels in vivo. Animals treated with Shield-1 (10 mg/kg) once every 48 hours maintained increased bioluminescence levels over the course of several weeks (Fig. 1d), suggesting that an infrequent and low-dose treatment regimen with Shield-1 is sufficient to maintain protein expression levels in vivo. Importantly, we observed no adverse effects on the animals (e.g., feeding behavior and body weight, overall activity), consistent with microarray analysis of mRNA levels in cells treated with Shield-1 which demonstrated no appreciable cellular response to treatment (22), as well as clinical studies of a structurally similar FKBP ligand in humans (23).
Conditional regulation of a secreted protein leads to reduction of tumor burden
We next applied this method to conditionally stabilize a biologically relevant protein in vivo to correlate protein levels with a physiological response. Interleukin-2 (IL-2) is a cytokine that is instrumental in inducing the differentiation and proliferation of a variety of lymphocyte populations (24). Recombinant IL-2 is used in a variety of cancer therapies; furthermore, IL-2 fusion proteins impart tumor-protective immunity in a number of therapeutic applications (25). Regulation of IL-2 would represent the first use of this technology for secreted proteins, a class of proteins controlling physiological processes ranging from metabolism to growth and differentiation. Our regulation of transmembrane glycoprotein levels at the cell surface in culture (19), and demonstrated conditional secretion of Gaussia luciferase (Supplementary Fig. 3) suggested that destabilizing domains might successfully regulate proteins that are trafficked through the secretory pathway.
The L106P destabilizing domain was inserted between the IL-2 signal peptide and the sequence encoding the secreted protein (L-L106P-IL-2) and stably introduced into HCT116 cells. Upon exposure to various concentrations of Shield-1, a dose-dependent increase in the L106P-IL-2 fusion protein was observed in the cell culture media (approximately 25-fold; Fig. 2a). L106P-IL-2 was further determined to be functionally active in an NK cell proliferation assay (Supplementary Fig 4). HCT116 L106P-IL-2 cells were subcutaneously xenografted into mice able to produce functional B and NK cells, and the ability of these cells to establish tumors and proliferate under Shield-1 treatment was monitored (Fig. 2b). Tumors failed to establish in mice that received HCT116 L-L106P-IL-2 cells that had been pretreated with Shield-1 and were subjected to Shield-1 treatment (10 mg/kg) every 48 hours (i.e., causing constitutive L106P-IL-2 secretion). To determine if regulated secretion of L106P-IL-2 from an established tumor might lead to a reduction in tumor burden, tumors were established in mice for 5 days. Mice were then treated with Shield-1 ip at either 5 or 10 mg/kg every 48 hours, and tumor volume was monitored over time. Tumor regression was observed in both treated groups, and by day 9 the tumor burden in these two groups was reduced to levels identical to that of mice pretreated with Shield-1. As expected, tumors in the untreated control group continued to increase in size over the course of the experiment. By day 16, all Shield-1 treated groups displayed significantly reduced tumor burden relative to untreated animals (p<0.05).
Figure 2.
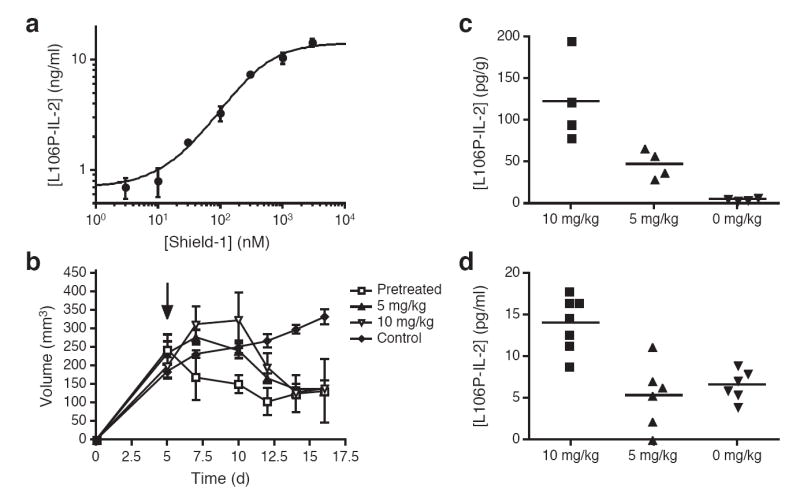
Conditional regulation of a secreted immunomodulatory protein leads to a reduction of tumor burden in vivo. (a) HCT116 L-L106P-IL-2 cells were treated with various concentrations of Shield-1 and culture media was assayed for the presence of IL-2. Data are represented as the average IL-2 concentration ± SEM (n=3). (b) CD1 nu-/nu- mice bearing subcutaneous HCT116 L-L106P-IL-2 tumors were either untreated (diamonds) or treated ip with Shield-1 at 5 mg/kg (triangles) or 10 mg/kg (inverted triangles) every 48 hr beginning 5 days post-transplantation (arrow). Alternatively, mice received HCT116 L-L106P-IL-2 cells that had been pre-treated with 1 μM Shield-1 for 24 hr, and were then treated with Shield-1 (10 mg/kg) every 48 hr beginning on day 0 (squares). Tumor volume was determined by caliper measurement and monitored over time. Data are represented as the average tumor volume ± SEM (n=5). At Day 16, all Shield-1 treated groups displayed significantly reduced tumor burden relative to controls (p=0.0019 for 10 mg/kg; p=0.0002 for 5 mg/kg and p=0.0046 for pre-treat group). (c) Tumors from mice treated with Shield-1 post-transplantation were collected 48 hr after the start of Shield-1 treatment. Tumors were weighed ex vivo and then homogenized, and the concentration of IL-2 per gram tumor tissue was determined by ELISA (n=4). Tumors from mice treated with Shield-1 at 10 mg/kg contained significantly higher levels of IL-2 than tumors from mice treated at 5 mg/kg (p=0.032), which in turn produced more IL-2 than tumors from untreated mice (p=0.0028). (d) Mice treated with Shield-1 post-transplantation were bled 48 hr after the start of Shield-1 treatment, and the concentration of IL-2 in the serum was determined by ELISA. Shield-1 treatment at 10 mg/kg (n=7) produced significantly higher levels of serum IL-2 relative to control mice (n=6, p=0.0004), whereas treatment at 5 mg/kg (n=6) did not produce any increase in serum IL-2 levels.
This technology allows dose-dependent regulation of protein levels in vivo, however we did not observe any appreciable difference in the reduction of tumor burden between mice treated at 5 or 10 mg/kg Shield-1. Analysis of L106P-IL-2 levels within the tumors of treated and untreated mice confirmed dose-dependent production of L106P-IL-2 at the tumor site (Fig. 2c) (10 mg/kg dose produced significantly greater L106P-IL-2 within the tumor than 5 mg/kg; p=0.032). Analysis of serum L106P-IL-2 levels showed significantly elevated levels in the 10 mg/kg Shield-1 group relative to tumor-bearing mice not receiving Shield-1 (p=0.0004), whereas L106P-IL-2 serum levels in mice treated with 5 mg/kg Shield-1 were no different than those observed for the control group (Fig. 2d). These observations suggest that treatment with Shield-1 results in tunable, dose-dependent secretion of a cytokine in vivo, and that different biological effects are achieved by varying the dose of Shield-1. A dose of 5 mg/kg Shield-1 was capable of producing the same robust anti-tumor response as 10 mg/kg Shield-1, without the potential for toxicity associated with systemic distribution in conventional high-dose recombinant IL-2 therapies (26). In addition, mice bearing tumors derived from HCT116 cells alone and treated with 10 mg/kg Shield-1 (n=5) displayed the same serum IL-2 levels as untreated mice bearing HCT116 L-L106P-IL-2 tumors, indicating that Shield-1 does not induce IL-2 production from other cells (data not shown).
Systemic delivery of a conditionally regulated protein enhances the efficacy of a targeted biological therapy
We previously described a viral gene delivery system for the systemic treatment of cancer based on a replication-selective (oncolytic) strain of vaccinia virus (hereafter vvDD), with deletions of viral thymidine kinase and growth factor genes restricting viral replication to cancer cells (27). Intravenous delivery of vvDD results in initial infection of both tumor cells and cells in other tissues, followed by rapid replication and spread of the viral vector (and any transgene in the viral genome) within the tumor and viral clearance from other tissues. Because vvDD can express transgenes in tumor cells following systemic delivery, it is an attractive model system to examine whether our technology might also be used to control protein function in an experimental model of a therapeutic application.
We established that infection of cultured cells with vvDD did not affect the kinetics or dynamic range of L106P-regulated protein stability (Supplementary Fig. 5), and that Shield-1 did not affect the replication of the virus (data not shown). We next constructed strains of vvDD expressing either an L106P-tsLuc or L106P-tomato fusion protein and verified Shield-1 dependent activity in cultured cells (Fig. 3a and Supplementary Fig. 6). While the dynamic range observed for L106P-tsLuc bioluminescence was modest, conditional regulation of virally delivered L106P-tomato resulted in greater than 100-fold increase in fluorescence levels upon treatment with Shield-1, with no background signal observed. We then assayed for Shield-1 dependent bioluminescence of virally delivered L106P-tsLuc in a mouse tumor model (Figs. 3b and 3c). By allowing 72 hours for viral infection to establish within the tumor and to be cleared from most other tissues before beginning Shield-1 treatment, we were able to selectively stabilize the protein of interest specifically in the tumor as the target tissue.
Figure 3.
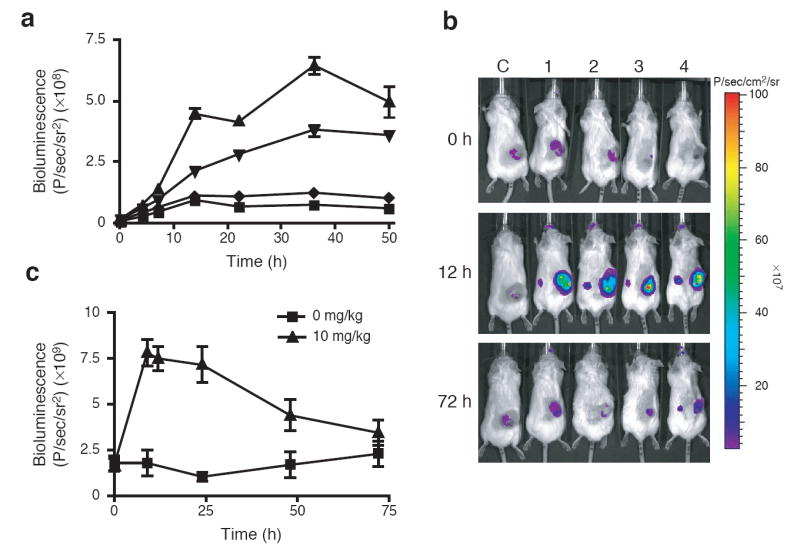
Systemic, targeted-delivery of a conditionally stabilized protein. (a) HCT116 cells were infected with vvDD L106P-tsLuc and then mock treated (squares) or treated with Shield-1 at 1 μM (triangles), 100 nM (inverted triangles), or 10 nM (diamonds). Data are represented as the average luminescence ± SEM (n=3). (b) SCID mice bearing subcutaneous HCT116 xenografts (50-100 mm3) received a single tail vein injection of vvDD L106P-tsLuc (1×108 PFU/mouse). After 72 hr, mice were either untreated (C, control) or treated with Shield-1 (10 mg/kg, #1-4). Bioluminescent signals were imaged over time. (c) Quantification of bioluminescent signal produced from regions of interest drawn around tumors as in panel b. Data presented are the average bioluminescence ± SEM (n=4).
To further test the ability of this approach to regulate therapeutic proteins, we applied this technology to the cytokine TNF-α. Despite its proven cytotoxic effects against primary tumors, systemic toxicity has limited the clinical use of TNF-α to local applications (e.g., isolated limb or organ perfusion) (28). In addition, TNF-α displays antiviral effects to which the vaccinia strain Western Reserve (the backbone for vvDD) is susceptible (29). An L-L106P-TNF-α chimeric gene was inserted into a strain of vvDD constitutively expressing luciferase, and dose-dependent control of the secreted L106P-TNF-α fusion protein from cultured cells was verified (Supplementary Fig. 7). Mice bearing large (150-250 mm3) subcutaneous HCT116 tumors were then treated with a single intravenous injection of this virus or a PBS control. Shield-1 (10 mg/kg) was delivered ip to the mice every 48 hours, starting either 1 day prior to vvDD treatment (resulting in constitutive viral expression and stabilization of TNF-α), 3 days post-treatment (resulting in stabilization of TNF-α only after vvDD gene expression is localized to the tumor), or not at all.
When Shield-1 dosing began prior to virus delivery (i.e., infected cells constitutively secrete TNF-α), we observed only modest levels of virus within the tumor (Fig. 4a) measured by constitutive viral luciferase expression. It is likely that secretion of TNF-α from infected cells rapidly targets these cells for destruction, limiting the ability of the virus to establish within the tumor. Despite reduced viral load in the tumors, TNF-α production resulted in significantly decreased tumor burden in this group relative to animals treated with virus alone (Fig. 4b, p<0.05), presumably due to the antitumor effects of the TNF-α. Shield-1 delivered alone had no effect on tumor burden. When Shield-1 administration was delayed until 3 days following virus treatment, initial delivery and persistence of viral infection measured by luciferase expression within the tumor were equivalent to animals treated with virus alone (Fig. 4a). In addition, the antitumor effects observed in this experimental group were significantly greater than those seen in any other treatment regimen (p<0.05). Seven of the eight animals demonstrated complete and durable responses (Fig. 4b), with no adverse physiological effects observed. The ability to regulate the stability of physiologically relevant or therapeutic proteins in this experimental setting illustrates the potential of this method to enhance a variety of biological therapies where reversible control of the timing and level of protein activity may be advantageous.
Figure 4.
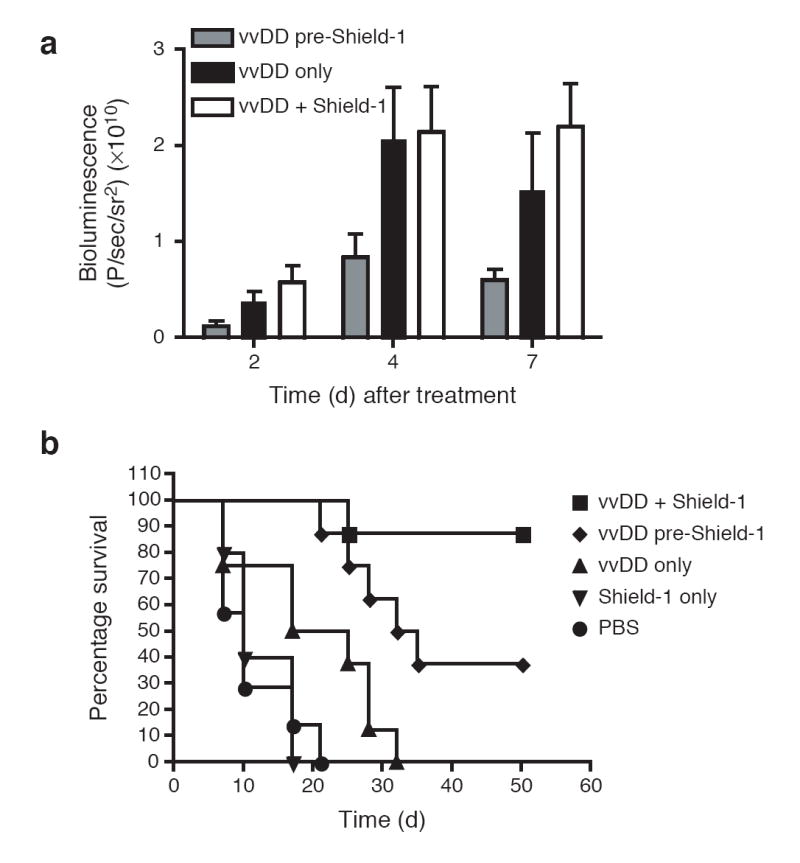
Antitumor benefit of conditional regulation of a targeted gene therapy vector. CD1 nu-/nu- mice bearing subcutaneous HCT116 tumors (150-250 mm3) were treated via a single tail vein injection with either PBS (circles) or the vaccinia strain vvDD expressing both luciferase as well as the L-L106P-TNF-α fusion protein (1 × 108 PFU/mouse). Treated mice also received Shield-1 (10 mg/kg) every 48 hr by three different protocols, starting (i) 24 hr prior to vvDD therapy (diamonds); (ii) 72 hr post-vvDD therapy (squares) or (iii) not at all (triangles) (n=8 mice/group). Tumor-bearing mice that did not receive vvDD therapy were also treated with Shield-1 (10 mg/kg) as a negative control (inverted triangles, n=5). (a) Viral load within the tumor was assayed by measuring constitutive viral gene expression (bioluminescence) for each group at the indicated time points after vvDD treatment. Shield-1 treatment starting 72 hr after vvDD (vvDD + Shield-1) resulted in significantly greater levels of viral gene expression in the tumor than when Shield-1 treatment was started prior to vvDD therapy (vvDD Pre-Shield-1; i.e., constitutive TNF-α expression) (p=0.035 at 2 days; p=0.035 at 4 days and p=0.002 at 7 days). (b) Tumor volume was determined by caliper measurement and mice were sacrificed once tumor volume reached 1.44 cm3. Kaplan-Meier survival graphs are shown, and all surviving mice at 50 days had no detectable tumor. Mice treated with Shield-1 prior to vvDD therapy (i.e., constitutive TNF-α expression) produced significantly enhanced survival relative to mice treated with vvDD alone (p=0.017). Addition of Shield-1 72 hr post-vvDD (vvDD + Shield-1) produced a further significant increase in survival relative to mice treated with Shield-1 prior to vvDD (vvDD pre-Shield-1) (p=0.031).
Conclusions
We have demonstrated that the FKBP-derived L106P destabilizing domain in concert with Shield-1 can confer conditional stability to a protein of interest in complex living organisms with excellent kinetics and dose control. We demonstrate regulation of secreted proteins in a dose-dependent manner, with conditional secretion of the physiologically relevant protein IL-2 resulting in biological effects (tumor regression) in vivo. By regulating the secretion of TNF-α, a protein of therapeutic value, we were able to demonstrate the benefits of controlling protein stability in a gene delivery vehicle, resulting in greatly enhanced anti-tumor effects once TNF-α was stabilized in the target tissue.
Central to any method of protein regulation involving the use of small molecules is the specificity of the perturbation. Our recent microarray analysis of cells treated with various concentrations of Shield-1 supports the notion that the small molecule causes little, if any, off-target cellular perturbation (22). Here, we demonstrate that treatment with Shield-1 alone does not improve the therapeutic outcome in tumor-bearing mice and Shield-1 treated mice maintain normal body weight, activity, and feeding behaviors. These observations suggest that any physiological responses observed are a direct result of the stabilization of a specific protein in a defined experimental context.
To date, we have successfully applied this approach to the regulated stability of more than 30 proteins, either in cultured cells or now, in vivo. We have used this technology to confer conditional stability to proteins of various subcellular localizations (19), and here we report the conditional regulation of secreted proteins. The broad applicability and simplicity of this approach suggest that this system may allow rapid and precise control of protein stability and function in a number of biological settings. The ability to regulate protein function in vivo with the precision and control conferred by this technology will likely prove useful for validating potential therapeutic targets in addition to advancing our understanding of basic biological processes and pathways.
Methods
Cell Culture and Transductions
Cell lines were cultured in DMEM with 10% fetal bovine serum (Invitrogen), 2 mM glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin. Thermostable luciferase (30) or the human IL-2 gene were cloned into pBMN L106P iBlasticidin and used to generate amphotropic retrovirus (19). HCT116 cells were incubated with retrovirus and polybrene (6 μg/mL) for 4 hrs at 37 °C and then selected with Blasticidin (5 μg/mL). Cells grown in 96-well plates (2 × 104 cells/well) were treated with Shield-1 as indicated and either bioluminescence measured using an IVIS 50 (Xenogen Product from Caliper Life Sciences) following luciferin addition (300 μg/mL), or media collected for ELISA.
Shield-1 Formulation and Delivery
Shield-1 was formulated in 50% N,N-dimethylacetamide and 50% of a 9:1 PEG-400:Tween-80 mixture. Shield-1 (40 μL) was administered intraperitoneally at 3, 6, or 10 mg/kg.
Mouse Models
SCID or CD1 nu-/nu- mice (Charles River Co.) received subcutaneous dorsal injections of ~1 × 107 cells, and tumors were allowed to establish as indicated. Animals were given an intraperitoneal injection of luciferin (225 mg/kg), anesthetized (2% isoflurane), and placed on the warmed stage (37 °C) of an IVIS 100 or IVIS 200 (Xenogen Product from Caliper Life Sciences) for imaging. Light produced was measured as photons/sec for designated regions of interest as described. Tumor volumes were also determined by caliper measurement, and mice were sacrificed when tumors reached 1.44 cm3 for survival assays. Serum samples were collected by retino-orbital bleedings and tumors collected post-mortem and homogenized for ELISA assay of cytokines as indicated. All experiments were run with institutional IACUC approval.
Vaccinia Virus Strains
CV1 cells were transfected with pSC-65 p7.5 L106P-tsLuc or pSC-65 p7.5 L106P-TNF-α pSE/L Luc and simultaneously infected with viral growth factor deleted Western Reserve Vaccinia (VSC20). Cassettes were integrated into the viral thymidine kinase gene by homologous recombination and selected by resistance to bromodeoxyuridine on 143B TK− cells. Single viral plaques were purified in 143B TK− cells.
vvDD Assays in Cultured Cells
HCT116 cells in a 96-well plate (2 × 104 cells/well) were incubated with vvDD carrying an L106P fusion (MOI>1) for 1 hr at 37 °C. Virus was removed and cells were treated with Shield-1. For luminescence assays, cells were incubated with luciferin (300 μg/mL) and imaged using an IVIS 50 (Xenogen product from Caliper Life Sciences). TNF-α in cell culture media was detected by ELISA.
Statistical Analyses
Two-tailed, unpaired Student’s T-tests were used, except for comparison of survival curves, when Gehan-Breslow-Wilcoxon test was used. Significance was achieved when p<0.05.
Shield-1
Reagent requests should be directed to T.J.W.
Supplementary Material
Acknowledgments
This work was supported by the NIH, through the Small Animal Imaging Resource Program, the In Vivo Cellular and Molecular Imaging Center (grant numbers R24 CA92862 and P50 CA114747) and GM073046, and the John A. and Cynthia Fry Gunn Research Fund. We thank T. Clackson for the Shield-1 formulation protocol.
References
- 1.Bockamp E, et al. Of mice and models: improved animal models for biomedical research. Physiol Genomics. 2002;11:115–132. doi: 10.1152/physiolgenomics.00067.2002. [DOI] [PubMed] [Google Scholar]
- 2.Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 3.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 4.Doetschman T, et al. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 5.Yamada G, et al. Targeted mutation of the murine goosecoid gene results in craniofacial defects and neonatal death. Development. 1995;121:2917–2922. doi: 10.1242/dev.121.9.2917. [DOI] [PubMed] [Google Scholar]
- 6.Poirier F, Robertson EJ. Normal development of mice carrying a null mutation in the gene encoding the L14 S-type lectin. Development. 1993;119:1229–1236. doi: 10.1242/dev.119.4.1229. [DOI] [PubMed] [Google Scholar]
- 7.Lewandowski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 8.Ryding ADS, Sharp MGF, Mullins JJ. Conditional transgenic technologies. J Endocrinol. 2001;171:1–14. doi: 10.1677/joe.0.1710001. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 10.Furth PA, et al. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc Natl Acad Sci USA. 1994;91:9302–9306. doi: 10.1073/pnas.91.20.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, et al. Calcineurin is a common target of cyclophillin-cyclosporin A and the FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 12.Brown EJ, et al. A mammalian protein targeted by G1-arrested rapamycin receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 13.Sabatini DM, Erdjument-Bromage H, Liu M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 14.Mayer TU, et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber SL. The small-molecule approach to biology: Chemical genetics and diversity-oriented organic synthesis make possible the systematic exploration of biology. Chem Eng News. 2003;81:51–61. [Google Scholar]
- 16.Godl K et al. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc Natl Acad Sci USA. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stankunas K, et al. Conditional protein alleles using knockin mice and a chemical inducer of dimerization. Moll Cell. 2003;12:1615–1624. doi: 10.1016/s1097-2765(03)00491-x. [DOI] [PubMed] [Google Scholar]
- 18.Liu KJ, Arron JR, Stankunas K, Crabtree GR, Longaker MT. Chemical rescue of cleft palate and midline defects in conditional GSK-3β mice. Nature. 2007;446:79–82. doi: 10.1038/nature05557. [DOI] [PubMed] [Google Scholar]
- 19.Banaszynski LA, Chen L-C, Maynard-Smith LA, Ooi AGL, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation, or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 21.Affar elB, et al. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maynard-Smith LA, Chen L-C, Banaszynski LA, Ooi AGL, Wandless TJ. A directed approach for engineering conditional protein stability using biologically silent small molecules. J Biol Chem. 2007;282:24866–24872. doi: 10.1074/jbc.M703902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iuliucci JD, et al. Intravenous safety and pharmacokinetics of a novel dimerizer drug, AP1903, in healthy volunteers. J Clin Pharmacol. 2001;41:870–879. doi: 10.1177/00912700122010771. [DOI] [PubMed] [Google Scholar]
- 24.Gaffen SL, Liu KD. Overview of interleukin-2 function, production and clinical applications. Cytokine. 2004;28:109–123. doi: 10.1016/j.cyto.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Lode HN, Reisfeld RA. Targeted cytokines for cancer immunotherapy. Immunol Res. 2000;21:279–288. doi: 10.1385/IR:21:2-3:279. [DOI] [PubMed] [Google Scholar]
- 26.Atkins MB, et al. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J Clin Oncol. 1999;17:2105–2116. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 27.Thorne SH, et al. Rational strain selection and engineering creates a broad-spectrum, systemically effective oncolytic poxvirus, JX-963. J Clin Invest. 2007;117:3350–3358. doi: 10.1172/JCI32727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas R, Kresse M, Latta M, Wendel A. Tumor necrosis factor: how to make a killer molecule tumor-specific. Curr Cancer Drug Targets. 2005;5:381–392. doi: 10.2174/1568009054863627. [DOI] [PubMed] [Google Scholar]
- 29.Alcami A, Khanna A, Paul NL, Smith GL. Vaccinia virus strains Lister, USSR and Evans express soluble and cell-surface tumor necrosis factor receptors. J Gen Virol. 1999;80:949–959. doi: 10.1099/0022-1317-80-4-949. [DOI] [PubMed] [Google Scholar]
- 30.Baggett B, et al. Thermostability of firefly luciferases affects efficiency of detection by in vivo bioluminescence. Mol Imaging. 2004;3:324–332. doi: 10.1162/15353500200403178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


