Abstract
Background
Mycobacterium avium subsp. paratuberculosis (Map) causes the chronic enteritis called paratuberculosis mainly in cattle, sheep and goats. Evidences that point out an association between Map and Crohn's Disease in humans are increasing. Strain differentiation among Map isolates has proved to be difficult and has limited the study of the molecular epidemiology of paratuberculosis. In order to asses the usefulness of the PCR based short sequence repeat (SSR) analysis of locus 1 and locus 8 in the epidemiological tracing of paratuberculosis strains we here compare for the first time the results of SSR and SnaBI-SpeI pulsed-field gel electrophoresis (PFGE) typing methods in a set of 268 Map isolates from different hosts (cattle, sheep, goats, bison, deer and wild boar).
Results
A total of nineteen different multi-locus SSR (SSR1_SSR8) types were identified amongst the 268 isolates compared to the 37 multiplex profiles differentiated by the SnaBI-SpeI PFGE. SSR type 7_4 was the predominant genotype (51.2% of all isolates and 54.3% of cattle isolates), but combined with PFGE results the abundance of the most prevalent genotype (7_4&{2-1}) dropped down to 37.7%. SSR types 7_3 and 14_3 were significantly spread amongst isolates recovered from small ruminants. The comparison of SSR1_SSR8 and SnaBI-SpeI PFGE typing of these isolates has shown that both methods perform at similar discriminatory level. These were 0.691 and 0.693, respectively for SSR and PFGE as indicated Simpson's Index of Diversity, and 0.82 when calculated for combined SSR and PFGE genotypes. Overall, SSR1_SSR8 analysis seemed to detect higher levels of within-farm strain diversity and seemed to give higher year-related information. Combination of both typing methods revealed 20 multi-type farms out of the 33 bovine farms studied with more than one isolate.
Conclusion
The particular SSR and PFGE typing approaches described here are in general agreement but they showed some discrepancies that might reflect differing evolutionary processes of Map strains. Both methods are able to reciprocally complement their results and neither should be replaced with the other if sufficient material and time is available. Overall, the results of our comparative analyses suggest that, based on current methodologies available, a combined approach that includes SSR and PFGE seems to provide the highest level of discrimination for Map strain typing with meaningful epidemiological information.
Background
Mycobacterium avium subsp. paratuberculosis (Map) is the causative agent of paratuberculosis, a chronic digestive disease affecting mainly bovine, ovine, caprine and cervine livestock. Although the aetiology of Crohn's Disease has been subject of strong controversy [1,2], recent information seems to confirm an association between Map and this chronic human disease [3,4]. This underlines the increasing interest the research of Map has gained during last years due to the worldwide distribution of paratuberculosis, to the economic losses attributed to this disease [5,6], and to the presence of viable bacteria in products ready for human consumption [7-10] as a potential hazard in relationship with human inflammatory bowel disease. Successful control strategies require a good understanding of the epidemiology of a disease. Strain differentiation is a useful tool in epidemiological studies of many pathogenic bacteria. But previous investigations have revealed a relative lack of genetic diversity amongst Map isolates (reviewed in references [11,12]). Combined with the slow growth of the organism in pure culture, strain differentiation among isolates has proved to be difficult and has limited the study of the molecular epidemiology of paratuberculosis. PCR based methods can interestingly reduce the amount of bacteria and time required for Map strain typing. We here compare for the first time a set of 268 isolates from different hosts (cattle, sheep, goats, bison, deer and wild boar) that have been previously characterized for IS1311 PCR-restriction endonuclease analysis and SnaBI-SpeI pulsed-field gel electrophoresis (PFGE) patterns [12] with the more recently described short sequence repeat (SSR) analysis of locus 1 and locus 8 [13].
Results and discussion
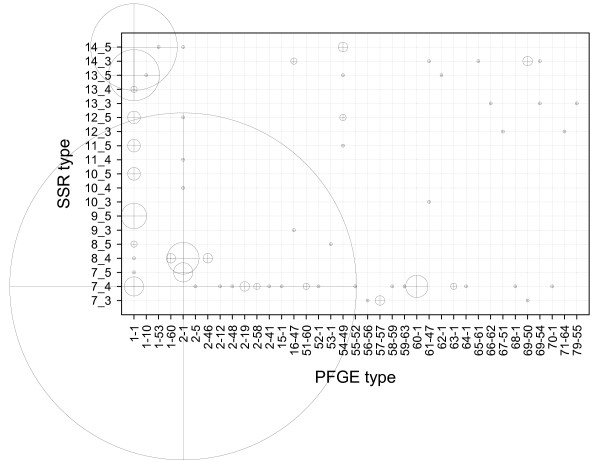
The results of SSR typing undertaken in the present work are summarized in Table 1. These results show that a total of nineteen different SSR1_SSR8 types were identified amongst the 268 isolates. In terms of host species distribution, there were 13 SSR types identified from cattle, 6 from sheep and 3 from goat isolates. Amongst isolates recovered from Spain, SSR type 7_4 accounted for the 54.3% of cattle isolates, while types 7_3 and 14_3 accounted for the 29% of sheep isolates each. Interestingly, amongst isolates recovered from goats, approximately the same proportion (43%) of isolates was typed as either cattle type 7_4 or sheep type 14_3. The remaining 14.3% of goat isolates were also sheep type strains and were identified as 9_3 type in SSR. Both deer and wild boar isolates belonged to the widest distributed type 7_4, in contrast they were {68-1} and {2-1} profiles in PFGE, respectively. Genetic homogeneity of Map isolates has been previously pointed out by other researchers using different typing methods [14-17]. Similarly and in agreement with our results, SSR method has demonstrated predominant type 7_4 to account for more than half the strains analyzed in previous studies [18,19]. The combination of SSR and PFGE types found in the present work made the prevalence of the most abundant genotype (7_4&{2-1}) drop down to 37.7%. None of the remaining combined types showed prevalences over 10%, except the combined type 14_5&{1-1}. The latter corresponds to the type assigned to MAP K10 strain and it was found in 10.07% of isolates under study. The amount of isolates showing particular genotypes in both techniques is graphically represented in Figure 1.
Table 1.
SSR1_SSR8 classification of Map strains.
| Country | Region Code | SSR1_SSR8 | Host sp | no. of isolates (%) | no. of farms (%) | IS1311 type |
| Spain | BC, As, CL, Cat, Can, An, Ga, Ar, Ma, Na, CM | 7_4 | Cattle | 126 (54.31) | 80 (61.07) | C type |
| BC, Ar, Na | 7_5 | Cattle | 7 (3.02) | 2 (1.53) | C type | |
| BC, Ex, CL, Can, Ga | 8_4 | Cattle | 17 (7.33) | 14 (10.69) | C type | |
| BC, Ar | 8_5 | Cattle | 3 (1.29) | 3 (2.29) | C type | |
| BC, Na | 9_5 | Cattle | 8 (3.45) | 3 (2.29) | C type | |
| BC | 10_4 | Cattle | 1 (0.43) | 1 (0.76) | C type | |
| BC, Na | 10_5 | Cattle | 4 (1.72) | 3 (2.29) | C type | |
| BC | 11_4 | Cattle | 1 (0.39) | 1 (0.76) | C type | |
| BC | 11_5 | Cattle | 5 (2.16) | 5 (3.82) | C type | |
| BC, Can, Cat | 12_5 | Cattle | 7 (3.02) | 7 (5.34) | C type | |
| BC | 13_4 | Cattle | 2 (0.86) | 2 (1.53) | C type | |
| As, BC, Na, Ar | 13_5 | Cattle | 19 (8.19) | 13 (9.92) | C type | |
| BC, Na, Ar | 14_5 | Cattle | 32 (13.79) | 22 (16.79) | C type | |
| BC, Ar, Na | 7_3 | Sheep | 5 (29.41) | 4 (40.0) | S type | |
| Ar | 7_4 | Sheep | 1 (5.88) | 1 (10.0) | C type | |
| BC | 10_3 | Sheep | 1 (5.88) | 1 (10.0) | S type | |
| BC | 12_3 | Sheep | 2 (11.76) | 2 (20.0) | S type | |
| BC | 13_3 | Sheep | 3 (17.65) | 3 (30.0) | S type | |
| BC, Na | 14_3 | Sheep | 5 (29.41) | 4 (40.0) | S type | |
| BC, An | 7_4 | Goat | 3 (42.86) | 2 (40.0) | C type | |
| CL | 9_3 | Goat | 1 (14.29) | 1 (20.0) | S type | |
| IB | 14_3 | Goat | 3 (42.86) | 3 (60.0) | S type | |
| CM | 7_4 | Deer | 1 (100.0) | 1 (100.0) | C type | |
| CM | 7_4 | Wild Boar | 1 (100.0) | 1 (100.0) | C type | |
| India | Mathura | 7_4 | Sheep | 2 (100.0) | 1 (100.0) | B type |
| Farah | 7_4 | Goat | 5 (100.0) | 1 (100.0) | B type | |
| USA | Montana | 7_4 | Bison | 3 (100.0) | 1 (100.0) | B type |
SSR types by region and host species. Name of SSR type: number of repeats in locus 1 (G residue) _ number of repeats in locus 8 (GGT residue). IS1311 PCR-REA classification of isolates is also included. Percentages in brackets are calculated according to the total number of isolates in each host species. Regions mentioned in the study are indicated as follows: An = Andalucia; Ar = Aragón; As = Asturias; BC = Basque Country; Can = Cantabria; Cat = Cataluña; CL = Castilla y León; CM = Castilla-La Mancha; Ex = Extremadura; Ga = Galicia; IB = Balearic Islands; Ma = Madrid; Na = Navarra.
Figure 1.
Bubble plot showing the distribution of Map strain genotypes studied by SSR1_SSR8 and SnaBI-SpeI PFGE. The diameter of bubbles corresponds to the number of isolates with particular SSR and PFGE types.
Cluster analysis with both PFGE and SSR based typing methods (not shown) confirmed that Map isolates are genetically divided into the cattle type and sheep type main groups, much as has been found in other works [20-25]. The agreement between this classification and the IS1311 groups C (including the less common B strains in this group) and S confirms the utility of IS1311 PCR-REA as a rapid and reliable method for preliminary typing of Map isolates, as indicated by results of previous works [12,26].
While the overall discriminatory power of both methods as calculated by Simpson's index of diversity (1-D) was almost the same (0.693 for PFGE and 0.691 for SSR), comparative analysis revealed that the most abundant PFGE {1-1}, {2-1} and {54-49} profiles (30%, 48% and 2.7% of all isolates, respectively) were subdivided into 11, 7 and 4 different types, respectively. Similarly, isolates representing the most abundant SSR type 7_4 (51% of all isolates) could be subdivided into 19 different PFGE profiles as shown in Table 2. As was to be expected, the overall 1-D value raised up to 0.82 if calculated considering the abundances of combined SSR and PFGE types (i.e. 7_4&{51–60}, 7_4&{2-1} ...). Other promising methods as the mycobacterial interspersed repetitive units/variable number tandem repeats (MIRU/VNTR) seem to have a high discriminatory power as well [27,28]. Amongst isolates recovered from sheep, there was a higher discrimination with PFGE (1-D = 0.865) than with SSR (1-D = 0.775), but such a difference was not noticed in the other host species (see Table 3).
Table 2.
Reciprocal complementation between SSR1_SSR8 and SnaBI-SpeI PFGE.
| PFGE type | subdivided by | SSR types |
| {1-1} | 11 | 7_4, 7_5, 8_4, 8_5, 9_5, 10_5, 11_5, 12_5, 13_4, 13_5, 14_5 |
| {2-1} | 7 | 7_4, 7_5, 8_4, 10_4, 11_4, 12_5, 14_5 |
| {16–47} | 2 | 9_3, 14_3 |
| {61-47} | 2 | 10_3, 14_3 |
| {54–49} | 4 | 11_5, 12_5, 13_5, 14_5 |
| {69-50} | 2 | 7_3, 14_3 |
| {69-54} | 2 | 13_3, 14_3 |
| SSR type | subdivided by | PFGE types |
| 7_3 | 3 | {56-56}, {57-57}, {69-50} |
| 7_4 | 19 | {1-1}, {2-1}, {2–41}, {15-1}, {52-1}, {60-1}, {2–5}, {2–12}, {2–48}, {2–19}, {2–58}, {51–60}, {55-52}, {58–59}, {59–63}, {63-1}, {64-1}, {68-1}, {70-1} |
| 7_5 | 2 | {1-1}, {2-1} |
| 8_4 | 4 | {1-1}, {1–60}, {2-1}, {2-–46} |
| 8_5 | 2 | {1-1}, {53-1} |
| 11_5 | 2 | {1-1}, {54-49} |
| 12_3 | 2 | {67-51}, {71-64} |
| 12_5 | 3 | {1-1}, {54-49}, {2-1} |
| 13_3 | 3 | {66-62}, {79-55}, {69-54} |
| 13_5 | 4 | {1-1}, {1–10}, {54–49}, {62-1} |
| 14_3 | 5 | {16–47}, {61-47}, {65-61}, {69-50}, {69-54} |
| 14_5 | 4 | {1-1}, {2-1}, {1–53}, {54-49} |
Number of SSR1_SSR8 types identified within each SnaBI-SpeI PFGE multiplex profile and vice versa, showing how each technique complements each other in subdividing the most prevalent types.
Table 3.
Genetic diversity and discriminatory power of SSR1_SSR8 and SnaBI-SpeI PFGE.
| PFGE | SSR | |
| number of different types | 37 | 19 |
| 1-D value | ||
| cattle | 0.621 | 0.669 |
| sheep | 0.865 | 0.775 |
| goat | 0.666 | 0.612 |
| Discriminatory power | 0.693 | 0.691 |
| combined | 0.817 | |
Estimation of the genetic diversity of isolates from Spanish cattle, sheep and goats, and the discriminatory power of both methods calculated as Simpson's Index of Diversity (1-D). Discriminatory power of the combined SSR&PFGE method was calculated taking into account all combinations found amongst Map isolates studied.
The polyclonal infection of one Holstein bull with three different strains earlier demonstrated by PFGE [12] was partially confirmed by SSR1_SSR8 typing. The isolate typed as {1-1} with PFGE was of 13_5 type by SSR, but the remaining two isolates classified as PFGE profiles {2-1} and {59-63} shared the same SSR type 7_4. None of the fingerprinting methods compared here detected any other polyclonal infections in the other three animals with more than one culture available included in the study. In general terms, SSR1_SSR8 analysis seemed to detect higher levels of within-herd strain variability than SnaBI-SpeI PFGE. With 33 bovine farms giving more than one isolate, SSR method used detected 17 farms with multi-type isolates, one farm with isolates belonging to five different types, two farms yielded isolates of four different types, another one gave isolates of 3 types, and finally 13 of these herds had isolates of two distinct SSR types (Table 4). On the other hand, PFGE typing detected up to 14 farms with multi-type isolates, three bovine herds with three different profiles and 11 herds carrying strains of two different profiles. Combination of both typing methods revealed 20 multi-type cattle farms. More than one isolate was recovered from four sheep flocks. In this case, a slightly higher level of intra-herd variability was detected by PFGE compared to SSR. Three flocks with 3 different strains were found by PFGE while SSR analysis identified two flocks with 3 different types and one flock with two.
Table 4.
Multi-type herds and sheep flocks detected by SSR1_SSR8 and SnaBI-SpeI PFGE.
| multi-type farm? | |||||||||
| Farm | Region | sp | Breed | SSR1_8 type | no. of isolates | SnaBI-SpeI PFGE type | no. of isolates | SSR | PFGE |
| BI5 | BC | Bov | Holstein | 8_4,9_5 | 1,2 | {1-1} | 3 | yes | no |
| BI6 | BC | Bov | Holstein | 7_4,9_5,10_5,14_5 | 1,3,2,1 | {1-1},{2-1} | 6,1 | yes | yes |
| SS5 | BC | Bov | Holstein | 8_5,11_5,13_5,14_5 | 1,1,3,1 | {1-1},{54-49} | 5,1 | yes | yes |
| SS15 | BC | Bov | Holstein | 11_5,14_5 | 1,1 | {1-1} | 2 | yes | no |
| SS20 | BC | Bov | Holstein | 7_4 | 3 | {2-1},{2–19} | 2,1 | no | yes |
| SS23 | BC | Bov | Holstein | 7_4,14_5 | 10,1 | {2-1},{54-49} | 10,1 | yes | yes |
| SS27 | BC | Bov | Holstein | 7_4,13_5 | 3,2 | {1-1},{2-1},{59-63} | 2,2,1 | yes | yes |
| SS28 | BC | Bov | Holstein | 7_4,12_5 | 1,1 | {2–19},{54-49} | 1,1 | yes | yes |
| SS38 | BC | Bov | Holstein | 13_5,14_5 | 1,1 | {1-1} | 2 | yes | no |
| SS43 | BC | Bov | Pyrenean | 12_5,14_5 | 1,1 | {1-1} | 2 | yes | no |
| SS45 | BC | Bov | Limousin | 7_4,14_5 | 1,1 | {2-1},{54-49} | 1,1 | yes | yes |
| SS52 | BC | Bov | Holstein | 7_4,13_5,14_5 | 1,1,1 | {1-1},{2-1} | 2,1 | yes | yes |
| S6 | Can | Bov | Holstein | 7_4,8_4 | 1,1 | {2-1},{2–46} | 1,1 | yes | yes |
| BU3 | CL | Bov | Holstein | 7_4,8_4 | 2,2 | {2-1} | 4 | yes | no |
| VA1 | CL | Bov | Holstein | 7_4 | 5 | {2-1},{2–41} | 4,1 | no | yes |
| NA1 | Na | Bov | Holstein | 9_5,10_5 | 3,1 | {1-1} | 4 | yes | no |
| NA2 | Na | Bov | Holstein | 13_5,14_5 | 4,9 | {1-1},{1–53},{62-1} | 11,1,1 | yes | yes |
| O4 | As | Bov | Holstein | 7_4 | 3 | {1-1},{2-1} | 2,1 | no | yes |
| SA2 | CL | Bov | Bullfight | 7_4,8_4 | 1,1 | {2-1},{2–46} | 1,1 | yes | yes |
| Z2 | Ar | Bov | Holstein | 7_4,7_5,8_5,13_5,14_5 | 1,6,1,1,3 | {1-1},{2-1},{53-1} | 5,6,1 | yes | yes |
| SS53 | BC | Ov | Latxa | 7_3,10_3,14_3 | 1,1,1 | {61-47},{69-50},{69-54} | 1,1,1 | yes | yes |
| SS54 | BC | Ov | Latxa | 12_3,13_3,14_3 | 1,1,1 | {67-51},{79-55},{69-50} | 1,1,1 | yes | yes |
| SS55 | BC | Ov | Latxa | 12_3,14_3 | 1,2 | {61-47},{69-50},{71-64} | 1,1,1 | yes | yes |
Farms with more than one isolate giving at least two different strain types in SSR and/or PFGE typing. SSR detected 17 multi-type bovine herds and 3 multi-type sheep flocks. In the other hand, PFGE identified 14 and 3 multi-type farms, respectively. Farm names indicate the name of the province (within a region) they belong to: BI = Bizkaia (BC); SS = Gipuzkoa (BC); S = Cantabria (Can); BU = Burgos (CL); VA = Valladolid (CL); NA = Navarra (Na); O = Asturias (As); SA = Salamanca (CL); Z = Zaragoza (Ar). Other abbreviations: As = Asturias; BC = Basque Country; Can = Cantabria; CL = Castilla y León; Na = Navarra; Bov = bovine; Ov = ovine.
A previous work suggested an apparent relation between particular G residue repeat alleles and host species in SSR analysis [29]. In the present study the previously described alleles 7G to 14Gs have been found, but 8Gs, 9Gs and 11Gs are almost restricted to isolates obtained from cattle. Interestingly, there seems to be a strong link between GGT residue alleles and host species. Thus, allele 3GGTs has been detected in all sheep (except one) and in 67% of goats while all cows analyzed were infected with strains showing 4 or 5GGTs repeats. Possession of 3GGT allele resembles possession of a cytosine at base pair position 223 that can be found in all copies of the IS1311 gene of typical sheep (S) type strains of Map [30].
The study of SSR1_SSR8/SnaBI-SpeI PFGE combined profiles from herds giving more than one isolate according to the date of cultures demonstrated the reliability and usefulness of these techniques for epidemiological tracing of paratuberculosis cases. Eleven Holstein farms giving at least two isolates from different years were identified. The SSR method appeared to give slightly higher year-related information. As shown in Table 5, strain type changed along the years during the follow-up period in farms SS5, SS27, SS28, SS38 and SS52 (the meaning of letters used to name farms under study is given in Table 5). On the contrary, farms BI2, BI9, HU1, LE1 and NA2 maintained the same strain types year after year. Herd SS23 showed an intermediate situation since it yielded two types in the first year, but maintained one of them afterwards. Further conclusions cannot be suggested due to a lack of information. A strain variation percentage was calculated for each of these farms dividing the number of different strain types minus one by the total number of isolates recovered minus one. Thus a 100% strain variation was observed for the first 3 farms while the last four showed no variation (Table 5). SS5, SS27, NA2 and SS23 showed intermediate strain variations of 60, 50, 25 and 10%, respectively. Collectively, our results indicate that SSR1_SSR8 analysis, helped by SnaBI-SpeI PFGE where possible, can offer very valuable epidemiologic information and indicate the existence of three models of strain type change in infected populations: stable, variable and intermediate. No obvious difference in the incorporation of new animals to the herd was observed between the different types of farms but the information on other management factors was very scarce. However, for the first time it has been shown that epidemiological patterns can vary according to cattle population and time. This observation requires further research by broadening the number of farms and extending the period of observation, as well as recording factors that might influence the strain shifting and determine its consequences in terms of severity of the disease, control measures effects and bacteria sources and reservoirs.
Table 5.
Circulation of Map strains in some bovine herds along time.
| SSR1_SSR8&SnaBI-SpeI PFGE profiles identified in different years (number of isolates in brackets) | ||||||
| Farm | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 |
| SS28 | 12_5/{54-49} (1) | - | - | 7_4/{2–19} (1) | - | - |
| SS38 | - | - | 13_5/{1-1} (1) | 14_5/{1-1} (1) | - | - |
| SS52 | - | - | 7_4/{2-1} (1) | 13_5/{1-1} (1) | 14_5/{1-1} (1) | - |
| SS5 | - | 8_5/{1-1} (1) | 11_5/{54-49} (1) 13_5/{1-1} (3) |
14_5/{1-1} (1) | - | - |
| SS27 | - | - | 13_5/{1-1} (2) | 7_4/{2-1} (2) 7_4/{59-63} (1) |
- | - |
| NA2 | - | - | - | - | 13_5/{1-1} (1) 14_5/{1-1} (1) |
13_5/{1-1} (2) 13_5/{62-1} (1) 14_5/{1-1} (7) 14_5/{1-53} (1) |
| SS23 | - | 7_4/{2-1} (1) 14_5/{54-49} (1) |
7_4/{2-1} (8) | - | 7_4/{2-1} (1) | - |
| BI2 | - | - | 7_4/{2-1} (1) | 7_4/{2-1} (2) | - | - |
| BI9 | - | - | 7_4/{2-1} (1) | - | 7_4/{2-1} (2) | 7_4/{2-1} (2) |
| HU1 | - | - | 7_4/{2-1} (1) | - | 7_4/{2-1} (1) | - |
| LE1 | - | 7_4/{2-1} (3) | 7_4/{2-1} (4) | - | - | - |
New strains turning up and within-herd spread of specific strains during time in some bovine Holstein farms as assessed by SSR/PFGE analysis combination. Strain variation along time seems to indicate three different epidemiologic situations: Farms with 100% strain variation (variable), farms with intermediate strain variation (intermediate) and farms carrying always the same types (stable).
Conclusion
These independent typing methods are in general agreement. However, they showed significant discrepancies indicating that each one might reflect differing evolutionary processes of Map strains. Since both SSR1_SSR8 and SnaBI-SpeI PFGE methods have the ability to reciprocally complement their results by subdividing the different genotypes identified in the other method, none of them should be used as a substitute for the other one if sufficient bacterial growth is available. Taken together, the results of our studies confirm the utility of the SSR approach as an easy and rapid method based on PCR and sequence analysis that requires only small amounts of sample to perform, compared to the big amount and good quality of DNA required for PFGE typing. The results also suggest that the addition of a third locus to SSR1_SSR8 typing may help in increasing the discriminatory power of this method. Overall, the results of our comparative analyses suggest that, based on current methodologies available, a combined approach that includes IS1311 PCR-REA, SSR and PFGE provides the highest level of discrimination for Map strain characterization. However, in practical terms, the use of IS1311 PCR-REA is not equivalent to the other two since it only provides broad group classification. The choice between PFGE and SSR, however, will be defined for the technical simplicity, lower DNA quality and quantity requirements and robustness for obtaining reliable epidemiologic information of SSR.
Methods
DNA from 232 isolates from cattle (Spain), 19 from sheep (17 from Spain and two from India), 12 from goats (seven from Spain and five from India), one from deer (Spain), one from wild boar (Spain) and three from bison (USA) grown on Herrold's egg yolk, Lowenstein-Jensen (Biomedics, Madrid, Spain) and/or Middlebrook media (Becton, Dickinson and Company, MD, USA) with or without supplements (mycobactin J and/or OADC enrichment) used in a previous work [12] was analyzed. In the previous PFGE study mentioned above isolates were classified as cattle (C), sheep (S) or bison (B) strains by IS1311 PCR-REA and subdivided into 37 different multiplex SnaBI-SpeI PFGE profiles (the PFGE nomenclature used earlier has been changed according to the instructions of the standardized database at http://www.mri.sari.ac.uk/bacteriology-current-project-01-01.asp. In the present paper square brackets have been replaced with curly brackets to distinguish between PFGE nomenclature and literature references, except in Figure 1). DNA was purified from proteinase K pre-treated agarose plugs previously prepared for PFGE. A piece of plug was cut and introduced into a 1.5 ml tube. QIAquick PCR purification kit (Qiagen, GmbH, Germany) was used according to the instructions of the manufacturer to remove the agarose and cell debris. One μl of purified DNA was used for PCR amplification of the most discriminatory SSR loci 1 (G residue) and 8 (GGT residue) as described earlier [13]. Afterwards, PCR products were sequenced by using standard dye terminator chemistry, and the sequences analyzed on a 3700 DNA Analyzer (Applied Biosystems, Foster City, CA, USA). All chromatograms were visually inspected, and sequences edited with the EditSeq program (DNASTAR, Madison, WI, USA) and then aligned by the use of the MegAlign program (DNASTAR). The number of G repeats in locus 1 and the number of GGT repeats in locus 8 separated by one underscore was used to designate different SSR genotypes (SSR1_SSR8). A bubble type plot was generated with SigmaPlot for Windows v10 software (Systat Software, Inc., San Jose, CA, USA) in order to show the number of isolates in a SSR type versus PFGE type matrix. Simpson's Index of Diversity (1-D) was calculated as follows in order to compare the genetic diversity of isolates between host species and to asses the discriminatory power of the typing methods used:
Simpson's index of diversity = 1-D = 1-[Σ(no. of isolates with a particular genotype/total no. of isolates)2]
Authors' contributions
IS and LL carried out the laboratory work, compiled and analysed information and data, and drafted the manuscript. AA, JMG and MVG helped to draft the manuscript. VK and RAJ conceived of the study, and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We wish to thank the farmers and veterinarians who provided the samples used in this study and JM Plazaola from the Diputación Foral of Gipuzkoa for his help with data collection. Thanks to V. Pérez and JF García-Marín (University of León, Spain), C Gortazar (IREC, Spain), S Rodríguez (University of Córdoba, Spain), A Verna (CONICET, Argentina), SV Singh (CIRG, India), R Whitlock (University of Pennsylvania, USA) and G Aduriz (NEIKER-Tecnalia) for isolation and information on some strains. We appreciate the kind help of I Heron and K Stevenson from the Moredun Research Institute for PFGE classification standardization. This study was supported with grants from the Dept. of Agriculture and Fisheries, the Dept. of Industry, the Dept. of Education, Universities and Research of the Basque Government and with EU-FEDER funds. The work of LL Li and V Kapur was supported by the Johne's Disease Integrated Program (JDIP), a USDA-CSREES-NRI-CAP program.
Contributor Information
Iker Sevilla, Email: isevilla@neiker.net.
Lingling Li, Email: lul17@psu.edu.
Alongkorn Amonsin, Email: Alongkorn.A@Chula.ac.th.
Joseba M Garrido, Email: jgarrido@neiker.net.
Maria V Geijo, Email: mgeijo@neiker.net.
Vivek Kapur, Email: vkapur@psu.edu.
Ramón A Juste, Email: rjuste@neiker.net.
References
- Behr MA, Kapur V. The evidence for Mycobacterium paratuberculosis in Crohn's disease. Curr Opin Gastroenterol. 2008;24:17–21. doi: 10.1097/MOG.0b013e3282f1dcc4. [DOI] [PubMed] [Google Scholar]
- Greenstein RJ. Is Crohn's disease caused by a mycobacterium? Comparisons with leprosy, tuberculosis, and Johne's disease. Lancet Infect Dis. 2003;3:507–514. doi: 10.1016/S1473-3099(03)00724-2. [DOI] [PubMed] [Google Scholar]
- Juste RA, Elguezabal N, Garrido JM, Pavon A, Geijo MV, Sevilla I, Cabriada JL, Tejada A, García-Campos F, Casado R, et al. On the prevalence of M. avium subspecies paratuberculosis DNA in the blood of healthy individuals and patients with inflammatory bowel disease. PLoS ONE. 2008;3:e2537. doi: 10.1371/journal.pone.0002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juste RA, Elguezabal N, Pavón A, Garrido JM, Geijo MV, Sevilla I, Cabriada JL, Tejada A, García-Campos F, Casado R, et al. Association between Mycobacterium avium subsp. paratuberculosis DNA in blood and cellular and humoral immune response in Inflammatory Bowel Disease patients and controls. Int J Infect Dis. 2008 doi: 10.1016/j.ijid.2008.06.034. [DOI] [PubMed] [Google Scholar]
- Kennedy DJ, Benedictus G. Control of Mycobacterium avium subsp. paratuberculosis infection in agricultural species. Rev Sci Tech. 2001;20:151–179. doi: 10.20506/rst.20.1.1274. [DOI] [PubMed] [Google Scholar]
- Whittington RJ, Sergeant ES. Progress towards understanding the spread, detection and control of Mycobacterium avium subsp. paratuberculosis in animal populations. Aust Vet J. 2001;79:267–278. doi: 10.1111/j.1751-0813.2001.tb11980.x. [DOI] [PubMed] [Google Scholar]
- Ellingson JL, Anderson JL, Koziczkowski JJ, Radcliff RP, Sloan SJ, Allen SE, Sullivan NM. Detection of viable Mycobacterium avium subsp. paratuberculosis in retail pasteurized whole milk by two culture methods and PCR. J Food Prot. 2005;68:966–972. doi: 10.4315/0362-028x-68.5.966. [DOI] [PubMed] [Google Scholar]
- Grant IR. Zoonotic potential of Mycobacterium avium ssp. paratuberculosis: the current position. J Appl Microbiol. 2005;98:1282–1293. doi: 10.1111/j.1365-2672.2005.02598.x. [DOI] [PubMed] [Google Scholar]
- Hruska K, Bartos M, Kralik P, Pavlik I. Mycobacterium avium subsp. paratuberculosis in powdered infant milk: paratuberculosis in cattle: the public health problem to be solved. Vet Med (Praha) 2005;50:327–335. [Google Scholar]
- Ikonomopoulos J, Pavlik I, Bartos M, Svastova P, Ayele WY, Roubal P, Lukas J, Cook N, Gazouli M. Detection of Mycobacterium avium subsp. paratuberculosis in retail cheeses from Greece and the Czech Republic. Appl Environ Microbiol. 2005;71:8934–8936. doi: 10.1128/AEM.71.12.8934-8936.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiwala AS, Li L, Kapur V, Sreevatsan S. Current understanding of the genetic diversity of Mycobacterium avium subsp. paratuberculosis. Microbes Infect. 2006;8:1406–1418. doi: 10.1016/j.micinf.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sevilla I, Garrido JM, Geijo M, Juste RA. Pulsed-field gel electrophoresis profile homogeneity of Mycobacterium avium subsp. paratuberculosis isolates from cattle and heterogeneity of those from sheep and goats. BMC Microbiol. 2007;7:18. doi: 10.1186/1471-2180-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amonsin A, Li LL, Zhang Q, Bannantine JP, Motiwala AS, Sreevatsan S, Kapur V. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J Clin Microbiol. 2004;42:1694–1702. doi: 10.1128/JCM.42.4.1694-1702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins DV, Williams SN, Hope A, Eamens GJ. DNA fingerprinting of Australian isolates of Mycobacterium avium subsp. paratuberculosis using IS 900 RFLP. Aust Vet J. 2000;78:184–190. doi: 10.1111/j.1751-0813.2000.tb10590.x. [DOI] [PubMed] [Google Scholar]
- Djonne B, Pavlik I, Svastova P, Bartos M, Holstad G. IS 900 restriction fragment length polymorphism (RFLP) analysis of Mycobacterium avium subsp. paratuberculosis isolates from goats and cattle in Norway. Acta Vet Scand. 2005;46:13–18. doi: 10.1186/1751-0147-46-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feizabadi MM, Robertson ID, Hope A, Cousins DV, Hampson DJ. Differentiation of Australian isolates of Mycobacterium paratuberculosis using pulsed-field gel electrophoresis. Aust Vet J. 1997;75:887–889. doi: 10.1111/j.1751-0813.1997.tb11259.x. [DOI] [PubMed] [Google Scholar]
- Motiwala AS, Strother M, Amonsin A, Byrum B, Naser SA, Stabel JR, Shulaw WP, Bannantine JP, Kapur V, Sreevatsan S. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: evidence for limited strain diversity, strain sharing, and identification of unique targets for diagnosis. J Clin Microbiol. 2003;41:2015–2026. doi: 10.1128/JCM.41.5.2015-2026.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadiali AH, Strother M, Naser SA, Manning EJ, Sreevatsan S. Mycobacterium avium subsp. paratuberculosis strains isolated from Crohn's disease patients and animal species exhibit similar polymorphic locus patterns. J Clin Microbiol. 2004;42:5345–5348. doi: 10.1128/JCM.42.11.5345-5348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiwala AS, Strother M, Theus NE, Stich RW, Byrum B, Shulaw WP, Kapur V, Sreevatsan S. Rapid detection and typing of strains of Mycobacterium avium subsp. paratuberculosis from broth cultures. J Clin Microbiol. 2005;43:2111–2117. doi: 10.1128/JCM.43.5.2111-2117.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerfeind R, Benazzi S, Weiss R, Schliesser T, Willems H, Baljer G. Molecular characterization of Mycobacterium paratuberculosis isolates from sheep, goats and cattle by hybridization with a DNA probe to insertion element IS 900. J Clin Microbiol. 1996;34:1617–1621. doi: 10.1128/jcm.34.7.1617-1621.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DM, Gabric DM, de Lisle GW. Identification of two groups of Mycobacterium paratuberculosis strains by restriction analysis and DNA hybridization. J Clin Microbiol. 1990;28:1591–1596. doi: 10.1128/jcm.28.7.1591-1596.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan L, Alvarez J, Aranaz A, Rodriguez A, Romero B, Bezos J, Mateos A, Dominguez L. Molecular epidemiology of Types I/III strains of Mycobacterium avium subspecies paratuberculosis isolated from goats and cattle. Vet Microbiol. 2006;115:102–110. doi: 10.1016/j.vetmic.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Pavlik I, Horvathova A, Dvorska L, Bartl J, Svastova P, du Maine R, Rychlik I. Standardisation of restriction fragment length polymorphism analysis for Mycobacterium avium subspecies paratuberculosis. J Microbiol Methods. 1999;38:155–167. doi: 10.1016/S0167-7012(99)00091-3. [DOI] [PubMed] [Google Scholar]
- Stevenson K, Hughes VM, de Juan L, Inglis NF, Wright F, Sharp JM. Molecular characterization of pigmented and nonpigmented isolates of Mycobacterium avium subsp. paratuberculosis. J Clin Microbiol. 2002;40:1798–1804. doi: 10.1128/JCM.40.5.1798-1804.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington RJ, Hope AF, Marshall DJ, Taragel CA, Marsh I. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: IS 900 restriction fragment length polymorphism and IS 1311 polymorphism analyses of isolates from animals and a human in Australia. J Clin Microbiol. 2000;38:3240–3248. doi: 10.1128/jcm.38.9.3240-3248.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla I, Singh SV, Garrido JM, Aduriz G, Rodríguez S, Geijo MV, Whittington RJ, Saunders V, Whitlock RH, Juste RA. PCR-REA genotype of paratuberculosis strains isolated from different host species and geographic locations. Rev – Off Int Epizoot. 2005;24:1061–1066. [PubMed] [Google Scholar]
- Mobius P, Luyven G, Hotzel H, Kohler H. High genetic diversity among Mycobacterium avium subsp. paratuberculosis strains from German cattle herds shown by combination of IS900 restriction fragment length polymorphism analysis and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing. J Clin Microbiol. 2008;46:972–981. doi: 10.1128/JCM.01801-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault VC, Grayon M, Boschiroli ML, Hubbans C, Overduin P, Stevenson K, Gutierrez MC, Supply P, Biet F. New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol. 2007;45:2404–2410. doi: 10.1128/JCM.00476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiwala AS, Amonsin A, Strother M, Manning EJ, Kapur V, Sreevatsan S. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis isolates recovered from wild animal species. J Clin Microbiol. 2004;42:1703–1712. doi: 10.1128/JCM.42.4.1703-1712.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington R, Marsh I, Choy E, Cousins D. Polymorphism in IS 1311, an insertion sequence common to Mycobacterium avium and Mycobacterium avium subsp. paratuberculosis, can be used to distinguish between and within these species. Mol Cell Probes. 1999;12:349–358. doi: 10.1006/mcpr.1998.0194. [DOI] [PubMed] [Google Scholar]