Abstract
Introduction
The genetic region coding for D-amino acid oxidase activator (DAOA) is considered an intriguing susceptibility locus for schizophrenia. However, association studies have often resulted in conflicting findings, and the risk conferring variants and their biological impact remain elusive. Our aim in this study was to investigate the relationship between DAOA variation and schizophrenia, and the influence of DAOA on cognitive performance.
Methods
We analyzed block structure and association patterns of a ~173 kb region on chromosome 13q33, applying genotype data of 55 SNPs derived from Caucasian North American sample (178 cases, 144 healthy controls). Haplotypes were assigned using the program PHASE and frequencies compared between cases and controls. We applied MANOVA to investigate the relationship between the identified risk haplotype on cognitive performance.
Results
We identified multiple haplotypes within the region containing the DAOA gene. Of these, one was significantly associated with schizophrenia, being over-represented in schizophrenia versus healthy controls. This haplotype was also associated with one aspect of cognitive performance, semantic fluency. Carriers of the risk haplotype showed better semantic fluency than non-carriers.
Conclusions
We report a significant effect of DAOA variation on risk for schizophrenia. Moreover, we identified a relationship between DAOA genetic variation and specific aspects of neurocognitive function. As the identified DAOA risk haplotype was associated with better performance on a semantic fluency measure, further work is required to identify the mechanism of DAOA action on CNS function, including the possibility of a role for balanced selection at this locus.
Keywords: DAOA, DAO, glutamate, genetics, neurocognition, schizophrenia
1. Introduction
Several lines of evidence suggest that the gene encoding D-amino acid oxidase activator (DAOA;MIM 607408, formerly referred to as G72) is involved in susceptibility to schizophrenia. First, linkage studies have implicated a large 68-Mb region spanning chromosomal regions 13q12 to 13q34 in several populations (Abecasis et al., 2004; Gershon and Badner, 2001; Levinson et al., 2000; Maziade et al., 2001). Second, Chumakov and colleagues (2002) dense-mapped a 5-Mb region from 13q22-33 with 191 single nucleotide polymorphisms (SNPs), and tested for association with schizophrenia in two independent samples. Significant associations to disease were reported within two clusters of SNPs in this region, several of which were located within DAOA. Finally, support for a role of DAOA in schizophrenia was provided by in vitro data suggesting that increasing concentrations of the G72 native protein, LG72, stimulated the oxidation of D-serine, an important activator at the glycine modulatory site of N-methyl-D-aspartate (NMDA) receptor (Chumakov et al., 2002), a glutamatergic receptor implicated in the pathophysiology of schizophrenia (Goff and Coyle, 2001; Krystal et al., 2002; Tsai and Coyle, 2002).
Subsequently, multiple association studies, investigating several of the previously associated SNPs, have also yielded significant results in case-control (Korostishevsky et al., 2004; Ma et al., 2006; Schumacher et al., 2004; Wang et al., 2004) and family-based designs (Mulle et al., 2005; Zou et al., 2005). Despite the number of positive association studies, however, the literature is marked by considerable allelic heterogeneity; few studies have reported compatible results for the same SNPs. Although a recent meta-analysis demonstrated highly significant p-values for several markers across multiple studies (Detera-Wadleigh and McMahon, 2006), the direction of effects often were inconsistent. Subsequently, an up-to-date meta-analysis maintained at the Schizophrenia Forum website (Allen et al. 2007; http://www.schizophreniaforum.org/res/sczgene/default.asp) demonstrates no SNPs with significant pooled odds ratios. Moreover, prior studies have typically genotyped a limited number of SNPs (mean=8.5), and have not systematically examined the haplotype structure of DAOA in association analysis. Similarly, the surrounding genomic region has not been comprehensively addressed, even though the original report (Chumakov et al., 2002) identified significant associations to schizophrenia more than 40 kb distal to the gene.
To date, only two studies have tested for the effects of DAOA and adjacent chromosomal regions on cognition. Goldberg et al. (2006) demonstrated a significant effect of several SNPs on cognitive measures, which are putatively dependent on NMDA signaling in a cohort of 600 subjects, including patients with schizophrenia, their unaffected siblings, and healthy controls. Further, these associations were also related to neurophysiological response parameters during those tasks, as measured by functional magnetic resonance imaging (fMRI). In a more recent study, Donohoe et al. (2007) reported that a functional polymorphism within DAOA (rs2391191), which results in an arginine-to-lysine substation, was associated with verbal memory performance in a cohort of 93 patients with schizophrenia. Although these data highlight one potential mechanism by which genetic variation in this chromosomal region might influence risk for schizophrenia, effects were modest and the functional variant remains elusive.
Our goal in the current study was to expand upon previous findings implicating DAOA as a candidate gene for schizophrenia and neurocognitive function. The present study assessed a large number of polymorphisms at the DAOA locus, allowing for the construction of haplotypes across the region. Relationship of haplotypes to schizophrenia risk was examined using a case-control association design; we then assessed for effects of the identified risk haplotypes on cognitive function in schizophrenia and healthy volunteers.
2. Materials and Methods
2.1 Subjects
The study sample consisted of 178 patients (113 males, 65 females) with schizophrenia-spectrum disorders (158 schizophrenia, 13 schizoaffective, 7 schizophreniform disorder), and 144 healthy controls (80 males, 64 females). Patients had a mean age of 37.8 (±10.5) years, a mean education level of 13.1 (±2.2) years, and the mean duration of illness was 16.0 (±11.2). Healthy controls had a mean age of 42.8 (±13.1) years and a mean education level of 15.7 (±2.9) years. All participants were self-identified as Caucasian, non-Hispanic, and provided written informed consent before participation. Population structure was tested by means of ancestry informative markers (AIMs) and the STRUCTURE program (Pritchard et al., 2000), which did not reveal significant stratification. Diagnoses were confirmed by a consensus diagnostic committee, based on results from the Structured Clinical Interview for DSM-IV Axis I disorders (SCID, version 2.0), administered by trained interviewers, and on all additional available information including medical records. Further detailed information on the study sample is provided by Lencz et al. (2007).
2.2 Genotyping
Genomic DNA was extracted from whole blood and hybridized to two chips containing ~262,000 and ~238,000 SNPs based on manufacturer’s (Affymetrix, Santa Clara, CA, USA) specifications. Patients and controls were proportionally distributed on each 96-well plate. Genotype calls were made using Bayesian Robust Linear Model with Mahalanobis distance classifier algorithm threshold at 0.5 applied to batches of 100 samples. Mean call rates <90% on both chips (or <85% on one chip) were rejected, resulting in a mean call rate for the retained sample of 97%. For an in depth description of preliminary steps and quality control procedures, see Lencz et al. (2007). The chromosomal region investigated included 55 SNPs (mean distance 3.21 kb, median 0.866 kb), spanning approximately 173 kb of chromosome 13q33 from chromosomal positions 104831385-105004420 (B35). The region extends beyond the DAOA gene region 85.2 kb upstream, and 63.0 kb downstream. Our primary focus was a haplotype-based approach to test for association with disease, as this provides more comprehensive information about the underlying genotype in the population than does single-marker association. Single markers were examined only subsequent to significant haplotype associations. Haplotype block structure and case-control haplotypic and allelic chi-square tests (χ2) were performed in Haploview v3.32. (Barrett et al., 2005). We subsequently assigned diplotypes to each individual subject using PHASE, version 2.1 (Stephens et al., 2001; Stephens and Donnelly, 2003).
2.3 Cognitive Testing
All subjects were administered a battery of standardized cognitive measures (Spreen and Strauss, 1998) including: Wide Range Achievement Test-3rd Edition-Reading Subtest (WRAT-III) for assessment of premorbid intellectual capacity; Wechsler Adult Intelligence Test-Revised (WAIS-R)-Digit Span for assessment of auditory attention and verbal working memory; Continuous Performance Test-Identical Pairs Version (CPT-I/P) to assess sustained attention and vigilance; California Verbal Learning Test (CVLT) to assess verbal learning and memory; and the Trail Making Tests A and B for assessment of visual-motor speed and executive control. Psychometricians with a Master's degree at least 5 years of testing experience, and high reliability in the administration of all cognitive tasks completed all of the neurocognitive testing.
2.4 Statistical procedure
The HAPLOVIEW program tests for association of alleles and haplotypes with disease by applying a Pearson goodness-of–fit χ2 test. Only haplotypes with a frequency ≥0.10 were included in the subsequent global p-value calculation for haploblocks, applying a Fisher’s Exact Test on the VassarStats website for statistical computation (http://faculty.vassar.edu/lowry/VassarStats.html). All subsequent analyses were performed using the Statistical Package for the Social Sciences, SPSS Version 13.0 for Windows (Chicago, IL). Differences between cases and controls, and between carriers and non-carriers of associated haplotypes, were computed with the Pearson χ2 tests for nominal variables, and with two-tailed t-tests or non-parametric tests for interval variables as appropriate. To assess the impact of carrier status on cognitive performance, we performed multivariate analyses of variance (MANOVA). To reduce the effects of multiple testing, only the identified risk haplotype was included in the MANOVA. Carrier status and affection status were included in the model as independent factors, age was used as a covariate, and select cognitive performance scores were included as dependent variables.
3. Results
3.1 Subject characteristics
There was no significant difference in sex distribution between cases and controls (63.5 % males in patients, 55.5 % males in controls, p=0.149), however, mean age differed significantly between groups (p=0.001), as did years of education (p<0.001). As expected, estimated premorbid intellectual capacity, assessed with the WRAT-III, also differed significantly between groups (p<0.001) with cases displaying lower scores as compared to healthy controls (96.5 ± 12.2 and 104.6 ± 9.2, respectively).
3.2 LD structure
After excluding markers displaying a minor allele frequency of <0.05, we had a total of 41 SNPs eligible for further analysis. None of the alleles of the single SNPs showed deviation from Hardy-Weinberg equilibrium (tested in the control sample, data not shown). The haploblock structure was also derived from the total sample and did not change when data from cases and controls were uploaded to HAPLOVIEW separately. The program identified eight blocks (termed blocks 1-8 herein) of high inter-marker LD using the default setting of the solid spine method, with D’ >0.80. The haploblock structure was comparable to that reported in the Hapmap release 21a /Phase II Jan 2007, on NCBI 35 assembly, dbSNPb125, derived from individuals with European descent (CEU) (Figure 1).
Figure 1.
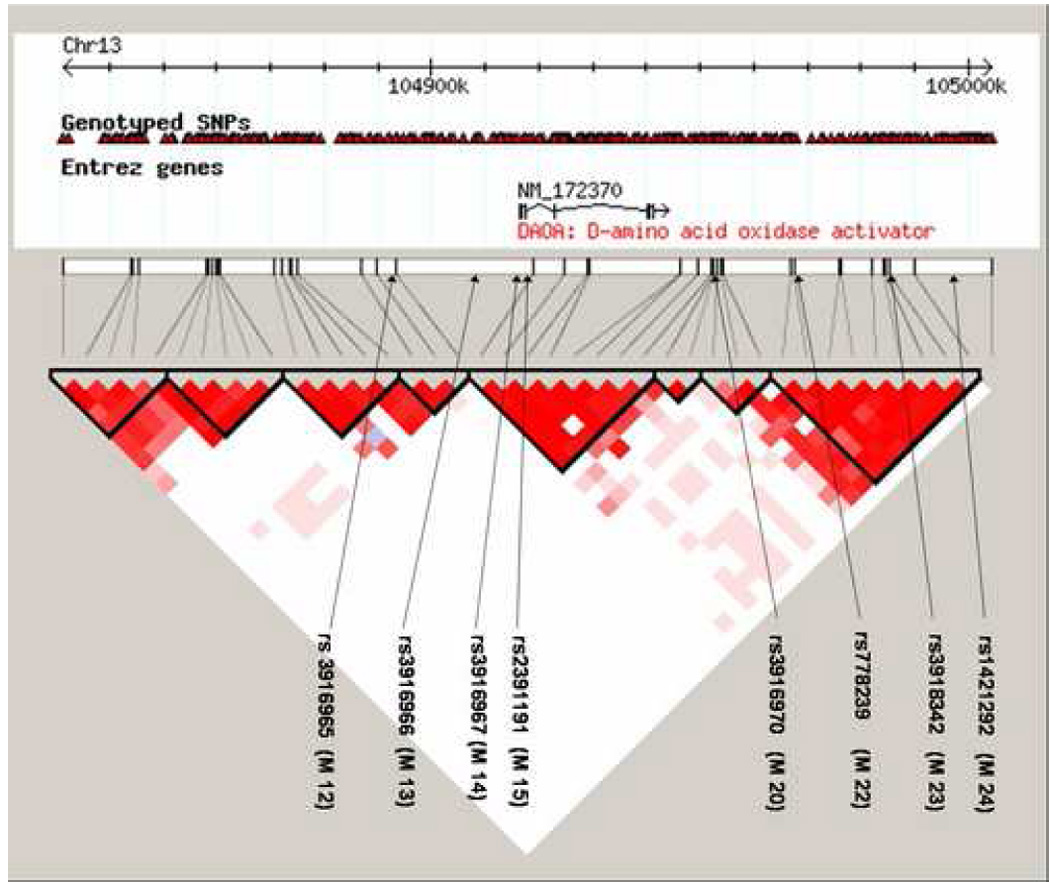
Schematic picture of haploblock structure of DAOA and flanking regions in the WGA sample (chromosomal region spanning from positions 104831385-105004420, 41 SNPs) in HAPLOVIEW. The haploblock structure was generated using solid spine of linkage disequilibrium (LD), with the default setting of D’ >.80. Four of the genotyped markers are located within the gene (rs12874006, rs10492528, rs2153674, rs11069595).Previously investigated markers are also depicted, along with their approximate position in the DAOA region and haploblocks, indicated by black arrows.
3.3 Genotype effects on risk for schizophrenia
As depicted in Table 1, Haploblock 1 yielded a significant global p-value for association to schizophrenia. HAPLOVIEW revealed that block 1, spanning approximately 26 kb from chromosomal positions 104831385-104858257, consisted of 5 SNPs (rs1570709, rs9586843, rs7324448, rs1575633, rs7329966). In PHASE, the second most common haplotype (GCGGC) was over-represented in patients with schizophrenia (0.21 frequency) vs. healthy controls (0.13 frequency) (p=0.013), and was considered a risk haplotype for disease. In addition, when carrier versus non-carrier status was assessed, the most common haplotype (ACGGT) was over-represented in cases (0.40 frequency) versus healthy controls (0.24 frequency) (χ2 = 8.75; p=0.003). SNP rs1570709 in block 1 was also significantly associated with disease, with the minor allele (G) identified as the risk allele (p=0.0039). No other haploblocks yielded significant evidence for association to illness.
Table 1.
DAOA and Risk for Schizophrenia: Haplotype and SNP Results
| Haplotype SNP Allele(s) | Global χ2 (df=2) | Global p-value | Frequency Total | Frequency Cases | Frequency Controls | Pearson χ2 (df=1) | P-value |
|---|---|---|---|---|---|---|---|
| Block 1 | 7.20 | 0.03 | -- | -- | -- | -- | |
| ACGGT | --- | --- | 0.482 | 0.454 | 0.516 | 2.405 | 0.121 |
| GCGGC | --- | --- | 0.168 | 0.201 | 0.127 | 6.179 | 0.013 |
| ATATC | --- | --- | 0.162 | 0.150 | 0.177 | 0.88 | 0.348 |
| rs1570709 G | --- | --- | 0.219 | 0.133 | 8.318 | 0.0039 | |
| Block 2 | 4.49 | 0.11 | --- | --- | --- | --- | --- |
| Block 3 | 0.87 | 0.65 | --- | --- | --- | --- | --- |
| Block 4 | 1.45 | 0.48 | --- | --- | --- | --- | --- |
| Block 5 | 1.07 (df=3) | 0.78 | --- | --- | --- | --- | --- |
| Block 6 | 2.23 | 0.33 | --- | --- | --- | --- | --- |
| Block 7 | 0.21 | 0.90 | --- | --- | --- | --- | --- |
| Block 8 | 3.62 | 0.16 | --- | --- | --- | --- | --- |
Note: Unless indicated otherwise (alleles in bold script), all alleles refer to the (-) strand relative to the human reference sequence, as provided by manufacturer’ s probe list annotations (https://www.affymetrix.com/analysis/netaffx/batch_query.affx?netaffx=mapping). Global p value and χ2 was calculated applying a Fishers Exact Test in VassarStats
As there are several approaches to data reduction in multi-SNP data given the presence of LD, we performed complementary analyses on the individual SNP data using two alternate approaches to estimate the amount of independent data represented by the 41 SNPs examined in our study. Using the tagging approach (implemented in Haploview, using "aggressive" multi-SNP tagging a threshold of r-squared>.8), 23 tagging SNPs were identified. However, this approach results in potentially arbitrary choices of tags. Therefore, we also utilized the method of Nyholt (2004) as modified by Li & Ji (2005) to determine the total informational content of our entire SNP dataset. Intriguingly, this method also revealed the effective number of independent loci was 23. Applying a significance threshold of p<0.05/23=0.0022, we then performed genotypic (dominant/recessive) tests on each of the individual SNPs in our dataset. The two SNPs highlighted in Table 1 (rs1570709 and rs9558546) demonstrated statistically significant genotypic p-values under a dominant model; rare allele carriers for each of these SNPs were significantly more likely to be cases than controls (p=0.000894 and p=0.00142, respectively).
3.2 Genotype effects on cognitive performance
We next examined the relationship between the DAOA risk haplotype (GCGGC) and neurocognitive function. There were no significant differences in demographics with respect to carrier vs. non-carrier status of the haplotype GCGGC, including age, sex, years of education, or premorbid intellectual capacity (Table 2). When analyzed separately, cases and controls (data not shown), again did not differ by genotype on any demographic variable.
Table 2.
Descriptive and demographic data for carriers and non carriers of the risk haplotype GCGGC (whole sample)
| Carrier | Non Carrier | p | |
|---|---|---|---|
| Mean (SD) | Mean (SD) | ||
| Age [years] | 39.4 (±11.7) | 39.5 (±11.7) | .912a |
| Sex [% males] | 59.4 | 60.1 | .897b |
| WRAT-III [Standard score] | 99.8 (±9.0) | 100.0 (±12.8) | .816a |
| Education [years] | 14.2 (±2.9) | 14.5 (±2.9) | .414 a |
Abbreviation: WRAT-III = Wide Range Achievement Test, Third Edition, Reading Subtest.
t-test, two-tailed
Pearson χ2 test
Multivariate analysis of variance (MANOVA) revealed a main effect of carrier status for the risk haplotype GCGGC in block 1 (F=2.072, df=8, p=.041) on cognitive performance (Table 3). We also found significant main effects for affection status (F=19.974, df=8, p<.001) and age (F=7.290, df=8, p<.001). Post-hoc testing revealed that the overall effect on cognition was driven by a significant main effect of GCGGC carrier status on semantic fluency, with carriers of the risk haplotype GCGGC demonstrating better performance than non-carriers. This pattern was consistent in both healthy controls and in patients with schizophrenia (Figure 2). The genotype x affection status interaction was not significant (F=.717, df=8, p=.676). As expected, age demonstrated a significant impact on performance in all cognitive domains, with the exception of phonemic and semantic fluency.
Table 3.
Impact of carrier status of risk haplotype GCGGC, affection status, interaction of affection and carrier status, and age on cognitive performance (N = 102 cases, 94 controls)
| MANOVA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Affection status | Carrier status | Affection*Carrier | Age | |||||||||
| F | df | p | F | df | p | F | df | p | F | df | p | |
| 19.974 | 8 | <.001 | 2.074 | 8 | .041 | .717 | 8 | .676 | 7.209 | 8 | <.001 | |
| ANOVA | ||||||||||||
| TMT-A | 82,439 | 1 | <.001 | .009 | 1 | .923 | .731 | 1 | .394 | 18.933 | 1 | <.001 |
| TMT-B | 84.179 | 1 | <.001 | .007 | 1 | .932 | .668 | 1 | .415 | 22.826 | 1 | <.001 |
| DSPN | 25.153 | 1 | <.001 | 2.020 | 1 | .157 | .154 | 1 | .695 | 4.564 | 1 | .034 |
| CVLT | 82.658 | 1 | <.001 | .085 | 1 | .771 | 1.968 | 1 | .162 | 21.970 | 1 | <.001 |
| CWAT | 28.183 | 1 | <.001 | .385 | 1 | .536 | .768 | 1 | .382 | .333 | 1 | .564 |
| FLUST | 58.586 | 1 | <.001 | 7.095 | 1 | .008 | .006 | 1 | .940 | .440 | 1 | .508 |
| CPT d’ F | 43.521 | 1 | <.001 | .206 | 1 | .650 | 1.103 | 1 | .295 | 18.854 | 1 | <.001 |
| CPT d’ S | 51.844 | 1 | <.001 | 1.076 | 1 | .301 | .432 | 1 | .512 | 20.363 | 1 | <.001 |
Abbreviations: TMT-A = Trail Making Test A, TMT-B = Trail Making Test B, DSPN = Wechsler Adult Intelligence Test-Revised (WAIS-R) - Digit Span, CVLT = California Verbal Learning Test (CVLT) - Abridged, CWAT = Semantic Fluency (3-letter-words, total number), FLUST = Verbal Fluency, (animals, total number), CPT d’ F = Continuous Performance Test-Identical Pairs Version, fast version (d prime), CPT d’ S = Continuous Performance Test-Identical Pairs Version, slow version (d prime).
Figure 2.
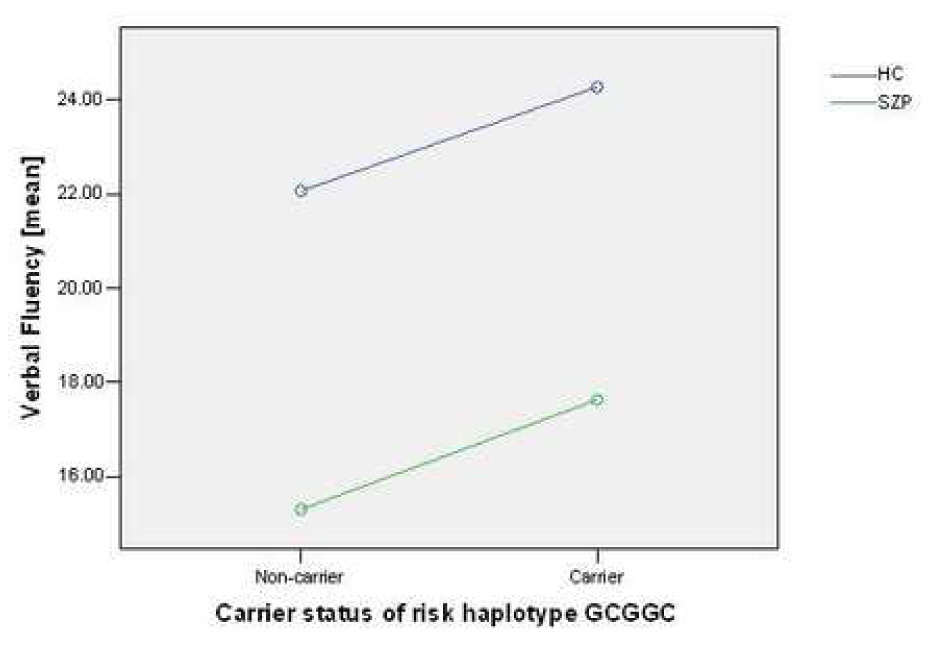
Impact of risk haplotype GCGGC on verbal fluency in subjects with schizophrenia-spectrum disorders (SZP) and healthy controls (HC).
4. Discussion
Converging evidence from multiple association and linkage studies has revealed a relationship between several loci on chromosome 13q and susceptibility for schizophrenia, with the most compelling evidence of linkage in the chromosomal region 13q31-q33. However, association studies have not converged on a single risk SNP within the DAOA locus. In the current study we attempted to extend previous associations with disease using extensive genotyping surrounding the locus, construction of haplotypes, and examination of the intermediate phenotype of cognition. In our American Caucasian cohort, we identified a haplotype block structure consistent with that reported in the Hapmap sample of individuals of European descent (CEU), including two large blocks of high inter-marker LD in the most intensively investigated region spanning from marker rs3916965 (M12) to rs1421292 (M24). In a haplotype-based investigation of this region, we report significant association with disease for a haplotype, located upstream of the presumed gene borders of DAOA. In addition, we found a significant impact of this haplotype on a measure of verbal fluency (semantic fluency) in both schizophrenia subjects and in healthy controls.
Our data supporting an association of variation in this chromosomal region with schizophrenia are at least partially consistent with a number of previous haplotype-based association (Chumakov et al., 2002; Goldberg et al., 2006; Korostishevsky et al., 2004; Ma et al., 2006; Schumacher et al., 2004; Zou et al., 2005). However, inconsistencies in markers genotyped and allelic heterogeneity are common findings in this region, as noted in a recent review by Abou Jamra et al. (2006). By using haplotypes, we have attempted to integrate previous findings. Specifically, the intensively investigated markers M22, M23, M24, M12 and M15 fall within haplotype blocks that were investigated in the present study; however, none of the blocks containing these markers, were significantly associated with disease in our study. Of note, the SNPs genotyped within this study were not identical with the previously investigated markers. Rather, our results suggest association of a region that is located ~ 80 kb upstream of the gene. One limitation of the current study in attempting to replicate previous findings, is that coverage was sparse in the immediate 5’ region of the actual known gene region, which includes the well-studied marker M12.
Although linkage findings in this region are among the most convincing for linkage across the genome, LOD scores have ranged considerably in schizophrenia, and this region has also been associated with other psychiatric disorders including bipolar disorder. DAOA represents one of a number of susceptibility genes that may act to increase risk for multiple neuropsychiatric disorders, challenging the once widely accepted Kraepelinian dichotomy for schizophrenia and bipolar disorder (Abou Jamra et al., 2006). These data are also consistent with the notion of gene action via an intermediate phenotype that is shared by both disorders. For example, cognitive dysfunction appears to be a valid and heritable phenotype common to both diseases, specifically in the domains of working memory, verbal learning, and executive function (Glahn et al., 2004).
To assess this, we investigated the potential impact of the associated haplotype on cognitive performance in patients with schizophrenia and in healthy controls. We found that the GCGGC risk haplotype, located in block 1, significantly influenced a measure of semantic fluency, in both subject groups. Interestingly, GCGGC carriers demonstrated better semantic fluency in our sample than did non-carriers, regardless of diagnostic status. There are several possible explanations for these seemingly counter-intuitive findings. First, it is possible that the GCGGC haplotype affects semantic fluency independent of its role in disease susceptibility. Supporting this possibility is the finding that semantic fluency was selectively spared in siblings of patients with schizophrenia, and did not correlate with other cognitive tests with greater familiality (Egan and Goldberg, 2003). Similarly, it is possible that DAOA haplotypes represent a non-cognitive risk factor for schizophrenia, such that patients with other genetic risk factors (e.g., DTNBP1, Burdick et al., 2006; Burdick et al., 2007) demonstrate greater cognitive burden. A more speculative explanation is derived from evolutionary theory. The proposed hypothesis is based on the argument of balanced selection, in which a deleterious allele (i.e. risk allele for schizophrenia) is maintained in a population as a result of some beneficial influence of the same allele on a different trait that is being positively selected for from a survival perspective (i.e. better cognition). A similar relationship was recently reported for PPP1R1B (Meyer-Lindenberg et al., 2007), in which a risk-conferring haplotype was associated with enhanced fronto-striatal neurofunctional performance. In that report, the authors suggested that enhanced connectivity may be detrimental in the context of poor prefrontal efficiency; similarly, enhanced semantic connectivity as indexed by semantic fluency performance may be deleterious in the context of other neurofunctional pathology in patients with schizophrenia. Finally, it is possible that these results represent a false positive, although the similarity of results across two samples (patients and controls) reduces the likelihood of this interpretation. Still, as with all genetic associations, independent replication will be useful to confirm the cognitive association.
To date, DAOA is the only candidate gene in the chromosomal region associated with schizophrenia and bipolar disorder that is believed to code for a protein. Messenger RNA expression of the gene has been shown to be increased in schizophrenic patients in postmortem studies (Korostishevsky et al. 2004). The presumed function of the gene, suggested by in vitro transcription / translation assays appears to involve the regulation of the NMDA-type glutamate receptor, which is intriguing in light of the proposed relationship between glutamatergic dysfunction and the pathophysiology of schizophrenia (Harrison and Owen, 2003). There is a substantial body of evidence suggesting that cognitive dysfunction is associated with abnormalities in the glutamatergic system in the brain. The glutamatergic system is mediated by multiple families of ion-channels, with the NMDA receptor site figuring prominently in synaptic plasticity (Liu et al., 2004). NMDA receptor-based long-term potentiation (LTP) is believed to represent the cellular basis of learning and memory (Cotman et al., 1988).
However, several findings serve to weaken the hypothesis that DAOA confers risk for schizophrenia via modulation of glutamatergic receptor function. In a recent study investigating LG72 in several mammalian cell lines (Kvajo et al., 2007), colocalization or interaction of DAO and LG72 could not be confirmed. Rather, LG72 was localized in mitochondria, where an overexpression of G72 led to mitochondrial fragmentation. Increased dendritic arborization in immature primary neurons due to overexpression of G72 suggested a substantial modulation of neuronal function by fission and fusion of organelles. The authors therefore concluded that, in the light of convincing association and linkage findings, an as yet unknown function of the gene and protein in modulating mitochondrial morphology might be accountable for the risk conferring property of the gene and chromosomal region. This view is supported by the hypothesis of mitochondrial dysfunction in neurodegenerative diseases, which has been applied to schizophrenia by several authors (e.g. Stork and Renshaw, 2005)
Given the lack of a native protein product of the DAOA gene, it has been suggested that the DAOA gene might be one example of a gene which consists entirely of repetitive elements, encoding mRNAs (Britten et al., 2004). It should be noted that several of the associated genetic variations in the region are located outside of the presumed borders of the DAOA gene. Thus unknown genes, or variants which might play a role in modifying gene expression or splicing, may be responsible for the disease associations reported to date.
More broadly, the study raises important questions about what constitutes a replication (NCI-NHGRI Working Group on Replication in Association Studies et al. 2007). One criticism of the work on DAOA is that there appears to be much allelic and locus heterogeneity. Meta-analyses that concluded that DAOA was a susceptibility gene for schizophrenia used statistics at the gene level, as opposed to the level of SNP or haplotype. This is further complicated by the fact that the majority of positive findings have been in extra-genic regions (including the findings discussed herein). DAOA’s influence on cognition may also be subject to these criticisms, as well as issues related to multiple comparisons. Until more progress is made in understanding the neurobiology of DAOA and the nature of the variants that confer risk or cognitive differences, be they causative mutations in the sequence, microRNAs, CNVs, or other unknown factors, our results should be accepted with circumspection. Finally, this study also raises the issue as to which follow-up analyses are considered appropriate in the context of statistically non-significant whole-genome association results. In the strictest statistical sense, analytic follow-up of results that do not meet the WGA threshold after correction (typically with p-values in the range of 10−7) are not considered valid. However, many researchers would argue that in the case of genes and/or gene regions with previous evidence of association/linkage, a lower threshold for purposes of confirmation or “replication” would be required. Alternatively, genes with prior data suggestive of specific biological plausibility related to the pathophysiology of a disorder might also warrant follow up regardless of WGA statistical significance. We acknowledge the possibility that our results could represent spurious findings, as the WGA threshold was not met; however, given both prior probabilities with regard to previous association and linkage findings, as well as the biological plausibility of DAOA, we believe that they are true positives which add further support to the link between DAOA and schizophrenia.
Acknowledgments
The authors would like to thank Drs T Vance Morgan and Raju Kucherlapati for their assistance in this work. In addition, we wish to thank all patients who participated in the study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Abecasis GR, Burt RA, Hall D, Bochum S, Doheny KF, Lundy SL, et al. Genomewide scan in families with schizophrenia from the founder population of Afrikaners reveals evidence for linkage and uniparental disomy on chromosome 1. Am. J. Hum. Genet. 2004;74:403–417. doi: 10.1086/381713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou JR, Schmael C, Cichon S, Rietschel M, Schumacher J, Nothen MM. The G72/G30 gene locus in psychiatric disorders: a challenge to diagnostic boundaries? Schizophr. Bull. 2006;32:599–608. doi: 10.1093/schbul/sbl028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balding DJ. A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Burdick KE, Hodgkinson CA, Szeszko PR, Lencz T, Ekholm JM, Kane JM, et al. DISC1 and neurocognitive function in schizophrenia. Neuroreport. 2005;16:1399–1402. doi: 10.1097/01.wnr.0000175248.25535.f6. [DOI] [PubMed] [Google Scholar]
- Burdick KE, Lencz T, Funke B, Finn CT, Szeszko PR, Kane JM, et al. Genetic variation in DTNBP1 influences general cognitive ability. Hum. Mol. Genet. 2006;15:1563–1568. doi: 10.1093/hmg/ddi481. [DOI] [PubMed] [Google Scholar]
- Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, et al. Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc. Natl. Acad. Sci. U S A. 2002;99:13675–13680. doi: 10.1073/pnas.182412499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, McMahon FJ. G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biol. Psychiatry. 2006;60:106–114. doi: 10.1016/j.biopsych.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Devlin B, Bacanu SA, Roeder K, Reimherr F, Wender P, Galke B, et al. Genome-wide multipoint linkage analyses of multiplex schizophrenia pedigrees from the oceanic nation of Palau. Mol. Psychiatry. 2002;7:689–694. doi: 10.1038/sj.mp.4001056. [DOI] [PubMed] [Google Scholar]
- Donohoe G, Morris DW, Robertson IH, McGhee KA, Murphy K, Kenny N, et al. DAOA ARG30LYS and verbal memory function in schizophrenia. Mol. Psychiatry. 2007;12(9):795–796. doi: 10.1038/sj.mp.4002026. [DOI] [PubMed] [Google Scholar]
- Dunham A, Matthews LH, Burton J, Ashurst JL, Howe KL, Ashcroft KJ, et al. The DNA sequence and analysis of human chromosome 13. Nature. 2004;428:522–528. doi: 10.1038/nature02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc. Natl. Acad. Sci. U S A. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Straub RE, Goldberg TE, Yakub I, Callicott JH, Hariri AR, et al. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc. Natl. Acad. Sci. U S A. 2004;101:12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainetdinov RR, Mohn AR, Caron MG. Genetic animal models: focus on schizophrenia. Trends Neurosci. 2001;24:527–533. doi: 10.1016/s0166-2236(00)01886-5. [DOI] [PubMed] [Google Scholar]
- Gershon ES, Badner JA. Progress toward discovery of susceptibility genes for bipolar manic-depressive illness and schizophrenia. CNS Spectr. 2001;6:965–968. 977. doi: 10.1017/s1092852900001073. [DOI] [PubMed] [Google Scholar]
- Goff DC, Coyle JT. The emerging role of glutamate in the pathophysiology and treatment of schizophrenia. Am. J. Psychiatry. 2001;158:1367–1377. doi: 10.1176/appi.ajp.158.9.1367. [DOI] [PubMed] [Google Scholar]
- Goff DC, Tsai G, Levitt J, Amico E, Manoach D, Schoenfeld DA, et al. A placebo-controlled trial of D-cycloserine added to conventional neuroleptics in patients with schizophrenia. Arch. Gen. Psychiatry. 1999;56:21–27. doi: 10.1001/archpsyc.56.1.21. [DOI] [PubMed] [Google Scholar]
- Goldberg TE, Straub RE, Callicott JH, Hariri A, Mattay VS, Bigelow L, et al. The G72/G30 gene complex and cognitive abnormalities in schizophrenia. Neuropsychopharmacology. 2006;31:2022–2032. doi: 10.1038/sj.npp.1301049. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am. J. Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Gur RE, Calkins ME, Gur RC, Horan WP, Nuechterlein KH, Seidman LJ, Stone WS. The consortium on the genetics of schizophrenia: neurocognitive endophenotypes. Schizophr. Bull. 2007;33:49–68. doi: 10.1093/schbul/sbl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SP, Fyer AJ, Durner M, Heiman GA, Baisre de LA, Hodge SE, et al. Further genetic evidence for a panic disorder syndrome mapping to chromosome 13q. Proc. Natl. Acad. Sci. U S A. 2003;100:2550–2555. doi: 10.1073/pnas.0335669100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Owen MJ. Genes for schizophrenia? Recent findings and their pathophysiological implications. Lancet. 2003;361:417–419. doi: 10.1016/S0140-6736(03)12379-3. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- Heresco-Levy U, Javitt DC, Ermilov M, Mordel C, Silipo G, Lichtenstein M. Efficacy of high-dose glycine in the treatment of enduring negative symptoms of schizophrenia. Arch. Gen. Psychiatry. 1999;56:29–36. doi: 10.1001/archpsyc.56.1.29. [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Jentsch JD, Roth RH. The neuropsychopharmacology of phencyclidine: from NMDA receptor hypofunction to the dopamine hypothesis of schizophrenia. Neuropsychopharmacology. 1999;20:201–225. doi: 10.1016/S0893-133X(98)00060-8. [DOI] [PubMed] [Google Scholar]
- Kelsoe JR, Spence MA, Loetscher E, Foguet M, Sadovnick AD, Remick RA, et al. A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc. Natl. Acad. Sci. U S A. 2001;98:585–590. doi: 10.1073/pnas.011358498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostishevsky M, Kaganovich M, Cholostoy A, Ashkenazi M, Ratner Y, Dahary D, et al. Is the G72/G30 locus associated with schizophrenia? single nucleotide polymorphisms, haplotypes, and gene expression analysis. Biol. Psychiatry. 2004;56:169–176. doi: 10.1016/j.biopsych.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Anand A, Moghaddam B. Effects of NMDA receptor antagonists: implications for the pathophysiology of schizophrenia. Arch. Gen. Psychiatry. 2002;59:663–664. doi: 10.1001/archpsyc.59.7.663. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Bennett A, bi-Saab D, Belger A, Karper LP, D'Souza DC, et al. Dissociation of ketamine effects on rule acquisition and rule implementation: possible relevance to NMDA receptor contributions to executive cognitive functions. Biol. Psychiatry. 2000;47:137–143. doi: 10.1016/s0006-3223(99)00097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal JH, Karper LP, Bennett A, D'Souza DC, bi-Dargham A, Morrissey K, et al. Interactive effects of subanesthetic ketamine and subhypnotic lorazepam in humans. Psychopharmacology (Berl) 1998;135:213–229. doi: 10.1007/s002130050503. [DOI] [PubMed] [Google Scholar]
- Lencz T, Morgan TV, Athanasiou M, Dain B, Reed CR, Kane JM, et al. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Mol. Psychiatry. 2007;12(6):572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- Levinson DF, Holmans P, Straub RE, Owen MJ, Wildenauer DB, Gejman PV, et al. Multicenter linkage study of schizophrenia candidate regions on chromosomes 5q, 6q, 10p, and 13q: schizophrenia linkage collaborative group III. Am. J. Hum. Genet. 2000;67:652–663. doi: 10.1086/303041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95(3):221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Liu C, Badner JA, Christian SL, Guroff JJ, Detera-Wadleigh SD, Gershon ES. Fine mapping supports previous linkage evidence for a bipolar disorder susceptibility locus on 13q32. Am. J. Med. Genet. 2001;105:375–380. doi: 10.1002/ajmg.1358. [DOI] [PubMed] [Google Scholar]
- Ma J, Qin W, Wang XY, Guo TW, Bian L, Duan SW, et al. Further evidence for the association between G72/G30 genes and schizophrenia in two ethnically distinct populations. Mol. Psychiatry. 2006;11:479–487. doi: 10.1038/sj.mp.4001788. [DOI] [PubMed] [Google Scholar]
- Malhotra AK, Kestler LJ, Mazzanti C, Bates JA, Goldberg T, Goldman D. A functional polymorphism in the COMT gene and performance on a test of prefrontal cognition. Am. J. Psychiatry. 2002;159:652–654. doi: 10.1176/appi.ajp.159.4.652. [DOI] [PubMed] [Google Scholar]
- Maziade M, Roy MA, Rouillard E, Bissonnette L, Fournier JP, Roy A, et al. A search for specific and common susceptibility loci for schizophrenia and bipolar disorder: a linkage study in 13 target chromosomes. Mol. Psychiatry. 2001;6:684–693. doi: 10.1038/sj.mp.4000915. [DOI] [PubMed] [Google Scholar]
- Meador-Woodruff JH, Healy DJ. Glutamate receptor expression in schizophrenic brain. Brain Res. Brain Res. Rev. 2000;31:288–294. doi: 10.1016/s0165-0173(99)00044-2. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Straub RE, Lipska BK, Verchinski BA, Goldberg T, Callicott JH, et al. Genetic evidence implicating DARPP-32 in human frontostriatal structure, function, and cognition. J. Clin. Invest. 2007;117:672–682. doi: 10.1172/JCI30413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle JG, Chowdari KV, Nimgaonkar V, Chakravarti A. No evidence for association to the G72/G30 locus in an independent sample of schizophrenia families. Mol. Psychiatry. 2005;10:431–433. doi: 10.1038/sj.mp.4001619. [DOI] [PubMed] [Google Scholar]
- NCI-NHGRI Working Group on Replication in Association Studies. Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447(7145):655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–7699. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MJ, Craddock N, O'Donovan MC. Schizophrenia: genes at last? Trends Genet. 2005;21:518–525. doi: 10.1016/j.tig.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Jamra RA, Freudenberg J, Becker T, Ohlraun S, Otte AC, et al. Examination of G72 and D-amino-acid oxidase as genetic risk factors for schizophrenia and bipolar affective disorder. Mol. Psychiatry. 2004;9:203–207. doi: 10.1038/sj.mp.4001421. [DOI] [PubMed] [Google Scholar]
- Spreen O, Strauss E. A Compendium of Neuropsychological Tests: Administration, Norms, and Commentary. 2nd ed. New York, NY: Oxford University Press; 1998. [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork C, Renshaw PF. Mitochondrial dysfunction in bipolar disorder: evidence from magnetic resonance spectroscopy research. Mol. Psychiatry. 2005;10:900–919. doi: 10.1038/sj.mp.4001711. [DOI] [PubMed] [Google Scholar]
- Tsai G, Coyle JT. Glutamatergic mechanisms in schizophrenia. Annu Rev Pharmacol Toxicol. 2002;42:165–179. doi: 10.1146/annurev.pharmtox.42.082701.160735. [DOI] [PubMed] [Google Scholar]
- Tsai G, Passani LA, Slusher BS, Carter R, Baer L, Kleinman JE, Coyle JT. Abnormal excitatory neurotransmitter metabolism in schizophrenic brains. Arch. Gen. Psychiatry. 1995;52:829–836. doi: 10.1001/archpsyc.1995.03950220039008. [DOI] [PubMed] [Google Scholar]
- Wang X, He G, Gu N, Yang J, Tang J, Chen Q, et al. Association of G72/G30 with schizophrenia in the Chinese population. Biochem. Biophys. Res. Commun. 2004;319:1281–1286. doi: 10.1016/j.bbrc.2004.05.119. [DOI] [PubMed] [Google Scholar]
- Williams NM, Green EK, Macgregor S, Dwyer S, Norton N, Williams H, et al. Variation at the DAOA/G30 locus influences susceptibility to major mood episodes but not psychosis in schizophrenia and bipolar disorder. Arch. Gen. Psychiatry. 2006;63:366–373. doi: 10.1001/archpsyc.63.4.366. [DOI] [PubMed] [Google Scholar]
- Wilson AF, Sorant AJ. Equivalence of single- and multilocus markers: power to detect linkage with composite markers derived from biallelic loci. Am. J. Hum. Genet. 2000;66:1610–1615. doi: 10.1086/302889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F, Li C, Duan S, Zheng Y, Gu N, Feng G, et al. A family-based study of the association between the G72/G30 genes and schizophrenia in the Chinese population. Schizophr. Res. 2005;73:257–261. doi: 10.1016/j.schres.2004.01.015. [DOI] [PubMed] [Google Scholar]


