Abstract
Background
The Oryza sativa L. indica subspecies is the most widely cultivated rice. During the last few years, we have collected over 20,000 putative full-length cDNAs and over 40,000 ESTs isolated from various cDNA libraries of two indica varieties Guangluai 4 and Minghui 63. A database of the rice indica cDNAs was therefore built to provide a comprehensive web data source for searching and retrieving the indica cDNA clones.
Results
Rice Indica cDNA Database (RICD) is an online MySQL-PHP driven database with a user-friendly web interface. It allows investigators to query the cDNA clones by keyword, genome position, nucleotide or protein sequence, and putative function. It also provides a series of information, including sequences, protein domain annotations, similarity search results, SNPs and InDels information, and hyperlinks to gene annotation in both The Rice Annotation Project Database (RAP-DB) and The TIGR Rice Genome Annotation Resource, expression atlas in RiceGE and variation report in Gramene of each cDNA.
Conclusion
The online rice indica cDNA database provides cDNA resource with comprehensive information to researchers for functional analysis of indica subspecies and for comparative genomics. The RICD database is available through our website http://www.ncgr.ac.cn/ricd.
Background
Rice is one of the most important crops feeding about half of the world's population. Indica and japonica are two major Asian cultivated rice subspecies, which show distinct divergence from sequence variations to phenotypic changes [1-3]. The rice indica varieties are the most widely cultivated in China, India and most Southeast Asia countries, occupying the largest area of rice production in the world. However, most cDNA resources of the publicly available databases were generated from japonica subspecies [4]. Comparison of genomic DNA sequences has showed a large number of variations in genic regions between indica and japonica [2]. So, collection and analysis of indica cDNAs are essential and urgent. Therefore, we collected over 20,000 full-length cDNAs and over 40,000 5' ESTs isolated from various cDNA libraries of two indica varieties Guangluai 4 and Minghui 63 during the last few years (Table 1), as part of the National Rice Functional Genomics Project of China [5-7]. The cultivar Guangluai 4 is a typical indica variety with the largest growing area during 1968 to 1987 in China [8,9], while the cultivar Minghui 63 is the restore line for lots of planted rice hybrids. Systematic analysis and utilization of the indica cDNA clones would be valuable to rice genetics and breeding. Therefore, a dedicated platform is required for search, characterization and retrieval of the indica cDNA clones.
Table 1.
Sources of indica cDNA sequences in the RICD database.
| Number of cDNAs | Number of cDNAs matching IRGSP 4* | Number of cDNAs matching TIGR Gene** | |
| Guangluai 4 FL-cDNA | 10,081 | 9,109 (90.4%) | 7,680 (76.2%) |
| Minghui 63 FL-cDNA | 12,727 | 12,317 (96.8%) | 8,209 (64.5%) |
| Guangluai 4 5'EST | 21,690 | 16,504 (76.1%) | 8,700 (40.1%) |
| Minghui 63 5'EST | 27,130 | 21,059 (77.6%) | 15,115 (55.7%) |
* IRGSP pseudomolecules Build 4.
** TIGR rice gene annotation version 5
RICD was developed to provide a handy way to access all the available indica cDNA sources and be integrated with comprehensive information, which includes encoded amino acid sequence information, the mapping information, the protein domain information, the results of similarity search, gene function annotation and so on. Several search and retrieval forms were developed to create a comprehensive data warehouse for rice functional genomics and comparative genomics research.
Construction and content
Database architecture
RICD is a relational database, and was developed using MySQL 4.1. The database was implemented on a server running Red Hat Enterprise Linux AS release 4 (×86). The web interface was constructed by using PHP scripts (PHP Version 4.3), and powered by an Apache server (Apache Version 2.0).
cDNA acquisition
The Oryza sativa L. indica Guangluai-4 cDNAs were generated from five types of cDNA libraries [7]. The libraries included: (1) Seedlings grown in water for two weeks with a cycle of 16 hours light and 8 hours dark at 30°C; (2) Panicles harvested from rice grown in paddy fields; (3) Root grown in water for two weeks with a cycle of 16 hours light and 8 hours dark at 30°C; (4) Two-day germinated shoots and roots collected when roots reached 1–2 cm long; (5) Two-week seedlings treated individually with various stresses, that is, high-salinity (100 mM NaCl, treated for 20 min, 3, 12, 24, 48 h, 3 days and recovered for 72 h), dehydration (15% PEG-4000, treated for the same time duration as high-salinity), cold (60°C for 1, 12, 24, 48 h, 3 days and recovered for 72 h), heat (45°C, for the same time duration as cold), or immersion under water (for 1, 12, 24, 48 h, 3, 5 days). The Oryza sativa L. indica Minghui-63 cDNAs were from a normalized whole-life-cycle cDNA library, which was constructed by use of 15 tissues collected from 9 developmental stages [5].
Sequence analysis
The cDNAs and TIGR rice genes were remapped to IRGSP pseudomolecules Build 4 [3,10] using the GMAP program [11]. Over 90% putative full-length cDNAs of Guangluai 4 and Minghui 63 could be matched to rice IRGSP pseudomolecules, and 76.2% of Guangluai 4 cDNAs and 64.5% of Minghui 63 cDNAs could be matched to TIGR gene loci, respectively. In addition, > 76% 5' ESTs of Guangluai 4 and Minghui 63 were matched to IRGSP pseudomolecules, and > 40% of them could be matched to TIGR gene loci. The cDNAs were annotated according to RAP-DB [12] and TIGR systems [13]. Other sequence analysis was performed as described previously [7].
Utility
RICD provides an interactive and user-friendly web interface for searching the cDNA clones and retrieving their sequences along with other detailed information (Figure 1). The panel on the right of the main page lists hyperlinks to the different search pages, online tools and web pages describing various information of the database. The cDNA component of the database can be searched by multiple ways as described below.
Figure 1.
Query and display of cDNA clones in Rice Indica cDNA Database. The query parts are in red, while the display parts are in blue.
BLAST search
The BLAST program (version 2.2) is integrated into RICD for sequence search. Either nucleotide sequence (BLASTN) or protein sequence (TBLASTN) can be used as query to search all indica cDNAs or any subset of the database, with an initial cut-off E value as 0.01 [14]. A typical BLAST result page displays cDNAs matching the query, with their clone ID and the sequence alignments.
Keyword search
The cDNA of the database can be searched directly using clone name, NCBI accession number, RAP gene locus ID, or TIGR gene locus ID. The NCBI accession number commonly starts with the two characters "CT". And two separate 'gene locus ID search' sections, for RAP gene locus ID and TIGR gene locus ID, were given. The querying result will show a list of entries that are linked to the individual cDNA clones.
Chromosome position search
Two tools were designed for users to look up cDNAs in the rice genome. One shows in table form, naming "Clone list". You can directly enter the appointed region to get a list of cDNAs in the region, each of which can be further entered individually. A separate web page is provided for listing the putative indica-specific cDNAs, which cannot be mapped into IRGSP pseudomolecules Build 4. You can also choose a browser tool "mapping view" to visualize the graphic display. The main browser display contains two main parts. On the top are the scales for users to specify a particular region in the genome. On the bottom shows a series of cDNAs in the database, and also RAP-DB and TIGR gene loci. Upon clicking one, users can enter into result display page of the cDNA clone.
Function search
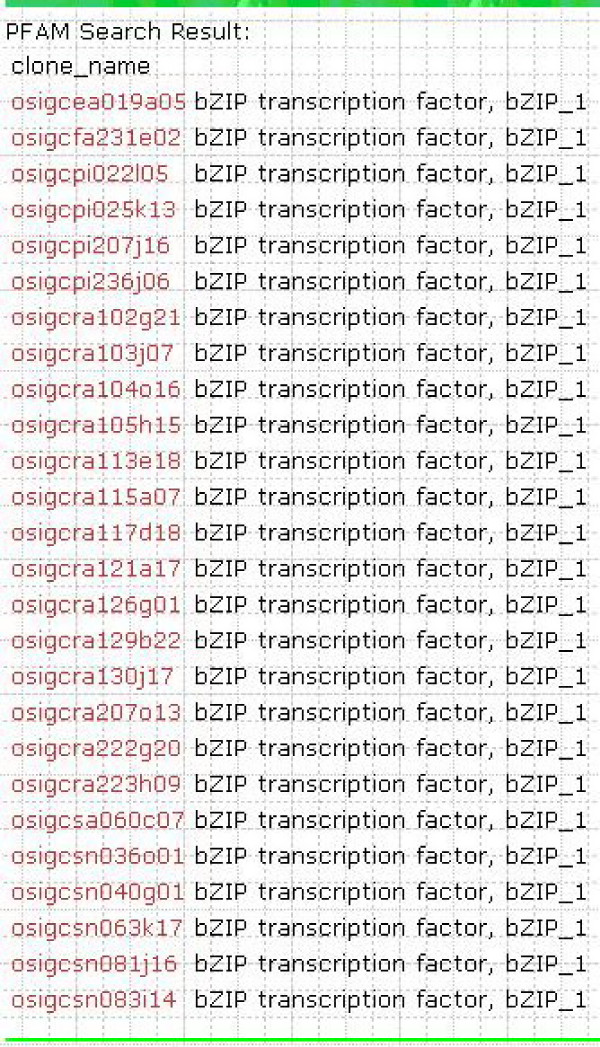
To query the cDNA clones, users can also enter a putative function (bZIP transcription factor, receptor protein kinase, and so on) to retrieve the responding cDNA clones (Figure 2). The putative functions rely on the protein domain Pfam descriptions [15] or RAP-DB gene annotations of any individual cDNA annotated.
Figure 2.
A example result page of a putative function query. The figure shows the page displaying Pfam search results for query term "bZIP transcription factor".
cDNA report display
For each cDNA clone, the result display page consists of three sections (Figure 3). The first section provides general information about the cDNA: genome position, clone name, accession number, library, RAP gene locus, TIGR gene locus, expression atlas in RiceGE [16] and variation report in Gramene [17]. The second section provides sequence of the cDNA and its ORF information. The last section includes the information of SNPs and InDels between japonica KOME and indica NCGR FL-cDNA (full-length cDNA) pairs and the results of similarity search with japonica and indica genome, KOME FL-cDNA and NCBI non-redundant databases.
Figure 3.
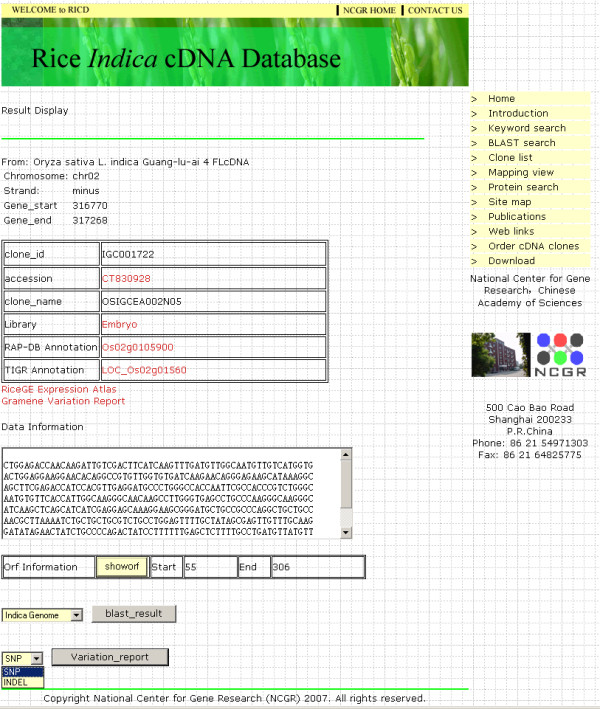
A typical result display page of a cDNA. The information of an indica cDNA clone includes the clone ID, GenBank Accession, library, genome position, gene annotation, expression atlas, variation report, cDNA sequence, ORF information. SNPs and InDels information and the results of similarity search will be shown after clicking corresponding buttons.
Conclusion
The initial aim of RICD is to provide a convenient way to search and retrieve incdia cDNA clone for research community. Via integrating with comprehensive information, it tries to grow to be a platform for broad applications to rice genetics, breeding and comparative genomics. Future expansion plans of the database include recruiting more indica cDNA clones, cataloging splice variants and adding gene expression data from indica cDNA microarray. The RICD database will be updated frequently if more information becomes available.
Availability and requirements
The RICD resource can be freely accessed via http://www.ncgr.ac.cn/ricd.
The cDNA clones in the database can be requested for research purpose by contacting Tingting Lu ttlu@ncgr.ac.cn
Authors' contributions
* The authors wish it to be known that, in their opinion, the first three authors should be regarded as joint first authors. TL designed the structure and layouts of database, provided the majority of annotations of sequences and partly drafted the manuscript. XH participated the designing of database, provided some annotation results and mostly drafted the manuscript. CZ developed the database and its web interface. TH and QZ provided the technical supports and discussion. KX, LX and QZ provided the batch sequences of Minghui-63 variety. BH conceived of the study, and participated in its design and helped to draft the manuscript. All coauthors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Lei Zhang (National Center for Gene Research) for offering the opinions of database construction. This research was supported by the grants from the Ministry of Science and Technology of China (The China Rice Functional Genomics Programs, grant numbers 2006AA10A102 and 2005CB120805), the Chinese Academy of Sciences (grant no. KSCW2-YW-N-024).
Contributor Information
Tingting Lu, Email: ttlu@ncgr.ac.cn.
Xuehui Huang, Email: xhhuang@ncgr.ac.cn.
Chuanrang Zhu, Email: crzhu@ncgr.ac.cn.
Tao Huang, Email: thuang@ncgr.ac.cn.
Qiang Zhao, Email: zqiang@ncgr.ac.cn.
Kabing Xie, Email: xiekabin@webmail.hzau.edu.cn.
Lizhong Xiong, Email: lizhongx@mail.hzau.edu.cn.
Qifa Zhang, Email: qifazh@mail.hzau.edu.cn.
Bin Han, Email: bhan@ncgr.ac.cn.
References
- Feng Q, Zhang Y, Hao P, Wang S, Fu G, Huang Y, Li Y, Zhu J, Liu Y, Hu X, Jia P, Zhang Y, Zhao Q, Ying K, Yu S, Tang Y, Weng Q, Zhang L, Lu Y, Mu J, Lu Y, Zhang LS, Yu Z, Fan D, Liu X, Lu T, Li C, Wu Y, Sun T, Lei H, Li T, Hu H, Guan J, Wu M, Zhang R, Zhou B, Chen Z, Chen L, Jin Z, Wang R, Yin H, Cai Z, Ren S, Lv G, Gu W, Zhu G, Tu Y, Jia J, Zhang Y, Chen J, Kang H, Chen X, Shao C, Sun Y, Hu Q, Zhang X, Zhang W, Wang L, Ding C, Sheng H, Gu J, Chen S, Ni L, Zhu F, Chen W, Lan L, Lai Y, Cheng Z, Gu M, Jiang J, Li J, Hong G, Xue Y, Han B. Sequence and analysis of rice chromosome 4. Nature. 2002;420:316–320. doi: 10.1038/nature01183. [DOI] [PubMed] [Google Scholar]
- Han B, Xue Y. Genome-wide intraspecific DNA-sequence variations in rice. Curr Opin Plant Biol. 2003;6:134–138. doi: 10.1016/S1369-5266(03)00004-9. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Profect (IRGSP) The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- The Rice Full-length cDNA Consortium Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- Xie K, Zhang J, Xiang Y, Feng Q, Han B, Chu Z, Wang S, Zhang Q, Xiong L. Isolation and annotation of 10828 putative full length cDNAs from indica rice. Sci China C Life Sci. 2005;48:445–451. doi: 10.1360/062004-90. [DOI] [PubMed] [Google Scholar]
- Zhang J, Feng Q, Jin C, Qiu D, Zhang L, Xie K, Yuan D, Han B, Zhang Q, Wang S. Features of the expressed sequences revealed by a large-scale analysis of ESTs from a normalized cDNA library of the elite indica rice cultivar Minghui 63. Plant J. 2005;42:772–780. doi: 10.1111/j.1365-313X.2005.02408.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Lu T, Yu S, Li Y, Huang Y, Huang T, Zhang L, Zhu J, Zhao Q, Fan D, Mu J, Shangguan Y, Feng Q, Guan J, Ying K, Zhang Y, Lin Z, Sun Z, Qian Q, Lu Y, Han B. A collection of 10,096 indica rice full-length cDNAs reveals highly expressed sequence divergence between Oryza sativa indica and japonica subspecies. Plant Mol Biol. 2007;65:403–415. doi: 10.1007/s11103-007-9174-7. [DOI] [PubMed] [Google Scholar]
- Lin S, Min S. Rice varieties and their genealogy in China. Shanghai: Shanghai Scientific and Technical Press; 1991. [Google Scholar]
- Monna L, Ohta R, Masuda H, Koike A, Minobe Y. Genome-wide searching of single-nucleotide polymorphisms among eight distantly and closely related rice cultivars (Oryza sativa L.) and a wild accession (Oryza rufipogon Griff.) DNA Res. 2006;13:43–51. doi: 10.1093/dnares/dsi030. [DOI] [PubMed] [Google Scholar]
- IRGSP Releases Build 4.0 Pseudomolecules of the Rice Genome http://rgp.dna.affrc.go.jp/E/IRGSP/Build4/build4.html
- Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- Rice Annotation Project The rice annotation project database (RAP-DB): 2008 update. Nucleic Acids Res. 2008;36:D1028–D1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Web address of RiceGE http://signal.salk.edu/cgi-bin/RiceGE
- Liang C, Jaiswal P, Hebbard C, Avraham S, Buckler ES, Casstevens T, Hurwitz B, McCouch S, Ni J, Pujar A, Ravenscroft D, Ren L, Spooner W, Tecle I, Thomason J, Tung CW, Wei X, Yap I, Youens-Clark K, Ware D, Stein L. Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 2008;36:D947–D953. doi: 10.1093/nar/gkm968. [DOI] [PMC free article] [PubMed] [Google Scholar]