Abstract
Many bacterial pathogens rely on a conserved membrane histidine sensor kinase, QseC, to respond to host adrenergic signaling molecules and bacterial signals in order to promote the expression of virulence factors. Using a high-throughput screen, we identified a small molecule, LED209, that inhibits the binding of signals to QseC, preventing its autophosphorylation and consequently inhibiting QseC-mediated activation of virulence gene expression. LED209 is not toxic and does not inhibit pathogen growth; however, this compound markedly inhibits the virulence of several pathogens in vitro and in vivo in animals. Inhibition of signaling offers a strategy for the development of broad-spectrum antimicrobial drugs.
A key challenge for medicine is to develop new drugs against pathogens that are resistant to current antimicrobial agents (1, 2). A promising strategy is to identify agents that inhibit microbial virulence without inhibiting growth, because these present less selective pressure for the generation of resistance (3–5). Many bacterial pathogens recognize the host environment by sensing and responding to the host adrenergic signaling molecules epinephrine and norepinephrine (NE) in order to promote the expression of virulence factors (6, 7). These pathogens appear to use the same membrane-embedded sensor histidine kinase, QseC (7), to recognize both host-derived adrenergic signals and the bacterial aromatic signal autoinducer-3 (AI-3) to activate their virulence genes (5, 6). Upon sensing any of these signaling molecules, QseC autophosphorylates and subsequently phosphorylates a transcription factor, QseB (Fig. 1A) (7), which initiates a relay to a complex regulatory cascade and leads to the transcription of key virulence genes (Fig. 1B) (5–8).
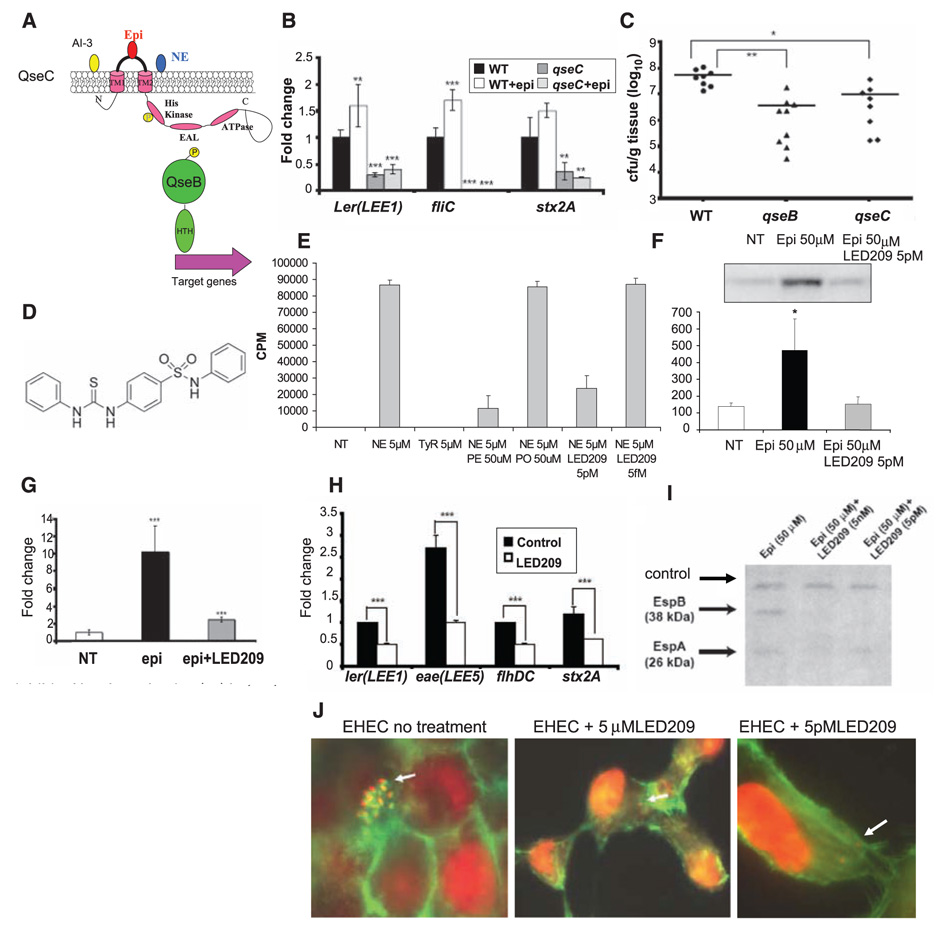
Fig. 1. LED209 inhibits EHEC virulence traits in vitro.
(A) Schematic of the autophosphorylation of QseC in response to signals and phosphotransfer to QseB. (B) QPCR of LEE1 gene ler, fliC, and stx2A in wild-type (WT) EHEC with self-produced AI-3 (black bars) and AI-3 plus 50 µM epinephrine (white bars), and the qseC mutant with self-produced AI-3 (dark gray bars) and AI-3 plus 50 µM epinephrine (light gray bars). (C) Colonization of the colon of infant rabbits by WT EHEC, the qseC and qseB mutants (CFUs/gram of tissue). (D) Chemical structure of LED209. (E) Binding of QseC to 5 µM tritiated NE and to 5 µM tyrosine (tyrosine is a negative control, not a signal to QseC). Binding of NE to QseC is inhibited by phentolamine (PE) but not propranolol (PO); 5 pM LED209 inhibits binding of 5 µM NE to QseC, but 5 fM LED209 does not. (F) QseC autophosphorylation in the absence of signal (NT, not treated), with 50 µM epinephrine (Epi) and with 50 µM epinephrine and 5 pM LED209. (G) QPCR of ler without signals (white bar), with 50 µM epinephrine (black bar), and with 50 µM epinephrine plus 5 pM LED209. (H) QPCR of LEE genes ler and eae, flagella genes flhDC and stx2A in WT EHEC with self-produced AI-3 (black bars) and AI-3 plus 5 pM LED209 (white bars). (I) Western blot of secreted proteins of EHEC (EspA and EspB) with 50 µM epinephrine, 50 µM epinephrine plus 5 nM LED209, and 50 µM epinephrine plus 5 pM LED209 (cross-reactive band as loading control). (J) Inhibition of the AE lesions by LED209 (5 µM and 5pM). Cell nuclei andbacterial cells are stained red (propidium iodide) and the cytoskeleton is stained green (fluorescein isothiocyanate–phalloidin). EHEC forms AE lesions atop the green-stained pedestals. There are no pedestals with LED209. * P < 0.05; ** P < 0.001; *** P < 0.0001.
QseC homologs are present in at least 25 important human and plant pathogens (table S1), and qseC mutants of enterohemorrhagic Escherichia coli (EHEC) (Fig. 1C and fig. S1) (7), Salmonella typhimurium (Fig. 2A) (8), and Francisella tularensis (9) are attenuated in infected animals. Because of the central role of the AI-3/epinephrine/NE QseC receptor in promoting the virulence of several important pathogens, we tested the effectiveness of inhibitors of this receptor as broad-spectrum antimicrobial agents.
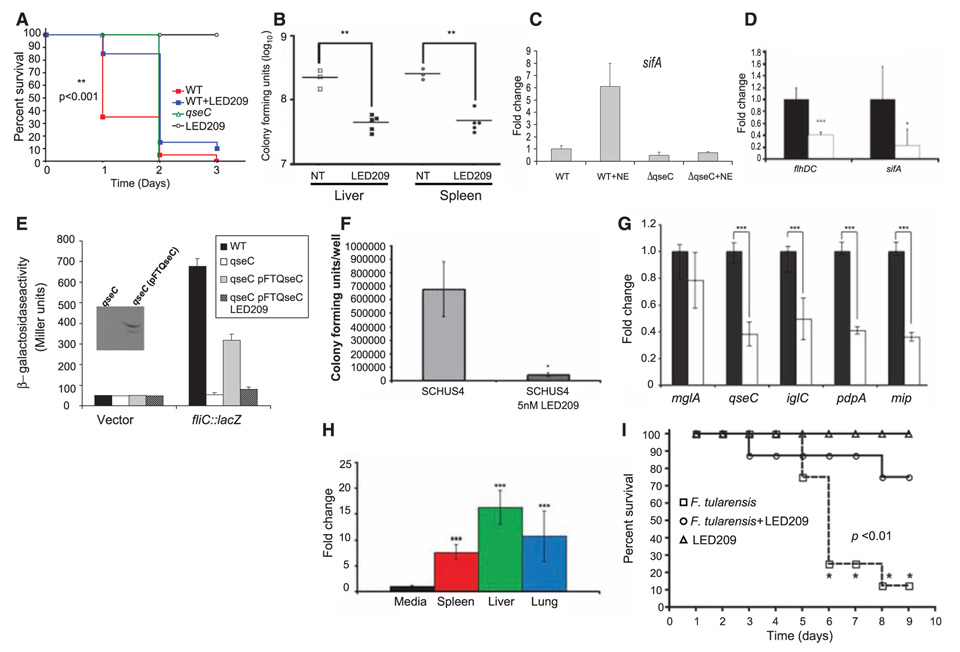
Fig. 2. LED209 inhibits S. typhimurium and F. tularensis virulence in vivo and in vitro.
(A) Survival plot of mice (strain 129×1/SvJ) on intraperitonial infection with 108 CFUs of WT S. typhimurium strain SL1344, oral treatment with LED209 (20mg/kg) alone, and intraperitonial infection with 108 CFUs of S. typhimurium strain SL1433 plus LED209 (20 mg/kg), and the qseC isogenic mutant. (B) CFUs of S. typhimurium harvested from liver and spleens of mice infected intraperitonially with 108 CFUs of S. typhimurium strain SL1344 and infected intraperitonially with 108 CFUs of S. typhimurium strain SL1433 plus LED209 (20mg/kg) 48 hours after infection. (C) QPCR of sifA in vitro in WT and a qseC mutant (ΔqseC) of S. typhimurium in the absence and presence of NE (50 µM). (D) QPCR of expression of the flagella regulator (flhDC) and sifA in vitro in the absence (black bars) and presence (5 pM) of LED209 (white bars). (E) The E. coli fliC::lacZ fusion was introduced into WT E. coli, the qseC E. coli mutant, and the qseC E. coli mutant complemented with F. tularensis qseC (qseC pFTQseC) in the absence and presence (5 pM) of LED209. The F. tularensis QseC was his-tagged and the inset shows that the F. tularensis QseC is expressed in the E. coli qseC mutant. (F) Infection of J774 murine macrophages with F. tularensis SCHU S4 in the absence and presence (5 nM) of LED209. (G) QPCR of F. tularensis virulence genes in the absence (gray bars) and presence (white bars) of LED209 (5 pM). (H) QPCR measuring expression of qseC in SCHU S4 during growth in vitro and in vivo (spleen, liver, and lungs). These data were collected from five C3H HeN mice intranasally infected with 30 CFUs of SCHU S4. QPCR of qseC was normalized against rpoA. (I) Survival plot of mice (C3H HeN) upon oral treatment with LED209 (20 mg/kg) alone, intranasal infection with 30 CFUs of SCHU S4, and intranasal infection with 30 CFUs of SCHU S4 plus LED209 (20 mg/kg). * P < 0.01; ** P < 0.001; *** P < 0.0001.
A library of 150,000 small organic molecules from the University of Texas (UT) Southwestern was screened at a 5 µM concentration in a high-throughput assay to identify inhibitors of QseC-dependent virulence gene activation in EHEC (fig. S2) (10). The first round of positive hits, which represented a diverse range of molecular architecture, was subjected to additional rounds of screening followed by preliminary evaluations for toxicity against bacterial cells in vitro. This yielded a pool of 75 potential inhibitors with median inhibitory concentration (IC50) values at or below 10−5 M. One compound was selected based on its potency as compared with other lead molecules, minimal toxicity toward bacterial and human cell lines, and potential for chemical modification. This molecule was subjected to structure-activity studies to identify ways to improve its potency. The result, LED209 [N-phenyl-4-{[(phenylamino) thioxomethyl]amino}-benzenesulfonamide] (Fig. 1D), was prepared in three steps (fig. S3).
New types of antimicrobial agents are needed to treat EHEC infections because treatments based on conventional antibiotics have been associated with worse clinical outcomes (11), probably because antibiotics induce an SOS response that enhances EHEC virulence (12). The genes encoding Shiga toxin are located within the late genes of a λ bacteriophage (a bacteriaphage that infects bacteria) and are transcribed when the phage enters its lytic cycle upon induction of an SOS response in EHEC (13). Shiga toxins are responsible for the morbidity and mortality of these infections. Transcription of EHEC virulence genes is induced by AI-3 and epinephrine. Neither AI-3 nor epinephrine had any effect on promoting virulence in a qseC mutant (Fig. 1B).
It was reported (7) and we confirmed that purified QseC in a liposome binds to tritiated NE and that phentolamine (an alpha-adrenergic antagonist) antagonizes this binding to block QseC autophosphorylation. In contrast, propranolol (a β-adrenergic antagonist) has no effect on QseC (Fig. 1E). We found that NE binding can be directly antagonized by 5 pM, but not 5 fM, LED209. Autophosphorylation of QseC in response to 50 µM epinephrine is also inhibited in the presence of 5 pM LED209 (Fig. 1F).
Consequently, QseC-dependent virulence gene expression in response to epinephrine (Fig. 1G) and AI-3 (Fig. 1H) was inhibited in the presence of 5 pM LED209. LED209-mediated inhibition of virulence gene expression also inhibited the secretion of EspA and EspB, two proteins encoded within the locus of enterocyte effacement (LEE) that are required for EHEC to translocate bacterial proteins into host cells and cause attaching-effacing (AE) lesions (Fig. 1I). At 5 pM, LED209 abolished EHEC AE lesion formation on cultured epithelial cells (Fig. 1J). Unlike conventional antibiotics, LED209 does not kill or hinder EHEC growth (fig. S4) or trigger the EHEC SOS response. Consequently, it does not promote the expression of Shiga toxin; indeed, it decreases the expression of the stxAB genes that encode this toxin (Fig. 1H).
However, administration of LED209 to infant rabbits before, after, or along with EHEC resulted in only a modest (and statistically nonsignificant) reduction in EHEC colonization of the intestine (fig S5). The failure of LED209 to reduce EHEC colonization in this animal may be attributable to rapid absorption from the gastrointestinal (GI) tract (see below and fig. S6). A nonabsorbable formulation may be required for noninvasive human pathogens such as EHEC.
S. typhimurium encodes a homolog of the EHEC QseC (87% similarity) sensor kinase that also controls virulence gene expression (14) (Fig. 2C). EHEC and S. typhimurium QseC are functionally interchangeable (14), and a S. typhimurium qseC mutant was found to be defective for colonization of the swine GI tract (8) and attenuated for systemic disease in mice (Fig. 2A). LED209 [20 mg per kilogram of body weight (mg/kg)] was given orally to mice 3 hours before and 3 hours after intraperitoneal injection of a lethal dose of S. typhimurium. Twenty-four hours after infection, only 30% of untreated mice remained alive, whereas 80% of the LED209-treated mice were still alive (Fig. 2A). All the S. typhimurium– infected mice died within 72 hours of infection, and 20% of LED209-treated mice survived up to 12 days (Fig. 2A and fig. S7). Even though LED209 did not influence S. typhimurium growth in vitro (fig. S4), ~10 times fewer S. typhimurium colony-forming units (CFUs) were recovered from the spleens and livers of treated animals (Fig. 2B). Addition of LED209 to cultures in vitro diminished expression of the sifA virulence gene (Fig. 2D), which is important for the development of systemic disease by S. typhimurium (15). Transcription of sifA is activated by NE in a QseC-dependent manner (Fig. 2C). Together, these observations suggest that LED209 inhibition of S. typhimurium virulence gene expression in vivo compromises the survival of the pathogen in the host.
F. tularensis has one histidine sensor kinase, QseC (57% similarity), encoded in its genome, and a qseC mutant is attenuated for mouse infection (9). The F. tularensis and EHEC QseC proteins are also functionally interchangeable, and the F. tularensis QseC can rescue expression of a QseC-dependent gene in an E. coli qseC mutant (Fig. 2E). LED209 reduced 10-fold the amount of F. tularensis strain SCHU-S4 recovered from macrophages (Fig. 2F). Furthermore, LED209 decreased the expression of several F. tularensis virulence genes in vitro (Fig. 2G), including qseC itself. Quantitative real-time fluorescence polymerase chain reaction (QPCR) assays demonstrated that qseC expression is up-regulated during infection of mice by SCHU-S4 (Fig. 2H). LED209 was administered orally in a single dose 3 hours after infection with SCHU-S4. Although 80% of LED209-treated mice remained alive 9 days after infection, only 10% of the untreated mice survived to this point (Fig. 2I). Thus, a single oral dose of LED209 can protect mice already exposed to aerosolized F. tularensis. There was no overt toxicity of the LED209, and none of the mice treated with LED209 alone died (Fig. 2I).
Because LED209 blocks epinephrine- and NE-triggering of QseC-mediated activation of virulence gene expression, we were concerned that this compound might also interfere with adrenergic signaling systems in the host. Using live cell-based assays (16), we showed that LED209 did not influence signaling through human β-adrenergic receptor isoforms (fig. S6). Pharmacokinetic studies suggested that there are no inherent structural features that might limit its bioavailability or biocompatibility and revealed that LED209 is approximately 70% bioavailable when administered orally to mice (fig. S6). Moreover, studies of LED209 in mice did not reveal any evidence of toxicity (figs. S8 to S10).
Bacterial histidine sensor kinases, such as QseC, should make attractive drug targets because mammals lack this class of signal-transduction molecules (17, 18). Because qseC is present in many important animal and plant pathogens, drugs that target this sensor kinase have the potential to have a broad spectrum. Because the QseC-dependent interkingdom-signaling system does not directly influence bacterial growth, inhibition of this signaling pathway may not exert a strong selective pressure for the development of resistance.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/321/5892/1078/DC1
Materials and Methods
Figs. S1 to S11
Tables S1 and S2
References
References and Notes
- 1.Levy SB, Marshall B. Nat. Med. 2004;10:S122. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 2.Palumbi SR. Science. 2001;293:1786. doi: 10.1126/science.293.5536.1786. [DOI] [PubMed] [Google Scholar]
- 3.Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. Science. 2005;310:670. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 4.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. Nat. Rev. Microbiol. 2008;6:17. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes DT, Sperandio V. Nat. Rev. Microbiol. 2008;6:111. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8951. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. Proc. Natl. Acad. Sci. U.S.A. 2006;103(27):10420. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bearson BL, Bearson SM. Microb. Pathog. 2008;44:271. doi: 10.1016/j.micpath.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Weiss DS, et al. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6037. [Google Scholar]
- 10.Materials and methods are available as supporting material on Science Online.
- 11.Tarr PI, Tsai HM, Chandler WL. N. Engl. J. Med. 2002;347:2171. [PubMed] [Google Scholar]
- 12.Zhang X, et al. J. Infect. Dis. 2000;181:664. doi: 10.1086/315239. [DOI] [PubMed] [Google Scholar]
- 13.Wagner PL, et al. J. Bacteriol. 2001;183:2081. doi: 10.1128/JB.183.6.2081-2085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merighi M, Carroll-Portillo A, Septer AN, Bhatiya A, Gunn JS. J. Bacteriol. 2006;188:141. doi: 10.1128/JB.188.1.141-149.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beuzon CR, et al. EMBO J. 2000;19:3235. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang LI, et al. J. Biol. Chem. 2007;282:10576. doi: 10.1074/jbc.M609695200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyon GJ, Muir TW. Chem. Biol. 2003;10:1007. doi: 10.1016/j.chembiol.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Roychoudhury S, et al. Proc. Natl. Acad. Sci. U.S.A. 1993;90:965. [Google Scholar]
- 19.We thank M. V. Norgard, E. Vitetta, V. Miller, and S. L. McKnight for comments and J. Richardson for pathological review of tissues from LED209-treated mice. This work was supported by NIH grants RO3 NS053582, RO1 AI053067, RO1 GM31278, PO1-AI055637-03, UO1-AI77853, and R21 AI067827; the Ellison Medical Foundation; the Burroughs Wellcome Fund; the Robert A. Welch Foundation; and a High Impact High Risk grant from UT Southwestern Medical Center. M.K.W. is a Howard Hughes Medical Institute investigator. This work yielded the provisional patent application UTSD:1854USP1.