Summary
The distal human gut is a microbial bioreactor that digests complex carbohydrates. The strategies evolved by gut microbes to sense and process diverse glycans have important implications for the assembly and operations of this ecosystem. The human gut bacterium Bacteroides thetaiotaomicron forages on host and dietary glycans. Its ability to target these substrates resides in 88 polysaccharide utilization loci (PULs), encompassing 18% of its genome. In this report, whole-genome transcriptional profiling and genetic tests are used to define the mechanisms underlying host glycan foraging in vivo and in vitro. PULs that target all major classes of host glycans were identified. Mucin O-glycans are the principal substrate foraged in vivo. Simultaneous deletion of five ECF-σ transcription factors that activate mucin O-glycan utilization produces defects in bacterial persistence in the gut and in mother-to-offspring transmission. Thus, PUL-mediated glycan catabolism is an important factor in gut colonization and likely impacts gut ecology.
Introduction
The human distal gut contains 10–100 trillion microbes, most belonging to the domain Bacteria. Like other mammals, the majority of our gut bacteria belong to just two phyla (divisions): the Bacteroidetes and the Firmicutes (Ley et al., 2008). The gut microbiota functions as a metabolic organ that extracts energy from dietary material that we cannot process on our own (Flint et al., 2008). Its members produce a vast arsenal of glycolytic enzymes that supplement our own 99-member collection of these activities (www.cazy.org) and enable fermentation of otherwise indigestible dietary glycans to host-absorbable compounds like short-chain fatty acids.
The molecular strategies evolved by members of this and other microbial communities to differentiate, capture, and degrade complex glycans are of interest to those seeking to understand the genetic and metabolic underpinnings of fitness and adaptations in human body-associated and other environmental habitats. Similarly, deciphering the nature and significance of evolved genetic solutions for glycan foraging by different microbial lineages has implications for those seeking to modulate or engineer energy harvest from complex polysaccharides by natural or synthetic microbial consortia. The human gut provides a rich, densely populated ecosystem for exploring these issues.
In addition to dietary sources, host-derived glycans represent a consistent nutrient foundation for members of the gut microbiota. Several gut species are capable of degrading these host substrates (Salyers et al., 1977a). One well-studied example is Bacteroides thetaiotaomicron, a prominent member of the human microbiota capable of growing on many different plant and host glycans (Salyers et al., 1977b) and endowed with 246 glycolytic enzymes (Xu et al., 2007). When B. thetaiotaomicron colonizes the distal guts of adult germ-free mice maintained on a diet rich in plant glycans, it increases expression of genes involved in catabolism of dietary substrates (Sonnenburg et al., 2005). In contrast, in adult mice fed a diet devoid of complex glycans, B. thetaiotaomicron alters its response to express genes involved in targeting host glycans. Transcriptional profiles in the guts of newborn mice consuming mother’s milk are similar to those observed in adult mice fed a glycan deficient diet, indicating bacterial reliance on endogenous glycan pools in the neonatal gut (Bjursell et al., 2006). These results indicate that some gut microbial species may rely on host glycans in the absence of dietary input and suggest that the endogenous carbohydrate ‘landscape’ of the host influences gut community structure and stability by selecting organisms capable of consuming these glycans. This notion raises two questions: which host glycan types are used in vivo and how are they sensed and differentiated by bacteria?
Comparative and functional genomic studies predict that the ability of gut Bacteroides spp. to utilize diverse glycans depends on a series of gene clusters that we have termed polysaccharide utilization loci (PULs) (Bjursell et al., 2006). PULs encode cell envelope systems that typically include glycolytic enzymes and homologs of two outer membrane proteins (SusC and SusD) that are part of the first described PUL, the Starch utilization system (Sus) locus (Fig. S2) (Reeves et al., 1997). SusC is a TonB-dependent transporter essential for energy-dependent import of starch oligosaccharides into the periplasm (Reeves et al., 1996). SusD is a secreted α-helical starch binding protein, which by virtue of its N-terminal lipidation, remains peripherally associated with the outer membrane. It is required for growth on starch molecules containing ≥6 glucose units (Koropatkin et al., 2008).
Bacteroides PULs universally contain homologs of susC and susD. However, both the genomic organization and gene content of these clusters can vary substantially (Xu et al., 2007). PULs frequently contain sensor regulators (extracytoplasmic function sigma/anti-sigma factor pairs, ‘hybrid’ two component phosphorelay systems, plus others), suggesting that each gene cluster encodes all of the requisite functions to act as a substrate-specific, glycan-sensing and acquisition system. Despite being abundant in gut Bacteroides species, the substrate specificities of all but the Sus PUL (Shipman et al., 2000), and a homologous starch utilization locus in Bacteroides fragilis (Spence et al., 2006), remain ill-defined.
We hypothesized that some PULs play key roles in targeting endogenous host glycans. If true, B. thetaiotaomicron could serve as a model to characterize the interactions between members of the gut microbiota and the host mucosa. Understanding these interactions could provide insights into host habitat features that affect initial colonization of the gut and subsequent assembly of a microbiota during the postnatal period (Palmer et al., 2007). Exploration of the host glycan-microbiota interface could also shed light on the pronounced interpersonal variations observed in human gut ecology (Eckburg et al., 2005; Ley et al., 2006), the determinants of how the microbiota is able to rebound from physiologic or pharmacologic perturbations, and the forces that regulate interactions between the microbiota and components of innate and adaptive immune systems in health and disease (An et al., 2007; Heazlewood et al., 2008; Peterson et al., 2008; Swidsinski et al., 2007).
Therefore, in this study we have investigated the biochemical identities of host glycans utilized by B. thetaiotaomicron in vivo as well as genes required for their metabolism. Our results reveal that that PUL-encoded Sus-like systems have remarkably broad glycan substrate specificities, spanning the range from host to plant glycans, and represent a general paradigm for how B. thetaiotaomicron and its relatives have solved the problem of competing for glycans in the gut.
Results and Discussion
In vitro model of the gut glycan landscape
To investigate how B. thetaiotaomicron targets host glycans in vivo, we first created an in vitro model. A heterogeneous mixture of glycans was prepared from porcine gastric mucosa. Monosaccharide analysis revealed the presence of all three classes of host glycans, although mucin O-glycans and GAGs were predominant (Table S1). When this mixture was supplied as the sole carbon source, B. thetaiotaomicron exhibited a diauxic growth profile (Fig. S4A), suggesting that some of its glycan components were catabolized preferentially over others.
To identify adaptations to growth on porcine mucosal glycans (PMG), we performed whole genome transcriptional profiling using custom B. thetaiotaomicron GeneChips that represent >98% of this organism’s protein coding genes. The transcriptome was analyzed near the midpoints of the two logarithmic growth phases on PMG (Fig. S4B; n=3 replicates/growth phase; 6 datasets total). Cultures that had been grown under identical conditions, but using glucose as a sole carbon source, served as reference controls (n=3 mid-log phase datasets). A total of 185 genes exhibited ≥10-fold changes in expression in response to in vitro growth on PMG: 173 (93.5%) of these were up-regulated.
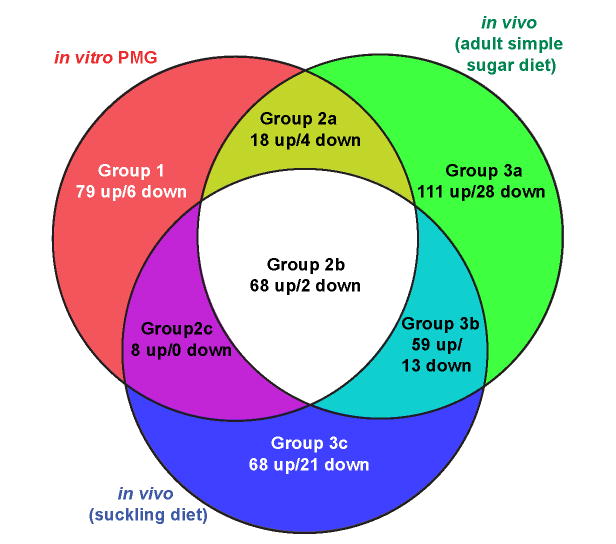
We next compared our in vitro data to two previously reported in vivo datasets obtained when B. thetaiotaomicron-colonized mice were fed diets lacking dietary glycans. One dataset was from the distal gut (cecal) contents of adult mice (n=3) fed a diet containing glucose and sucrose as the sole carbohydrates (‘simple sugar diet’) (Sonnenburg et al., 2005); the second was from the distal gut contents of 17 day old suckling mice (n=6; ‘suckling diet’) (Bjursell et al., 2006). By referencing the transcript signals from each of these in vivo datasets to the same minimal medium (MM) plus glucose control dataset used for the MM-PMG analysis, we were able to directly compare all three datasets in which B. thetaiotaomicron utilizes host glycans (Fig. 1). As with in vitro growth on PMG, most genes with ≥10-fold expression change in each of the in vivo datasets were up-regulated relative to MM-glucose: 256 of 303 genes (84.5%) in adult mice fed the simple sugar diet; 203 of 239 genes (84.9%) in 17d old suckling mice (see Table S2 for a list).
Figure 1. Comparison of in vivo and in vitro transcriptional profiles.
Genes from each of the three host glycan datasets that exhibited ≥10-fold increased (‘up’) or decreased (‘down’) expression relative to growth on MM-glucose are represented by individual circles in the Venn diagram. Regulated genes from both in vitro growth phases on MM-PMG were combined into one group, but are separated by growth phase in Fig. 2 and Table S2. Regions of overlap indicate inclusion of regulated genes from multiple lists.
The comparison shown in Fig. 1 yielded three groups of regulated genes: those specific to in vitro growth on PMG (Group 1, 85 genes); those shared between in vivo and in vitro growth (Group 2, 100 genes); and those specific to in vivo growth (Group 3, 300 genes). Identification of group 2 genes suggested that our in vitro model replicated some features of the glycan environment experienced by B. thetaiotaomicron in vivo. We also categorized genes from Groups 2 and 3 into three sub-groups (A, B and C) based on whether the genes were contributed from just one of the in vivo datasets (A and C subgroups) or both (B sub-groups). Partitioning of genes into these seven ‘sectors’ likely reflects bacterial responses to differences in host glycan biochemistry and/or availability. These differences could be species-specific (porcine versus murine), tissue-specific (gastric versus cecal mucosa) or age-specific (recently weaned versus adult hosts). They could also reflect variations between glycan use in vitro and in vivo (chemically extracted glycans versus those luminally exposed in the healthy gut).
Regulation of PUL expression during foraging on mucosal glycans in vitro
Based on coordinated transcription of the genes comprising several different PULs in vivo, and the similarities between these loci and others in B. thetaiotaomicron, we identified 88 putative PULs, which encompass 866 genes and represent 18.1% of the genome (see Table S3 for a list of these genes and Fig. S5 for a map of their locations). The minimum feature used to define each PUL was a single pair of susC/D genes (there are 101 such gene pairs in the B. thetaiotaomicron genome). However, the majority of PULs we delineated also encode other functions: 91% contain at least one gene in addition to the susC/D homolog pair; 68% contain one or more glycolytic enzymes; 72% contain a transcriptional regulator; and 61% contain both glycolytic enzymes and a transcriptional regulator (average PUL size is 10 genes).
Of the 485 genes regulated during growth on host glycans, 211 (44%) were associated with PULs (total of 43 different PULs). All of these PUL-associated genes showed increased expression relative to MM-glucose (range, 10.1 to 2,017-fold; mean, 114; median, 34; Fig. S6) and were distributed among all three groups of host glycan-responsive genes delineated in Fig. 1.
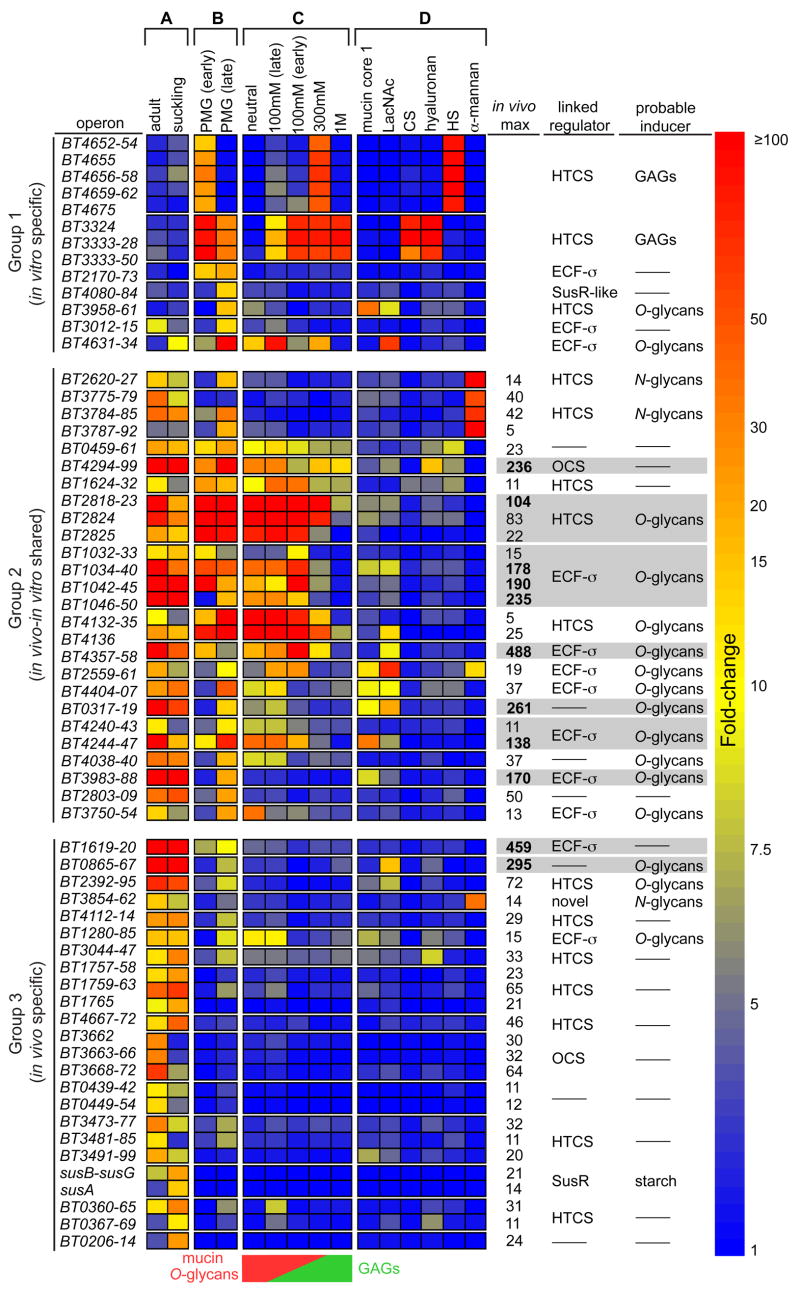
In many cases, not all genes within a predicted PUL were present in our list generated using a ≥10-fold cutoff. To more accurately portray the responses of entire PULs to fractionated mucosal glycans, we included all of the genes in the 43 PULs exhibiting host glycan responses in our next analysis: 213 remaining genes from these 43 PULs (excluding regulators, which typically did not show changes in expression) were added to our original list resulting in 424 genes (Fig. S6). To assist viewing gene induction trends across multiple bacterial growth conditions, we then sub-grouped the genes within each PUL into their predicted operons (Westover et al., 2005) and averaged the induction values for each operon into a single value. Sixty-four operons in 39 PULs, encompassing 264 genes, exhibited an average induction value ≥10-fold in response to the in vitro and/or in vivo growth conditions. These were retained for further expression analysis under more defined conditions (Table S4 and Fig. 2).
Figure 2. Responses of host glycan-induced PULs to fractionated PMG and purified glycans.
A heat map showing PUL operon induction by various host glycans. Each box represents the average fold-change (relative to MM-glucose) of all genes within the indicated operon (gene numbers listed vertically at left), and is calibrated according to the vertical bar at the extreme right. Individual PULs are separated by horizontal breaks in the heat map. Only PUL operons with average fold-change values ≥10 are shown. The gene coordinates of several PULs listed in the text are more expansive. Growth conditions are separated into four groups: (columns A,B) the original in vivo and in vitro datasets, respectively, outlined in Fig. 1; (column C) PMG glycan fractions; and (column D) pure forms of various host glycans or disaccharide components. The mucin O-glycan and GAG distribution in fractionated PMG glycans is schematized at the bottom of column ‘C’ as a reference. Columns adjacent to the vertical scale bar on the right describe the following notable characteristics of each PUL/operon: ‘in vivo max’, the maximum induction observed in either in vivo dataset for Group 2 and Group 3 loci (gray bars are used to highlight PULs with operons showing maximum induction >100-fold); ‘linked regulator’, the type of regulator associated with each PUL; ‘probable inducer’, the class of host-glycan that appears to induce each PUL (dashes indicate the lack of a conspicuously associated regulator or an inability to clearly predict the type of inducing host glycan from the data). Abbreviations: N-acetyllactosamine (LacNAc), chondroitin sulfate (CS), heparin sulfate (HS), hybrid two-component system (HTCS); extracytoplasmic function sigma/anti-sigma pair (ECF-σ, homologs of the inner membrane-spanning maltose sensor SusR (SusR-like), and cytoplasmic one-component system (OCS).
To determine which host glycan types stimulated PUL expression, we separated the PMG mixture into four fractions by anion exchange chromatography (mucin O-glycans and N-glycans contain more neutral sugars and less sulfation than GAGs, which contain higher negative charge due to both uronic acid and sulfate content). The resulting fractions were named according to the NaCl concentration used for their elution: neutral (flow-through fraction), 100mM, 300mM and 1M (Fig. S3). Monosaccharide analysis verified that individual fractions were enriched for either mucin O-glycans (mostly in neutral and 100mM, but some in 300mM fraction) or GAGs (100mM, 300mM and 1M; Table S1). The presence of abundant mannose in one fraction (100mM) indicated that it also contained N-glycans. B. thetaiotaomicron grew on all four fractions (see Fig. S4).
Two PULs are required for GAG utilization
We performed transcriptional profiling of B. thetaiotaomicron during growth on each of the four PMG fractions, as well as on a series of pure polysaccharides representing the linkages they contain (columns C and D in Fig. 2). Bacteria were harvested near the midpoint of logarithmic growth in each condition (n=2 per condition, sampling points for each are indicated in Fig. S4C–F). For growth on the 100mM glycan fraction, which produced a polyphasic profile, bacteria were sampled within the first prominent growth phase (‘100mM early’) as well as from three later phases: equivalent amounts of bacterial RNA from each of these later phases were pooled into one sample (‘100mM late’).
Many of the 39 ‘host glycan’-inducible PULs exhibited substrate-specific responses (Fig. 2). Two PULs belonging to Group 1, encompassed within BT4652-75 and BT3324-50, were highly induced during growth on the PMG fractions containing GAGs, as well as on pure forms of this class of glycans (columns C and D in Fig. 2). Each of these loci was specific for different GAGs: the BT4652-75 PUL was expressed exclusively in response to heparin sulfate while the BT3324-50 PUL responded to both chondroitin sulfate and hyaluronan (column D in Fig. 2; Fig. S7). Each of these two GAG-responsive PULs was most highly expressed during the first phase of growth on unfractionated PMG, and decreased expression during the second growth phase (column B in Fig. 2), consistent with our observation that B. thetaiotaomicron prioritizes GAGs relative to other mucosal glycans in vitro (Fig. S4H). Despite this apparent preference for GAGs in vitro, neither of these loci was comparably induced in vivo (column A in Fig. 2), suggesting that GAGs are not a major host glycan used in the mouse cecum, and may not be exposed in sufficient quantities in the healthy mucosa to be a useful nutrient foundation.
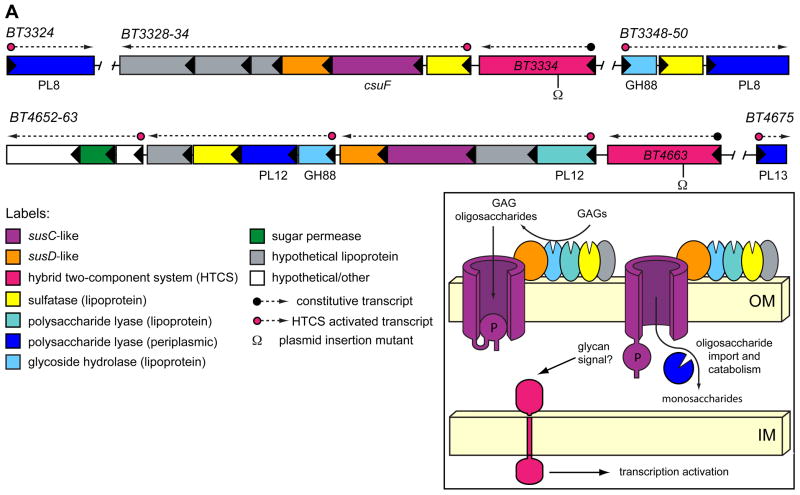
We made the unanticipated observation that, in addition to a core set of immediately adjacent genes, each of the GAG-induced PULs has one or more co-regulated genes that are not contiguously linked, but rather are positioned nearby in the genome (Fig. 3). Interestingly, the genomic regions that separate some of these displaced genes from the core PUL are occupied by different PULs (BT3344-47 and BT4667-73, Fig. S5). Thus, PUL gene displacements may reflect a legacy of intra- or inter-genomic events, such as homologous recombination or horizontal gene transfer, that involve multiple PULs resorting themselves near the same genomic regions.
Figure 3. GAG utilization PULs.
The genomic architecture of two PULs required for GAG utilization. Enzymatic functions are labeled according to their assigned CAZy glycoside hydrolase (GH) and polysaccharide lyase (PL) family membership. The susC homolog, BT3332 was previously identified in a transposon screen for chondroitin utilization mutants and named csuF (Cheng et al., 1995), although the surrounding sequences were not reported. The inset shows a model depicting the putative cellular locations of GAG-utilizing functions, and is based on similarities to the prototypic Sus PUL (Fig. S2). Inferred locations of constitutive and HTCS-activated promoters are indicated above PUL genes. Omega (Ω) symbols below HTCS genes indicate sites of plasmid insertions to disrupt these genes.
Like the Sus PUL, each GAG-responsive locus encodes a complement of enzymatic functions that equips it with the requisite activities to process its target substrate (Fig. 3). In the prototypic Sus model (Fig. S2), the endo-amylase lipoprotein, SusG, is positioned extracellularly where it plays an indispensable role in processing starch to malto-oligosaccharides (Shipman et al., 1999). Six of the enzymes encoded in the GAG-utilization PULs contain similar secretion and lipidation sequences, suggesting that they too are positioned outside the cell to perform initial glycan processing steps. Alternatively, some of these lipoproteins may remain periplasmically-oriented and catalyze downstream degradation of imported oligosaccharides. Three of these enzymes are sulfatases while the remaining three are glycolytic (two predicted unsaturated glucuronyl hydrolases and a heparinase; Fig. 3 and Table S5). In the prototypic Sus PUL, imported oligosaccharides undergo additional processing by periplasmic enzymes (Fig. S2). Similar processing of GAG oligosaccharides likely occurs through the action of four additional enzymes that contain N-terminal secretion signals, but no apparent lipidation site. These products, which encode chondroitin lyases and heparinases, may therefore be retained in the periplasm to catalyze the final degradation of imported oligosaccharides to simpler sugars (Fig. 3 and Table S5).
Each GAG-responsive PUL contains a single transcriptional regulator belonging to the hybrid two-component system (HTCS) class. This group of sensor/regulators is prominent in Bacteroides species: each contains the domains present in a classic two-component system (sensor histidine kinase and response regulator) but fused into a single polypeptide (Sonnenburg et al., 2006). Neither of these HTCS genes exhibited a noticeable expression change during growth on GAGs, indicating that if they are responsible for activating gene expression in response to GAGs, they are not auto-regulated (Fig. S7).
To test whether these regulators activated their adjacent PULs, we constructed separate plasmid-insertion mutants in HTCS genes BT3334 and BT4663 and evaluated the ability of these two mutants to grow in minimal medium with one of four different GAGs (chondroitin sulfate, hyaluronan, dermatan sulfate or heparin sulfate; Table S6). Disruption of BT3334 eliminated the ability to grow on chondroitin sulfate, hyaluronan and dermatan sulfate, but did not affect growth on heparin sulfate. Conversely, mutation of BT4663 only impaired growth on heparin sulfate. Thus, B. thetaiotaomicron contains two discrete HTCS-regulated PULs that are responsible for catabolizing four chemically distinct GAG species.
The requirement of one system (BT3324-50) for metabolism of three different GAGs is notable because this system’s substrates are varied chemically in two ways: (i) in contrast to chondroitin sulfate and dermatan sulfate, hyaluronan is devoid of sulfation; and (ii) each of the three GAGs varies one of the monosaccharides contained in its repeating region (Table S6 and Fig. S1). The observation that these systems encode lipidated sulfatases that may be surface-exposed, is consistent with extracellular GAG de-sulfation, as this process would render all three substrates free of sulfate prior to import. Recognition of these three different GAGs requires that the binding and catalysis functions associated with this system are sufficiently promiscuous to accommodate all three epimeric compounds. The observed association of HTCS BT3334 with chondroitin sulfate sensing is also insightful with respect to earlier work, which revealed that the minimum chondroitin sulfate oligosaccharide length capable of inducing B. thetaiotaomicron chondroitinase activities is an octasaccharide (Salyers and Kotarski, 1980). Thus, some piece of the machinery required for binding, sensing and importing chondroitin sulfate must interact with glycan derivatives ≥8 sugars long for activation to occur.
N-glycan foraging in vivo
To determine whether B. thetaiotaomicron forages N-glycans in the mouse gut, we also profiled bacteria grown in vitro on α-mannan derived from the cell wall of Saccharomyces cerevisiae. This substrate, composed of an α1,6-mannose backbone substituted with α1,2- and α1,3-linked side chains (Cawley and Ballou, 1972), is a chemical proxy for the same α-mannosidic linkages that make up the core region of nascent and/or mature mammalian N-glycans (Fig. S1).
Growth on α-mannan stimulated expression of genes within two Group 2 PULs (BT2620-27 and BT3773-92) and one Group 3 PUL (BT3854-62) (see column D in Fig. 2). The fact that all three of these systems are expressed in vivo in mice fed a diet lacking exogenous α-mannans suggests that these PULs are deployed in the mouse gut in response to N-glycans. However, these systems were only modestly induced compared to PULs showing the highest activity in vivo, suggesting that N-glycans are not the most abundant host glycans foraged in the cecum (see column labeled ‘in vivo max’ in Fig. 2, plus Fig. S8). Additional descriptions of the enzyme content and associated regulators for these three α-mannan PULs are presented in Supplemental Results and Discussion.
Mucin O-glycans are a prominent host substrate foraged in vivo
Most of the remaining Group 1 and Group 2 loci depicted in Fig. 2 exhibit induction patterns consistent with recognition of mucin O-glycans (hereafter referred to as ‘O-glycans’). This idea is supported by the observation that these loci show maximum induction in response to the neutral and 100mM NaCl PMG fractions, which contain the highest O-glycan concentrations. Many of these loci also respond to disaccharide components of O-glycans: mucin core 1 (Galβ1,3GalNAc) and/or N-acetyllactosamine (Galβ1,4GlcNAc) (columns C and D in Fig. 2). The fold levels of induction of some O-glycan PULs are among the largest exhibited by B. thetaiotaomicron in vivo: among the nine PULs that contain one or more operons exhibiting ≥100-fold induction in vivo (highlighted in gray in Fig. 2), seven show in vitro expression profiles consistent with responsiveness to O-glycans.
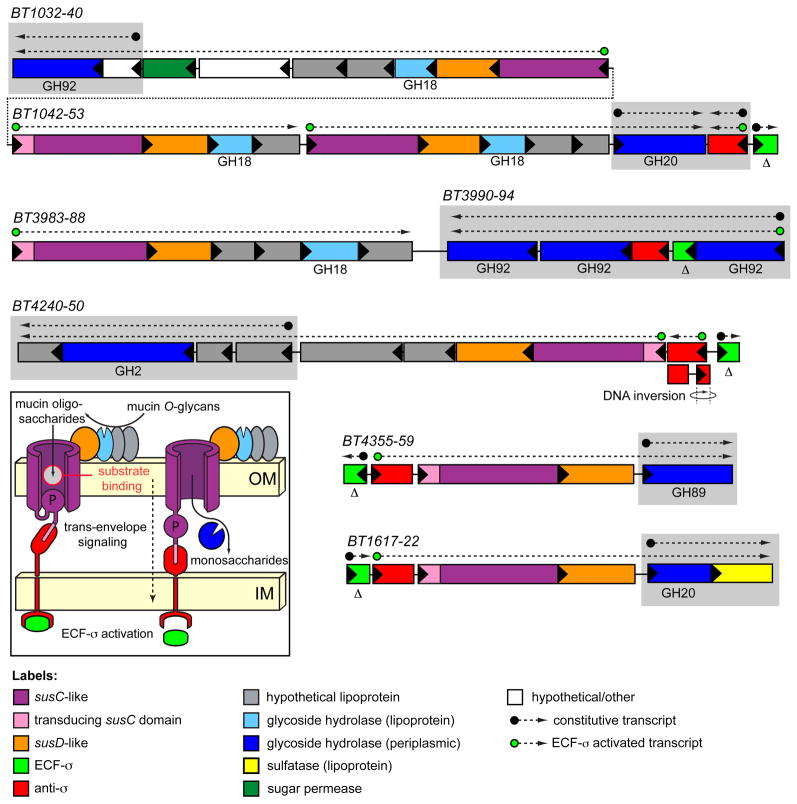
The architecture of O-glycan responsive PULs is similar to those noted for the GAG- and N-glycan responsive loci (Fig. 4 and Fig. S9). However, some distinctive features are apparent. The most striking is that among 16 PULs with probable specificities for O-glycans, nine are coupled to extracytoplasmic function sigma (ECF-σ) factor/anti-σ factor pairs in place of the previously described sensor/regulator types. Of the remaining seven loci, four are coupled to HTCS regulators and three are not conspicuously associated with any regulator. Based on similarity to regulatory systems in other bacteria, it is likely that the ECF-σ/anti-σ regulators control PUL gene expression via a mechanism termed ‘trans-envelope signaling’, which was first described for the Escherichia coli ferric dicitrate (Fec) iron acquisition complex (Braun and Mahren, 2005; Koebnik, 2005). In trans-envelope signaling, a specialized TonB-dependent outer membrane receptor signals, via protein-protein interactions, to the periplasmic domain of an inner membrane spanning anti-σ factor, which in turn controls the activity of a cytoplasmic ECFσ factor (Fig. 4 inset).
Figure 4. Mucin O-glycan utilization PULs.
Four loci induced during growth in the presence of mucin O-glycans, and a fifth in vivo-specific system (BT1617-22). Note the presence of an extra N-terminal trans-envelope signaling domain on one of the susC homologs associated with each system (Koebnik, 2005). The anti-σ factor associated with the BT4240-50 locus was assembled in the VPI-5482 genome as two divergent genes (BT4248-49) but undergoes a DNA inversion that positions BT4249 upstream of BT4248, resulting in an intact anti-σ factor gene (see Fig. S12 for details). Gray boxes indicate genes for which elevated basal transcription was observed (see Supplemental Data). Delta (Δ) symbols below ECF-σ genes indicate that they were targeted for in-frame deletions for in vivo competition experiments.
The enzyme content of O-glycan responsive PULs is consistent with metabolism of O-glycans (Table S5). Prominently represented in these loci are glycoside hydrolases belonging to CAZy family 18 (GH18), which includes endo-β-N-acetylglucosaminidases. Each of these GH18 members is predicted to be lipidated, indicating potential surface-exposure like SusG. This notion is consistent with a model in which these activities cleave within the N-acetyllactosamine (LacNAc) repeats of O-glycan chains (Fig. S1). A single PUL (BT2818-26; Fig. S9) also contains a family16 glycoside hydrolase (endo-β-1,4-galactosidase). In contrast to the GH18 enzymes, this activity is predicted to make an adjacent cut (β1,3Gal instead of β1,4GlcNAc) within the O-glycan LacNAc extension. Thus, these two activities may act in concert to degrade larger O-glycans into smaller oligosaccharides by targeting two of their most typical glycosidic linkages. Liberated oligosaccharides are likely further degraded in the periplasm prior to import of the resulting mono- and disaccharides (the substrate specificities and transcriptional responses of the enzymes that likely facilitate this conversion are discussed in Supplemental Results and Discussion).
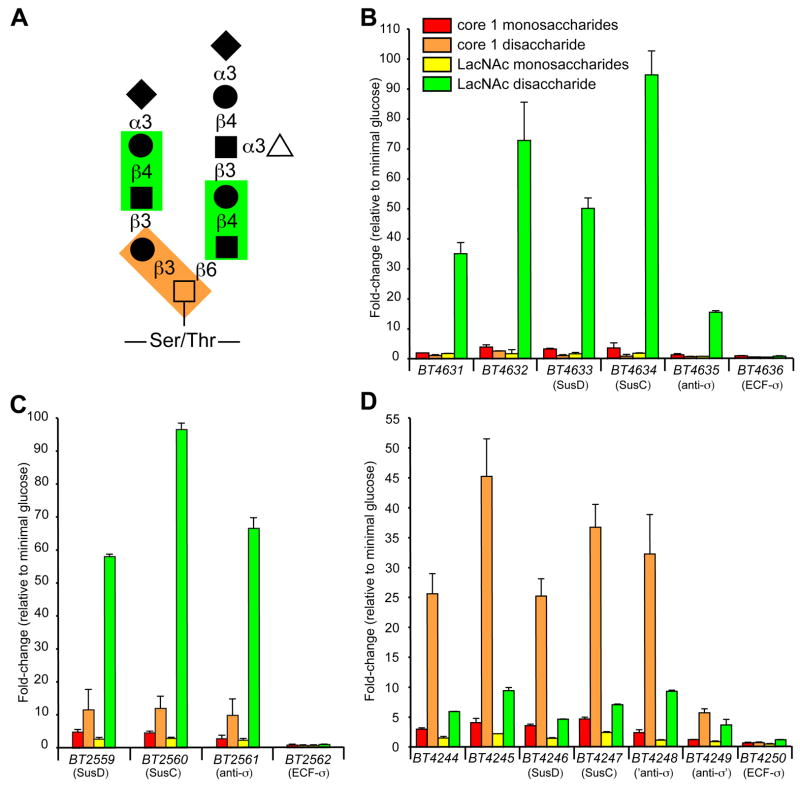
The specific responses of some ECF-α-linked PULs to individual substrates (e.g., core 1 vs. LacNAc in Fig. 2) led us to conclude that some systems were responding to different glycosidic linkages that could potentially occur within a single glycan molecule. To explore this further, we generated transcriptional profiles of B. thetaiotaomicron grown on equimolar amounts of the monosaccharides contained in core 1 and LacNAc (Gal/GalNAc and Gal/GlcNAc, respectively; Fig. 5). We then compared the responses of individual ECF-σ-linked loci that were induced by either core 1 or LacNAc disaccharides to growth on their respective monosaccharide components. The results for three loci provide clear evidence that these systems are sensitive to these differences in glycosidic linkage: two systems (BT4631-36 and BT2559-62) show strong responses to LacNAc but not core 1 (Fig. 5B,C); a third system (BT4244-4250) shows the opposite specificity for core 1 and not LacNAc (Fig. 5D). Furthermore, all three systems respond better to their cognate disaccharide compared to monosaccharide controls, indicating that both carbohydrate composition and glycosidic linkage contribute to the activation of these PULs.
Figure 5. Specificity of ECF-σ-linked PULs for O-glycan components.
(A) Representative core 2 mucin O-glycan structure. Two common disaccharides, core 1 and N-acetyllactosamine (LacNAc), are highlighted by orange- and green-shaded boxes, respectively. Monosaccharide symbols: GalNAc (open squares); Gal (closed circles); GlcNAc (closed squares); Fuc (open triangles); and NeuNAc (closed diamonds). (B) GeneChip-based induction values for genes in the BT4631-36 locus during growth on equimolar amounts of core 1 monosaccharides, core 1 disaccharide, LacNAc monosaccharides, and LacNAc disaccharide. Values are expressed as fold-difference relative to growth on MM-glucose. Color codes are the same in B–D. (C) Responses of genes in the BT2559-62 locus to growth on O-glycan mono- and disaccharide components. (D) Responses of genes in the BT4244-50 locus to growth on O-glycan mono- and disaccharide components. Note the differing substrate specificities for LacNAc and core 1 disaccharides. Values represent the mean±range of two biological replicates performed for each substrate.
O-glycan foraging contributes to fitness in vivo
The notion that B. thetaiotaomicron principally targets O-glycans in vivo suggests that this host nutrient niche may help stabilize the bacterial population despite perturbations in dietary glycan intake by the host. Therefore, we constructed a B. thetaiotaomicron mutant (‘ΔECF-σ’) that contains deletions of five ECF-σ genes associated with use of O-glycans (see Fig. 4), anticipating that this mutant would have reduced ability to forage O-glycans in vivo. We then colonized three groups of germfree NMRI mice, each with one of three strains: the Δ5ECF-σ mutant, its isogenic parent (‘wild-type’), or a complemented mutant strain (‘complement’) that harbors a restored chromosomal copy of all deleted ECF-σ genes with their native promoters. Mice were maintained on a simple sugar diet for 10d and then sacrificed. Bacterial density was 5.4±3.4×1011 cfu ml−1 for wild-type (n=4); 3.9±2.3×1011 cfu ml−1 in the case of Δ5ECF-σ (n = 5); and 3.8±3.2×1011 cfu ml−1 for the complemented strain (n = 5). Thus, in the absence of competing strains, the Δ5ECF-σ mutant colonizes similarly to wild-type.
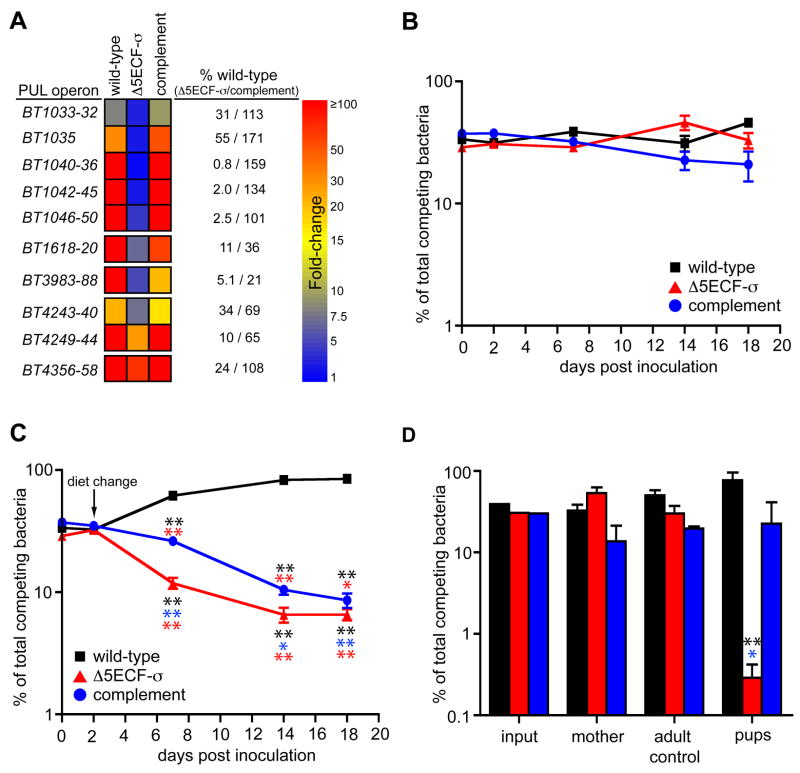
To determine the effects of the ECF-σ deletions on PUL expression in vivo, we obtained whole genome transcriptional profiles using bacterial RNA extracted from the cecal contents of these mice (n= 3 animals surveyed for each bacterial strain). In vivo expression data was compared to the same MM-glucose reference used in Fig. 1. As expected, the mutant harboring five ECF-σ gene deletions exhibited reduced expression of the O-glycan responsive PULs associated with the disrupted ECFσ genes (Fig. 6A), demonstrating that these transcription factors regulate PUL expression. In two cases, ECF-σ deletion did not completely reduce expression of adjacent PUL genes to the level observed in MM-glucose. This was most clearly illustrated with the BT4356-58 system, which sustained a 76% reduction relative to wild-type, but still exhibited 72-fold induction relative to MM-glucose. This phenomenon may reflect cross-activation from non-mutated ECF-σ systems that are also activated under these in vivo conditions.
Figure 6. Effects of multiple ECF-σ deletions on mucin O-glycan foraging, fitness and transmission in vivo.
(A) Expression of five ECF-σ-linked mucin O-glycan PULs in isogenic wild-type, mutant (Δ5ECF-σ) and complemented B. thetaiotaomicron strains. Fold-differences in expression are calibrated to the same scale as Fig. 2. Numbers to the right of the GeneChip-based heat map indicate the percent of wild-type expression observed in the Δ5ECF-σ and complement strains. Note that in Δ5ECF-σ cells, some PUL systems still exhibit substantial induction in vivo, indicating that other mechanisms are partially responsible for their activation. Likewise, not all systems are restored to wild-type levels by complementation. (B) Competitive colonization of adult germfree mice fed a standard plant glycan-rich diet by wild-type (black squares), Δ5ECF-σ (red triangles) and complemented strains (blue circles) (values plotted are mean ± one standard deviation). (C) Colonization of mice that were started on the same plant glycan-rich diet as in panel B, and then switched to a simple sugar diet on day 2, by wild-type (black squares), Δ5ECF-σ (red triangles) and complemented strains (blue circles). (D) Relative proportions of each competing strain in the input inoculum, mother, a co-housed adult female germfree control, and pups at various points in the natural transmission experiment. Values shown for the mother and adult female control are the average of samples taken at days −8, 0, 4, 11 and 15, or days 4, 11 and 15 relative to delivery, respectively. Values shown for the pups are the average of eight individually assayed animals at postnatal day 15. Error bars in panels B–D represent 1 standard deviation. Instances where a given strain’s population differs significantly from another strain(s) at the same time point are indicated with asterisks, (color-matched to the strain from which they differ). Later time points where the Δ5ECF-σ population differed significantly from its own 2d level (red asterisks) are also indicated in panel C (*, P≤0.05; **, P≤0.01 by Student’s t-test). For statistical comparisons in panel C, population values for the wild-type and mutant strains were normalized according to their initial abundance relative to Δ5ECF-σ at day 2.
To determine if the Δ5ECF-σ mutant had reduced fitness as a result of its O-glycan PUL defects, we performed competitive colonization experiments with this strain and its isogenic wild-type and complemented strains. Each of the three strains was labeled with a unique 24bp genomic signature-tag that allowed it to be differentiated from its competitors, and then mixed in equal proportions prior to inoculation of ten adult male germfree mice (~108 cfu total per animal by oral gavage). For the first 2d of the experiment, all 10 mice were fed a plant glycan-rich diet. After 2d, the colonization level of each strain was assessed in each mouse by assaying the proportion of each genomic signature-tag present in fresh feces. All three strains showed similar abundance, indicating that each was able to initiate colonization in mice fed this diet.
After verifying colonization at 2d, one group of 5 mice was switched to the simple-sugar diet used in previous experiments while the remaining 5 were retained on the plant glycan rich diet. Bacterial colonization of all 10 mice was monitored up to an endpoint of 18d post colonization. Over the time course, bacterial populations in the guts of mice fed the glycan-rich diet remained relatively stable: the maximum population fold-changes for each strain were +1.5 for the wild-type, +1.5 for the Δ5ECF-σ and −1.8 for the complemented strain relative to 2d levels (Fig. 6B). However, in mice that were switched to a simple-sugar diet at 2d, the population of wild-type bacteria consistently increased at later times, whereas both the Δ5ECF-σ and complement strains continually declined, suggesting that they were less fit than wild-type (Fig. 6C). From these data, we concluded that the Δ5ECF-σ mutant exhibits a diet-specific fitness defect: this strain’s diminished capacity to forage O-glycans in vivo causes it to be less persistent under conditions that force reliance on this host nutrient niche. Although the complemented strain also declined over time, the Δ5ECF-σ population decreased significantly faster, suggesting it harbors a more substantial defect. Our ability to only partially restore wild-type transcription levels of some genes in the complemented strain (e.g., the BT1618-20, BT3983-88 and BT4240-BT4249 PULs, Fig. 6A) likely accounts for its residual defect, and may indicate that the PULs for which only partial expression was restored are more critical for persistence in the adult mouse.
To further explore the contribution of mucin O-glycan foraging to colonization of the mouse gut, we assayed the abilities of the wild-type, Δ5ECF-σ and complemented strains to be naturally transmitted from an adult mother to her offspring in the days immediately after their birth. A pregnant germ-free mother, consuming the plant glycan-rich diet, was colonized 10d prior to delivering her litter with a similar mixture of the three competing strains used in our previous competition experiment. Fresh fecal pellets were sampled 2d after inoculation (−8d relative to parturition), as well as on the day her litter was born (0d), and at days 4, 11 and 15 post-delivery.
Our previous competition experiment indicated that each of the competing strains was adept at colonizing the adult mouse gut when artificially inoculated by gavage. As a control for natural adult-to-adult transmission, we added a single germ-free adult female mouse to the cage in which the colonized mother was housed on the same day her pups were born. This germ-free female was exposed to the same maternal fecal microbiota as the newborn pups. However, in contrast to the pups, which were nursing on mother’s milk, the adult co-housed female consumed a diet rich in plant glycans. Fresh fecal pellets from this adult control were sampled at days 4, 11 and 15 after delivery of the pups. Levels of the three strains remained stable in the mother and in the co-housed female: over the time course, all strains colonized to levels that were within 2.1-fold of the mother’s microbiota on the day of birth (Fig. 6D). Thus, each of the three strains tested has the ability to be naturally transmitted between adult mice consuming a plant-rich diet via the fecal-oral route and to persist there for at least 15d.
Finally, we assayed the ability of each of the competing B. thetaiotaomicron strains to be transmitted from the colonized mother to her pups. Eight pups were born to the colonized mother and these were sacrificed at 15d after delivery. Although the small size of the neonatal mice did not allow reliable sampling of fecal material throughout the 15d time course, we obtained a fresh colonic fecal sample from each mouse at the time of sacrifice. The population fold-change of each competing strain after 15d were +2.8 for wild-type (relative to the abundance of each strain in the mother’s microbiota on the day of birth), -201 for Δ5ECF-σ and +1.6 for the complemented strain (Fig. 6D). Thus, in a fecal-oral transmission model that accounts for the infant animals’ consumption of mother’s milk, the ability of B. thetaiotaomicron to forage on mucin O-glycans is a major contributor to its ability to colonize and/or persist in the distal gut. Furthermore, loss of these five systems is not compensated for by the bacterium’s other glycan acquisition functions, including several different PULs, which remain intact and are expressed by our mutant strain in the mouse gut.
Prospectus
The factors influencing microbial community structure in the human gut are poorly understood, but likely are manifold. Based on the diversity of complex glycans that human gut Bacteroides spp. can ferment (Salyers et al., 1977a), the large repertoire of glycan acquisition genes in their genomes (Xu et al., 2007), and the results of the experiments reported in this study, we conclude that the capacity for dynamic glycan foraging is an important evolved component of their adaptation to the gut habitat. A thorough understanding of how glycan foraging contributes to microbial colonization and persistence is contingent upon being able to deconstruct the complex gut ecosystem and to understand the contributions of its component parts. We were able to specifically examine the role of host glycans in B. thetaiotaomicron colonization using gnotobiotic animals. In this vastly simplified model of the distal gut ecosystem, this bacterium principally forages on mucin O-glycans although it is capable of using all major types of host glycans. The importance of mucin O-glycan foraging by organisms like B. thetaiotaomicron remains to be fully investigated in the context of both increasing dietary complexity, and competition between the thousands of species, each with its own glycolytic repertoire, that are also competing to occupy this niche.
The sequenced human gut-derived B. thetaiotaomicron type strain VPI-5482 devotes 18% of its genome to PULs. Our finding that fewer than half of its 88 putative PULs are deployed in response to host glycans suggests that the others are responsive to a wide variety of plant-derived substrates: e.g., recent work has indicated that different PULs from B. thetaiotaomicron and other gut species such as Bacteroides ovatus can target plant pectic and hemicellulosic glycans (E. C. Martens, D. N. Bolam and J. I. Gordon, unpublished). Moreover, Sus-like PUL systems are detectable in all but one lineage in the Bacteroidetes for which there are draft or finished genome sequences (Fig. S13): the one exception is Candidatus sulcia muelleri, an endosymbiont of Hemipteran insects that has a markedly streamlined 250 kb genome reflecting its evolved adaptation to its specialized intracellular habitat (Moran et al., 2005).
In some respects, Bacteroidetes Sus-like systems are reminiscent of another microbial paradigm for glycan utilization, the cellulosome (Bayer et al., 2004): each is a multi-protein cell-envelope system that contains both carbohydrate binding and glycolytic functions. However, despite some previous data supporting interactions between the SusC and SusD proteins (Shipman et al., 2000), more detailed studies are needed to determine whether the other outer membrane components in these systems either participate in a multi-protein complex, or function as separate entities. In contrast to microbial cellulosomes, which to date have been shown to degrade cellulose, hemicelluloses and pectin (Bayer et al., 2004), the multi-component Sus-like systems may enable broader substrate plasticity.
Finally, the present study provides a starting point for further defining the factors that shape microbial ecology in health and disease, and that influence interactions between the microbiota and the host immune system. Limited surveys of patients with inflammatory bowel diseases have revealed alterations in mucus glycans and more intimate interactions between gut bacteria and the mucus layer that overlies the intestinal epithelium (Pullan et al., 1994; Swidsinski et al., 2007). In this view, the repertoire of PULs present in members of the Bacteroidetes, together with host glycan content and diet, may under certain circumstances conspire to determine whether a microbiota has pathologic potential.
Experimental Procedures
Bacterial strains and culture conditions
Bacterial strains and plasmids are summarized in Table S8. B. thetaiotaomicron was routinely grown in tryptone-yeast extract-glucose (TYG) medium (Holdeman et al., 1977) or on brain-heart infusion (BHI; Beckton Dickinson) agar supplemented with 10% horse blood (Colorado Serum Co.). Antibiotics were added as appropriate: erythromycin (25 μg ml−1), gentamicin (200 μg ml−1), tetracycline (2 μg ml−1) and 5-fluoro-2′-deoxyuridine (FUdR, 200 μg ml−1). Minimal medium (MM) contained 100mM KH2PO4 (pH 7.2), 15mM NaCl, 8.5mM (NH4)2SO4, 4mM L-cysteine, 1.9μM hematin/200μM L-histidine (prepared together as a 1,000x solution), 100μM MgCl2, 1.4μM FeSO4·7H2O, 50μM CaCl2, 1μg ml−1 vitamin K3, and 5ng ml−1 vitamin B12. All carbon sources were added to MM [final concentration 0.5% (w/v) except unfractionated PMG glycans which were 1.0% (w/v)]. Media were filter sterilized using a Millipore Express filter unit (0.22μm pore diameter). Additional culture details are provided in Supplemental Data.
Preparation and analysis of porcine mucosal glycans
Porcine mucosal glycans (PMG) were purified from porcine gastric mucin (Type III, Sigma) as previously described (Manzi et al., 2000) using a combination of proteolysis and alkaline β-elimination to break down mucin glycopeptides and release O-and N-linked glycans. Liberated glycans were dialyzed against water, lyophilized, dissolved in ddH2O (20 mg ml−1) and stored at −20°C. A portion of the prepared glycans was fractionated using anion exchange chromatography (Manzi et al., 2000), according to the scheme outlined in Fig. S3. The relative masses of fractionated material recovered after ion-exchange chromatography were 4:1:4:1 for the neutral, 100mM NaCl, 300mM and 1M fractions, respectively. The monosaccharide compositions of unfractionated and fractionated PMG glycans were analyzed at the University of California San Diego Glycotechnology Core Resource by high pH anion exchange chromatography with pulsed amperometric detection.
Whole genome transcriptional profiling
Transcriptional profiling was performed using custom Affymetrix GeneChips containing probesets representing >98% of 4,779 predicted protein-coding genes in the B. thetaiotaomicron genome (Sonnenburg et al., 2005). GeneChip targets were prepared from whole bacterial RNA as previously described (Bjursell et al., 2006) and hybridized to the microarrays according to standard Affymetrix protocols (www.affymetrix.com). Data were normalized using Microarray Suite 5 software (Affymetrix) by adjusting the average B. thetaiotaomicron transcript signal on each GeneChip to an arbitrary value of 500. Subsequent GeneChip data comparisons were performed using GeneSpringGX 7.3.1 software (Agilent) and the following workflow: (i) raw intensity values ≤1.0 were adjusted to 1 prior to calculating fold-changes; (ii) genes with average fold change of ≥10 in each experimental data set were identified; and (iii) the list of genes exhibiting ≥10-fold changes was restricted to include only those for which the fold-difference had a P-value ≤0.01 (t-test), a ‘Present’ or ‘Marginal’ call in the majority of GeneChips, and an intensity value in the higher expression state ≥100. Operons used in Fig. 2 were delineated based on a previous analysis (Westover et al., 2005) that assigned each gene a probability (between 0 and 1, with 1 being most probable) of being transcriptionally linked to its adjacent genes based on intergenic distance and functional similarity. Adjacent genes with a probability ≥0.5 were included in the same operon.
All of the GeneChip data used in this study are available from the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/projects/geo/). Additional details of bacterial growth conditions, experimental parameters and GEO accession numbers for each experiment are provided in Supplemental Data and Table S7.
Genetic manipulation of B. thetaiotaomicron
B. thetaiotaomicron mutants were constructed either by insertion-duplication mutagenesis (IDM) using the Bacteroides suicide plasmid pGERM (Salyers et al., 2000), or by allelic exchange resulting in unmarked, in-frame gene deletion (Koropatkin et al., 2008). For mutant complementation and strain signature-tagging, we used two different NBU2-based (Wang et al., 2000) genomic insertion vectors (pNBU2-ermGb and pNBU2-tetQb) that differ only in the antibiotic resistance marker that each encodes. Complementation of the Δ5ECF-σ mutant was accomplished by amplifying wild-type alleles encoding each ECF-σ gene and promoter region using primers listed in Table S9. In the case of BT3993, which does not have a discernable σAB promoter (Vingadassalom et al., 2005), we created a hybrid allele in which the BT1053 promoter (mapped using 5′ RACE; data not shown) was fused to the start codon of BT3993. Each of the five complementing alleles was ligated in tandem in the pNBU2-ermGb MCS site and the resulting construct inserted into NBU2 att1 (tRNASer between BT4680 and BT4681). The corresponding uncomplemented and wild-type strains, used in competition experiments, each harbored an empty copy of pNBU2-ermGb at the same site to control for any effects of NBU2 insertion. Each competing strain was also signature-tagged with a pNBU2-tetQb construct containing a unique 24bp sequence, which allowed each, individually labeled strain to be quantitatively identified by quantitative real-time PCR (qPCR, described below).
Competitive colonization of gnotobiotic mouse
All mice were from the NMRI-KI inbred line and were reared in gnotobiotic isolators as previously described (Bjursell et al., 2006). Six week-old germfree animals were used for the competitive colonization experiment. Mice were gavaged with 100μl of a mixture containing approximately equal proportions of each of the three competing strains (~108 cfu ml−1 total; an aliquot of the input inoculum was frozen for bacterial enumeration and corresponds to the day 0 time points in Fig. 6B,C). All subsequent community enumerations from mice were determined from freshly collected fecal pellets. Controls performed at the endpoint of the colonization experiment indicated that community enumerations in fecal pellets and cecal contents were indistinguishable (data not shown).
For the maternal transmission experiment, a single pregnant 8 week-old female was inoculated with competing strains, as described above, and housed in isolation until her delivery day. On the same day that she delivered her litter of 8 pups, a single germfree 5 week-old female mouse was also added to the cage to serve as an adult-to-adult transmission control. Fresh fecal pellets from the mother and adult control were assayed at the indicated time points.
qPCR enumeration of competing strains in vivo
Competing strains were enumerated using qPCR to quantify the relative amounts of signature-tagged genomic DNA from each strain contained in total fecal DNA. Total DNA was isolated from fecal pellets by performing an initial bead-beating/phenol:chloroform extraction (Ley et al., 2005) followed by purification using the DNeasy Blood and Tissue Kit (Qiagen). For each mouse sampled at each time point, 10ng of fecal DNA was assayed in duplicate in a Stratagene Mx3000P machine, using previously described parameters (Bjursell et al., 2006) at an assay temperature of 78°C. Purified genomic DNA standards prepared from each tagged strain were included in each qPCR run (range, 100, 20, 2, 0.4, 0.08 and 0.01ng). A standard curve was generated from these standards and used to calculate the relative representation of each strain in each sample. Control experiments in which individual tagged genomes were omitted, or reduced in concentration among pools of other tagged genomes, indicated that our method achieved both tag-specific sensitivity and a linear response over the DNA concentrations employed as standards (data not shown).
Additional details of each of the procedures described here are provided in Supplemental Data.
Supplementary Material
Acknowledgments
We thank Sabrina Wagoner, Maria Karlsson and David O’Donnell for superb technical assistance and Laura Kyro for help with graphics. The University of California at San Diego Glycotechnology Core Resource skillfully performed all host glycan analyses. Jeremiah Faith, Andrew Goodman, Nicole Koropatkin, Federico Rey and Justin Sonnenburg provided many helpful suggestions during the course of these studies. This work was supported in part by a grant from the NIH (DK30292). E.C.M. is the recipient of NIH post-doctoral fellowships T32 HD07409 and F32 AI073060. H.C was supported by Washington University Medical Scientist Training Program (GM07200) and by NIH training grant HG00045.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An G, Wei B, Xia B, McDaniel JM, Ju T, Cummings RD, Braun J, Xia L. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J Exp Med. 2007;204:1417–1429. doi: 10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EA, Belaich JP, Shoham Y, Lamed R. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Ann Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- Braun V, Mahren S. Transmembrane transcriptional control (surface signalling) of the Escherichia coli Fec type. FEMS Microbiol Rev. 2005;29:673–684. doi: 10.1016/j.femsre.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Cawley TN, Ballou CE. Identification of two Saccharomyces cerevisiae cell wall mannan chemotypes. J Bacteriol. 1972;111:690–695. doi: 10.1128/jb.111.3.690-695.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Yu MC, Reeves AR, Salyers AA. Identification and characterization of a Bacteroides gene, csuF, which encodes an outer membrane protein that is essential for growth on chondroitin sulfate. J Bacteriol. 1995;177:3721–3727. doi: 10.1128/jb.177.13.3721-3727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science (New York, NY. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nature Rev. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Gilbert HJ. Cellulosomes: microbial nanomachines that display plasticity in quaternary structure. Mol Microbiol. 2007;63:1568–1576. doi: 10.1111/j.1365-2958.2007.05640.x. [DOI] [PubMed] [Google Scholar]
- Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, Thornton DJ, Png CW, Crockford TL, Cornall RJ, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS medicine. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdeman LV, Cato ED, Moore WEC. Anaerobe Laboratory Manual. Blacksburg, Va: Virginia Polytechnic Institute and State University Anaerobe Laboratory; 1977. [Google Scholar]
- Koebnik R. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 2005;13:343–347. doi: 10.1016/j.tim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1–11. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Nat Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of mammals and their gut microbes. Science (New York, NY. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Manzi AE, Norgard-Sumnicht K, Argade S, Marth JD, van Halbeek H, Varki A. Exploring the glycan repertoire of genetically modified mice by isolation and profiling of the major glycan classes and nano-NMR analysis of glycan mixtures. Glycobiology. 2000;10:669–689. doi: 10.1093/glycob/10.7.669. [DOI] [PubMed] [Google Scholar]
- Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005;71:8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host & Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullan RD, Thomas GA, Rhodes M, Newcombe RG, Williams GT, Allen A, Rhodes J. Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut. 1994;35:353–359. doi: 10.1136/gut.35.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AR, D’Elia JN, Frias J, Salyers AA. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J Bacteriol. 1996;178:823–830. doi: 10.1128/jb.178.3.823-830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AR, Wang GR, Salyers AA. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol. 1997;179:643–649. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Bonheyo G, Shoemaker NB. Starting a new genetic system: lessons from Bacteroides. Methods (San Diego, Calif. 2000;20:35–46. doi: 10.1006/meth.1999.0903. [DOI] [PubMed] [Google Scholar]
- Salyers AA, Kotarski SF. Induction of chondroitin sulfate lyase activity in Bacteroides thetaiotaomicron. J Bacteriol. 1980;143:781–788. doi: 10.1128/jb.143.2.781-788.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, West SE, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl Environ Microbiol. 1977a;34:529–533. doi: 10.1128/aem.34.5.529-533.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Vercellotti JR, West SE, Wilkins TD. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol. 1977b;33:319–322. doi: 10.1128/aem.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman JA, Berleman JE, Salyers AA. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J Bacteriol. 2000;182:5365–5372. doi: 10.1128/jb.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman JA, Cho KH, Siegel HA, Salyers AA. Physiological characterization of SusG, an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1999;181:7206–7211. doi: 10.1128/jb.181.23.7206-7211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Sonnenburg JL, Manchester JK, Hansen EE, Chiang HC, Gordon JI. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc Nat Acad Sci USA. 2006;103:8834–8839. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science (New York, NY. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Spence C, Wells WG, Smith CJ. Characterization of the primary starch utilization operon in the obligate anaerobe Bacteroides fragilis: Regulation by carbon source and oxygen. J Bacteriol. 2006;188:4663–4672. doi: 10.1128/JB.00125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Sydora BC, Doerffel Y, Loening-Baucke V, Vaneechoutte M, Lupicki M, Scholze J, Lochs H, Dieleman LA. Viscosity gradient within the mucus layer determines the mucosal barrier function and the spatial organization of the intestinal microbiota. Inflamm Bowel Dis. 2007;13:963–970. doi: 10.1002/ibd.20163. [DOI] [PubMed] [Google Scholar]
- Vingadassalom D, Kolb A, Mayer C, Rybkine T, Collatz E, Podglajen I. An unusual primary sigma factor in the Bacteroidetes phylum. Mol Microbiol. 2005;56:888–902. doi: 10.1111/j.1365-2958.2005.04590.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Shoemaker NB, Wang GR, Salyers AA. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J Bacteriol. 2000;182:3559–3571. doi: 10.1128/jb.182.12.3559-3571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westover BP, Buhler JD, Sonnenburg JL, Gordon JI. Operon prediction without a training set. Bioinformatics (Oxford, England) 2005;21:880–888. doi: 10.1093/bioinformatics/bti123. [DOI] [PubMed] [Google Scholar]
- Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, et al. Evolution of Symbiotic Bacteria in the Distal Human Intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.