Abstract
The anticipated therapeutic uses of neural stem cells depend on their ability to retain a certain level of developmental plasticity. In particular, cells must respond to developmental manipulations designed to specify precise neural fates. Studies in vivo and in vitro have shown that the developmental potential of neural progenitor cells changes and becomes progressively restricted with time. For in vitro cultured neural progenitors, it is those derived from embryonic stem cells that exhibit the greatest developmental potential. It is clear that both extrinsic and intrinsic mechanisms determine the developmental potential of neural progenitors and that epigenetic, or chromatin structural, changes regulate and coordinate hierarchical changes in fate-determining gene expression. Here, we review the temporal changes in developmental plasticity of neural progenitor cells and discuss the epigenetic mechanisms that underpin these changes. We propose that understanding the processes of epigenetic programming within the neural lineage is likely to lead to the development of more rationale strategies for cell reprogramming that may be used to expand the developmental potential of otherwise restricted progenitor populations.
Keywords: neural stem cell, progenitor, development, plasticity, differentiation, epigenetic
1. Introduction
A diverse range of neurological disorders affecting the central nervous system (CNS) have been considered as targets for stem cell therapies. These range from acute ischaemic insults to chronic neurodegenerative disorders, such as Parkinson's disease (PD), Huntington's disease (HD) and multiple sclerosis (MS). Critically, each disorder requires the development of different cell phenotypes from grafted cells, for example midbrain dopaminergic neurons for PD, medium spiney gamma aminobutyric acid (GABA)-ergic projection neurons for HD, diverse motor neuron classes for motor neuron disease and oligodendrocytes for MS (Bjorklund & Lindvall 2000; Zhang 2003; Lindvall et al. 2004; Chandran & Compston 2005). The ability of a neural stem cell population to be directed to differentiate towards different neuronal pathways requires a certain level of developmental plasticity.
In vivo, the specification of diverse neural and glial subtypes occurs within a longitudinal programme of nervous system development. The developmental processes involved are reviewed extensively elsewhere (Jessell 2000; Wilson & Edlund 2001; Rallu et al. 2002a; Zaki et al. 2003), but for the purposes of this review, these involve progressive changes in the developmental potential of cells (figure 1). Briefly, in response to neural-inducing signals, pluripotent primitive ectoderm differentiates through a multipotent stage of neural competence, to form neuroectoderm that comprises pseudostratified epithelial cells committed to the neural lineage. Early neural progenitor cells (NPCs) proliferate and are highly neurogenic but not gliogenic. The decision to retain a progenitor cell phenotype or commit to cell cycle exit and neural differentiation is temporally regulated and partly controlled by opposing activities of negative and positive transcriptional regulators of differentiation (SoxB1 (Sox1/2/3) and SoxB2 (Sox14/21), and Hes and proneural bHLH genes) in large part determined by Notch signalling (Bylund et al. 2003; Sandberg et al. 2005; Yoon & Gaiano 2005). Regulation of neural differentiation per se does not, however, generate neural diversity. The extensive neural diversity within the nervous system and the spatial organization of different neural subtypes are established initially by a process of neural patterning. The patterning process is mediated by a combination of morphogen (e.g. HH, WNT, BMP, FGF) and cell contact (e.g. Notch/delta, integrin)-mediated signalling that confers upon progenitor cells distinct rostrocaudal and dorsoventral positional identities (Briscoe & Novitch 2007) by regulating the expression of specific profiles of transcription factor genes. A key aspect to the patterning process is its temporal regulation. At early developmental stages, the neuroectoderm is multipotent, with cells from different positions being similarly responsive to different patterning cues. However, once progenitor domains have been established, inhibitory, or repressive, mechanisms appear to alter or limit the responses of cells to a given patterning cue. Mechanisms that limit the temporal responsiveness to growth factors and morphogens with respect to regulating the expression of fate-determining genes would be vital to prevent unwanted secondary patterning of cells as the context of growth factor and morphogen exposure changes with time, for example as cells patterned within one progenitor domain migrate through another. Additionally, placing a temporal limit on the patterning functions of growth factors and morphogens allows their signalling pathways to be reused for different purposes as development proceeds, for example the regulation of progenitor pool cell number and brain growth (Wechsler-Reya & Scott 1999; Agarwala et al. 2001; Lai et al. 2003; Tannahill et al. 2005).
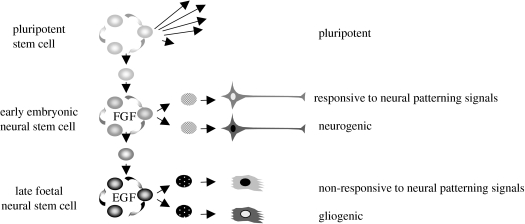
Figure 1.
Developmental potential of neural progenitor cells is temporally regulated. Early-stage embryonic and ES cell-derived neural progenitor cells proliferate in response to FGF2. They are neurogenic and responsive to morphogen patterning cues to generate a diversity of neural subtypes. Later-stage neural progenitors that are EGF responsive are non-responsive to patterning cues and undergo a switch from neurogenic to gliogenic differentiation.
It has been established for some time that gliogenesis and the differentiation of astrocytes and oligodendrocytes are under further temporal control and follow the period of spatial fate specification and neurogenesis (Temple 2001). Oligodendrocyte precursors (OPCs) are specified sequentially to motor neuron precursors in the ventrally restricted pMN domain of the neural tube, under the regulation of sonic hedgehog (SHH; Rowitch 2004; Wu et al. 2006). In contrast, astrocyte differentiation occurs throughout the CNS in an apparently position-independent manner. Despite the widespread origin of astrocytes, there is evidence for regional non-equivalence in their functions that could reflect positional programming (Castelo-Branco et al. 2005), and recent work has identified a regionally restricted transcriptional programme necessary for astrocyte development in the ventral spinal cord (Muroyama et al. 2005).
At present, the mechanisms that regulate a cell's temporal responsiveness to patterning cues and differentiation are poorly understood; however, it is evident that it involves the integration of both intrinsic and extrinsic mechanisms (Edlund & Jessell 1999; Lillien & Raphael 2000; Muhr et al. 2001; Shirasaki & Pfaff 2002). In particular, cell-intrinsic mechanisms are vital to determine a state of competence that is required for a cell to respond to extrinsic patterning and differentiation cues (Edlund & Jessell 1999; Corbin et al. 2000; Kobayashi et al. 2002; Rallu et al. 2002a,b). Furthermore, intrinsic mechanisms involving epigenetic programming control the heritability of induced phenotypes over subsequent cell generations (Fisher 2002; Li 2002; Arney & Fisher 2004). Epigenetic programming encompasses covalent modifications to DNA and chromatin that are heritable with cell division, and act to determine the accessibility, and therefore the potential, of genes to be transcribed. Epigenetic control of chromatin regulates both gene activation and silencing. Here, we briefly introduce major epigenetic mechanisms involved and discuss their roles in regulating neural lineage progression.
2. Epigenetic mechanisms of gene regulation and chromatin structure
Study of gene regulation through epigenetic control is one of the most rapidly increasing areas of research and is the subject of a number of excellent reviews (Li 2002; Jaenisch & Bird 2003; Stillman & Stewart 2004; Fuks 2005). Gene expression is not only dependent on the presence of the appropriate transcription factors that interact with enhancer or promoter elements in a cell-type or tissue-specific manner, but also dependent on the availability of binding sites regulated by higher orders of chromatin structure.
Chromatin is described as either heterochromatic or euchromatic. Compared with euchromatin, heterochromatin is characterized as being highly condensed, inaccessible to DNA nucleases, late-replicating and transcriptionally silent. Within euchromatic domains, the degree of condensation or DNA accessibility varies depending on gene activity; thus genes with potential for expression are often referred to as being in a ‘poised’ chromatin state (figure 2).
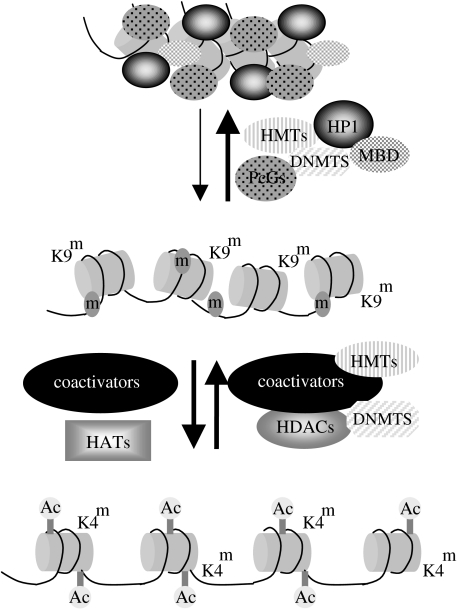
Figure 2.
Epigenetic modifications regulate the accessibility of genes for transcription by modifying chromatin structure. Expressed genes are present in chromatin with an open configuration and are characterized by histones that are acetylated (Ac) and have activity-related patterns of histone methylation, such as methylated lysine-4 on histone H3 (K4m). Transition to more repressed and silent chromatin states involves dynamic changes in the histone code, including histone deacetylation by HDACs, and changes in the histone methylation status, including demethylation of H3-K4 (e.g. by the demethylase LSD1) and gain in methylation of H3-K9 (K9m) by histone methyltransferases (HMTs). Repression and silencing are also mediated by DNA methylation (m) catalysed by DNA methyltransferases (DNMTS). Modified DNA and histones provide the substrate for the recruitment of further chromatin-modifying proteins (MBDs, HMTs), and non-histone chromatin proteins involved in chromatin compaction and long-term gene repression (e.g. HP1, PcGs, HMGs).
(a) Histone modification and a histone code for epigenetic states
Chromatin structure is founded on the nucleosome, i.e. 146 base pairs of DNA wrapped around an octamer of core histones, comprising an H3 : H4 tetramer and two H2A : H2B dimers with linker histones (H1) binding the DNA present between the nucleosomes. In recent years, it has become evident that the functions of histones extend far beyond that of providing a structural scaffold for chromatin assembly, but rather they play vital roles in many aspects of gene regulation (Fischle et al. 2003). Local chromatin structure and, consequently, the potential for gene expression are directly regulated by a number of post-translational covalent modifications of histone amino-terminal tails, including acetylation, methylation, phosphorylation, ADP-ribosylation, sumoylation and ubiquitinylation. The diversity of modifications and their potential combinatorial complexity led to the concept of a ‘histone code’ that determines chromatin structure (Strahl & Allis 2000; Turner 2000). Histone acetylation and methylation are the best-studied modifications. Histone acetylation modifies chromatin structure directly by effecting a change in electrostatic charge. In contrast, methyl groups added to lysine or arginine residues of histone tails act as binding sites for non-DNA-binding chromatin proteins. Typically, poised or open chromatin and gene expression are associated with histone acetylation and monomethylation of lysine K4 on histone H3, and closed chromatin and gene repression with histone deacetylation and trimethylation of K4 and K9.
This concept of the histone code is now well established; however, the potential modifications, their consequences for chromatin remodelling and gene regulation, and the dynamics of the mechanisms involved are becoming increasingly complex; for example, there are 17 known sites of lysine methylation and 7 known sites of arginine methylation on histones H3 and H4, which, in all combinations, would potentially give 3×1011 distinct methylation states of histone proteins (Fischle et al. 2003; Bannister & Kouzarides 2005).
Histone-modifying enzymes function by recruitment to their targets as part of chromatin-remodelling complexes. The complexity of histone modification is also evident by the large number of identified enzymes that catalyse reactions. Enzymes with histone acetyltransferase (HAT) activity include the MYST, GNAT, p300/CBP classes, as well as basal transcription factors and nuclear receptor cofactors (Roth et al. 2001; Carrozza et al. 2003). There are four classes of histone deacetylases (HDACs): class I (HDACs 1–3 and 8); class II (HDACs 4–7, 9 and 10); class III (SIRT 1–7); and class IV (HDAC 11) (Thiagalingam et al. 2003; Yang & Gregoire 2005). In humans and mice, 14 histone methyltransferases with differing lysine and arginine target specificities have been characterized (Bannister & Kouzarides 2005), and most recently, the first histone demethylases have been identified (Shi et al. 2004; Metzger et al. 2005; Shi et al. 2005; Tsukada et al. 2005).
Clearly, resolving the usage and potential complexity of the histone code presents a major challenge. Despite the identification of the large number of enzymes involved in histone modification, the relative roles of each enzyme, specifically, in neural development are very poorly understood. Comparison of mRNA levels for HDACs 1–10 in proliferating and differentiating cultured neuroblastoma cells identified steady levels of class I HDACs 1–3 and an upregulation of the class II HDACs 5–7 and 9, while HDACs 4, 8 and 10 were not expressed (Ajamian et al. 2003); however, there is currently little information concerning the spatial or temporal regulation of the histone-modifying enzymes during CNS development.
(b) DNA methylation
DNA methylation undoubtedly has a major role to play in gene silencing and thereby restricting the developmental potential of neural progenitor cells. Although, at present, the methylation status of the majority of genes expressed or repressed in different neural progenitor states is largely unknown, it is clear that regulation of DNA methylation is a dynamic process and that genes involved in its regulation are widely expressed during neural lineage development. Genomic methylation at CpG dinucleotides is essential for development and is catalysed by DNA methyltransferases (DNMTS; Li et al. 1992; Li 2002; Goll & Bestor 2004). DNMT1 is primarily a maintenance methyltransferase that uses hemi-methylated DNA as a template and copies the methylation pattern from the parental to daughter DNA strands during replication. De novo methylation, to establish new methylation patterns, is primarily catalysed by DNMT3a and DNMT3b (Robertson & Wolffe 2000; Goll & Bestor 2004). Dnmt1 is ubiquitously expressed, whereas Dnmt3a and Dnmt3b show a greater degree of differential regulation during CNS development that reflects their roles in de novo methylation. Dnmt3b may be more important for de novo methylation in the early phase of neurogenesis, since its expression is limited to a period of early foetal development (E10–14) in cells dispersed across the ventricular zone of the CNS. This would coincide with the major period of neurogenesis and after the period of fate specification by neural patterning. In contrast, Dnmt3a expression continues for longer and persists postnatally in postmitotic neurons, suggesting a role for de novo methylation and gene silencing during neuronal maturation (Feng et al. 2005). The importance of the maintenance activity of Dnmt1 in neural development has been shown in mice using the cre/lox system to conditionally delete Dnmt1 in the neural lineage. Surprisingly, hypomethylated NPCs made by Dnmt1 deletion using a nestin-Cre transgene did not have a major effect on neurogenesis itself; however, hypomethylated neurons that formed were functionally impaired and showed poor postnatal survival. Neonates with high numbers (95%) of hypomethylated neurons died at birth due to a failure to start respiration. Mosaic animals with approximately 30% hypomethylated neurons were able to survive, and in these animals, the hypomethylated neurons were selectively eliminated from the brain in the first few weeks after birth (Fan et al. 2001).
The mechanisms by which methyltransferases identify target sequences are currently being elucidated. The maintenance methylation activity of DNMT1 is coupled to DNA replication by its interaction with proliferating cell nuclear antigen at the replication fork (Sarraf & Stancheva 2004). For de novo methylation and methylation in non-proliferating cells, DNMTs are recruited to DNA by interaction with chromatin-remodelling complexes, and specificity is conferred by interaction with sequence-specific DNA-binding proteins. Major characterized interactions are with ATP-dependent remodelling complexes and, importantly, chromatin repression by DNA methylation is mechanistically very closely coupled to repression by histone deacetylation and histone methylation (Robertson 2002; Fuks et al. 2003a,b).
Methylation of CpG dinucleotides can have direct effects on gene expression, as the presence of methyl groups in the major groove of the double helix of DNA affects its structure and can directly interfere with the DNA binding of transcription factors (Takizawa et al. 2001). More pervasive effects of DNA methylation are mediated by methyl-CpG-binding proteins, such as MECP2, MBD1, MBD2 and KAISO. MECP2, MBD1 and MBD2 are members of a family of transcriptional repressors; each possesses a conserved methyl-CpG-binding domain (MBD; Hendrich & Tweedie 2003), and KAISO is a Bric-a-brac Tramtrack Broad (BTB) domain protein that binds to the methylated sequence mCGmCG or the non-methylated sequence CTGCNA to modulate transcription (Daniel et al. 2002). For MECP2, methylation-dependent repression of transcription can occur by interacting with the basal transcription machinery, for example by interaction of its transcription repressor domain with TFIIB (Kaludov & Wolffe 2000); however, more robust repression of methylated DNA is mediated by modifying chromatin structure. MECP2, MBD1 and MBD2 are all components of chromatin-remodelling complexes that provide mechanisms for crosstalk between repression by DNA methylation and histone modification (Jones et al. 1998; Nan et al. 1998; Feng & Zhang 2001; Burgers et al. 2002; Fuks et al. 2003b; Sarraf & Stancheva 2004). Despite the severe post-implantation failure of methyltransferase-deficient embryos defective in global DNA methylation, phenotypes associated with loss of methyl-CpG-binding proteins are comparatively mild. Loss of Mecp2 is most critical, which when mutated is responsible for the mental retardation disorder Rett's syndrome, and has been extensively reviewed (Ballestar & Wolffe 2001; Hendrich & Tweedie 2003; Caballero & Hendrich 2005; Fan & Hutnick 2005). In the mouse models of Rett's syndrome, loss of Mecp2 has no major effect on development. Instead, null mice develop apparently normally until five to six weeks of age, after which the severe neurological symptoms begin to emerge. Mutation of a transcriptional repressor was expected to result in widespread dysregulation of gene expression; however, extensive microarray gene expression analyses revealed only subtle changes in overall gene expression (Tudor et al. 2002). Nevertheless, one consequence of Mecp2 mutation was identified to be partial dysregulation of neuronal activity-dependent brain-derived neurotrophic factor (BDNF; Chen et al. 2003), and ectopic expression of BDNF, independent of Mecp2 regulation, can partially rescue the phenotypes seen in Mecp2 null mice (Chang et al. 2006).
Targeted mutations of Mbd1, Mbd2 and Kaiso exhibit surprisingly milder phenotypes. Mbd1 mutant mice appear healthy throughout life; however, closer examination found that they had decreased adult neurogenesis and that neural stem cells exhibited reduced neuronal differentiation and increased genomic instability (Zhao et al. 2003). Mbd2 mutant mice also developed normally, but were found to have a neurological phenotype that affected maternal nurturing behaviour (Hendrich et al. 2001), and no developmental defects in mice have been reported for Kaiso (Prokhortchouk et al. 2006). Although the mild phenotypes of mice mutant for Mbd1, Mbd2 and Kaiso might be expected to be due to genetic redundancy, this does not appear to be the case and this discrepancy remains unresolved. Evidence for extensive functional redundancy has not been supported by biochemical evidence, and further examination of the DNA-binding specificities of methyl-CpG-binding proteins suggests that different MBDs recognize methyl CpG in different DNA contexts (Klose et al. 2005).
3. Neural patterning and inheritance of progenitor cell fates: a role for Groucho family co-repressors
The expression patterns of fate-determining transcription factors are restricted to progenitor domains and are maintained within tight boundaries. In the spinal cord, homeodomain transcription factors (TCFs) can be classified as either class I or class II depending on their regulation by SHH signalling, where SHH either represses or activates their expression, respectively. A major discovery of Briscoe and colleagues (Briscoe et al. 2000; Briscoe & Novitch 2007) was that class I and II genes function as cross-regulatory pairs, and that domains of expression are established by mutual cross-repressive interactions between the class I and the class II factors (figure 3a). The class I and II proteins include members of the NKX, PAX, IRX and DBX families. Clues to their mechanisms of action came from recognizing that eight out of ten known homeodomain TCFs involved in dorsoventral patterning possess conserved transcriptional repressor domains with homology to eh-1, the repressor domain of the TCF engrailed (Muhr et al. 2001). In the case of the class II NKX proteins, the eh-1 homology resides within the conserved NK decapeptide or TN domain. In Drosophila, the eh-1 domain mediates its repressive effects by interaction and recruitment of the co-repressor Groucho (Jimenez et al. 1997). Muhr and colleagues showed that patterning and fate specification by Nkx genes were dependent on recruitment of Groucho family co-repressors by interaction with their TN domains (figure 3b; Muhr et al. 2001). The mammalian homologues of Groucho are the transducin-like Enhancer of split (TLE1–4) proteins in humans and mice. TLE co-repressors do not themselves bind DNA, but are recruited to their target loci by interaction with repressor domains of DNA-binding transcription factors, including the eh-1 domain (Chen & Courey 2000; Gasperowicz & Otto 2005).
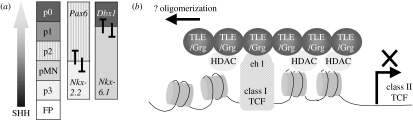
Figure 3.
Responsiveness to morphogen and neural patterning involves repressive functions of fate-determining genes. (a) The graded activity of morphogens, such as SHH expressed in the floor plate (FP) of the neural tube, establishes progenitor domains of cells committed to differentiate with restricted neural fates (e.g. motor neurons derive from the pMN domain and domains p0–p3 derive four distinct classes of interneuron). In the ventral spinal cord, progenitor domains are established through the cross-repressive actions of transcription factors (TCFs) that are either activated (class II genes, e.g. Nkx2.2, Nkx6.1) or repressed (class I genes, e.g. Pax6, Dbx1) by SHH. (b) Repressive effects of TCFs possessing eh1 domains are mediated by Groucho family co-repressors (TLE). TLEs recruit repressor complexes, including HDAC activity, and may promote a spread of repressor activity in cis through auto-oligomerization.
It is unlikely that the mechanism of defining regional identity by cross-repression of fate-determining TCFs is restricted to dorsoventral patterning of the neural tube; for example, Six3 and Irx3 have been identified to mutually repress each others' expression and are the key determinants of rostrocaudal identity, contributing to the definition of forebrain and midbrain territories (Kobayashi et al. 2002). Furthermore, it has been shown that SIX3 functions as a TLE-dependent repressor in forebrain and eye development (Kobayashi et al. 2001; Zhu et al. 2002).
The regulation and mode of action of TLE co-repressors is only partly understood. A major activity involves chromatin remodelling by HDAC recruitment; however, TLEs are subject to a variety of post-translational modification, including phosphorylation, ADP-ribosylation and ubiquitinylation, and their actions will be context dependent. Several roles for TLEs in dynamic control of gene expression have been characterized (Chen & Courey 2000; Gasperowicz & Otto 2005). In the case of neural patterning, while TLEs play a role in establishing cross-repressive domains, it is not yet clear whether TLEs also play a role in the long-term maintenance of gene repression, acting beyond their initial interaction with remodelling complexes. One property of TLE proteins of interest is their potential capacity for long-range gene silencing by oligomerization through interaction between their Q domains, and thereby their potential to extend complex formation in cis along the chromosome (Chen & Courey 2000; Song et al. 2004). Commonly, stable heritability of a differentiated state uses two major mechanisms, DNA methylation and higher-order chromatin assembly that is in part dependent on histone methylation. Understanding how positional information is retained in the neural tube and how this may relate to TLE function will require further analysis of the chromatin states, or configurations, of fate-determining genes over developmental time.
4. Co-repressor regulation and epigenetic control of neurogenesis
The decision made by progenitor cells to exit proliferation and commit to neural differentiation is regulated at many levels (Ohnuma & Harris 2003; Gotz & Huttner 2005). Of particular importance is the role played by Notch signalling (Louvi & Artavanis-Tsakonas 2006; Yoon & Gaiano 2005). Effectors of Notch signalling help to maintain cells in a proliferative progenitor state, for example the transcription repressor HES1 actively inhibits the expression of neurogenic bHLH genes, such as Ascl1 (Cau et al. 2000; Kageyama et al. 2005). HES1 like other repressors acts in concert with co-repressor complexes. Vital to HES1 function is its interaction with the co-repressor TLE1 (Nuthall et al. 2002; Nuthall et al. 2004). TLE1 activity is central to the switch in fate that is made when neural progenitors commit to neural differentiation. In proliferating progenitor cells, a HES1/TLE1 complex represses Ascl1 by SIN3a and HDAC recruitment. The repression complex includes poly(ADP-ribose) polymerase 1 (PARP-1), and derepression of Ascl1 involves a regulated dismissal of the TLE co-repressor complex by its ADP-ribosylation. PARP-1 acts as a sensor of extrinsic platelet-derived growth factor (PDGF) signalling, being activated by increased Ca2+ and CaMKIIδ phosphorylation. After the TLE1 dismissal, PARP-1 modifies HES1 that remains bound to the promoter and switches its activity from repressor to activator, allowing subsequent recruitment of histone acetylase to modify chromatin ready for transcription (Ju et al. 2004).
The neurogenic competence of progenitor cells is also regulated by the activity of the nuclear receptor co-repressor N-CoR. N-CoR acts as a co-repressor for a number of transcription factors including RBPSUH, the mammalian homologue of Drosophila Suppressor of Hairless [Su(H)], a further downstream effector of Notch signalling (Kao et al. 1998). Like other non-DNA-binding co-repressors, N-CoR participates in a number of HDAC-containing repressor complexes (Jepsen & Rosenfeld 2002), and it could therefore function in early-stage neurogenic progenitors to repress later-stage gliogenic gene expression. Indeed, in the absence of N-CoR, foetal cortical progenitors failed to undergo FGF-dependent self-renewal in vitro and spontaneously differentiated into astrocytes (Hermanson et al. 2002). Further analysis showed that in wild-type cells, the nuclear localization and therefore the activity of N-CoR are determined by their phosphorylation status. Protein phosphatase-1 activity confers nuclear localization of N-CoR, whereas ciliary neurotrophic factor (CNTF) signalling that triggers glial differentiation leads to phosphatidyl-inositol-3-OH kinase/Akt1 kinase-dependent phosphorylation of N-CoR and its relocation to the cytoplasm (Hermanson et al. 2002).
While neurogenic programmes promote neural differentiation and the expression of neural genes, in non-neuronal cells, the expression of neuron-specific genes is actively maintained in transcriptionally repressed states by the actions of another repressor complex orchestrated by REST. REST is a zinc-finger DNA-binding protein with two repressor domains that is expressed in non-neuronal cells, including neural progenitor cells, and represses genes that contain a conserved 23 bp repressor element, RE1. The amino-terminal repressor domain recruits the SIN3/HDAC complex and the carboxy-terminal domain interacts with CoREST, a co-repressor that mediates more long-term gene silencing by recruitment of HDACs, histone H3-lysine-9 histone methyltransferases and the histone H3-lysine-4 demethylase LSD1. The CoREST complex is also found to interact with MECP2 and the heterochromatin protein HP1 (Ballas & Mandel 2005). Although REST occupies RE1 silencer elements in both non-neuronal cells and neural progenitors, recent studies have found that its mode of action is distinct. In non-neuronal cells, the repressive activities of REST confer closed and compact chromatin configurations on neural genes; in contrast, in ES cells and neural progenitor cells, REST mediates gene repression, but the chromatin of the neural-specific gene is maintained in an open or poised chromatin state; thus upon neural differentiation of progenitor cells neural-specific genes are already poised for expression and gene activation does not require a more complex process of reprogramming (Ballas et al. 2005).
5. Temporal regulation of gliogenesis: interaction of extrinsic and epigenetic mechanisms establishes competence for glial-specific gene expression
Following neural patterning and the generation of neurons with regionally specified identities, a subsequent major change in developmental plasticity of neural progenitor cells is the so-called gliogenic switch. It is well established that neurogenesis precedes gliogenesis (Qian et al. 2000). As with earlier stages of neural lineage progression, the differentiation of multipotent neural progenitors is determined by their underlying competence for glial gene expression. Extrinsic mechanisms that regulate gliogenesis are partially understood (Rajan & McKay 1998). Two pathways that have been studied in detail are those activated by interleukin-6 (IL-6) family of neurotrophic cytokines (including leukaemia inhibitory factor (LIF), CNTF and cardiotrophin-1 (CT-1)) and bone morphogenic protein (BMP) signalling that act synergistically to promote glial differentiation (Bonni et al. 1997; Rajan & McKay 1998; Nakashima et al. 1999; Sun et al. 2001; Barnabe-Heider et al. 2005). Cytokines including CNTF, LIF and CT-1 activate gp130 and the JAK-STAT pathway after binding the LIF receptor (Stahl & Yancopoulos 1994), and BMPs signal to regulate nuclear translocation of SMAD transcriptional co-activator proteins (Massague 2000). The importance of cell competence for gliogenesis was recognized following observations that while late-stage NPCs differentiated into glia after CNTF addition, early-stage NPCs failed to respond, despite the fact that early NPCs expressed gp130 and the LIF receptors, and were able to activate the JAK-STAT pathway by STAT1/3 phosphorylation (Bonni et al. 1997; Nakashima et al. 1999; Takizawa et al. 2001). This suggested that intrinsic mechanisms acting in early-stage NPCs act to inhibit the gliogenic pathway. Further analyses have now shown that signalling by the activated JAK-STAT and SMAD pathways is in fact-regulated at multiple levels (figure 4). Activation of glial gene expression by STAT1/3 involves association with the transcriptional co-activators CBP/p300 and SMAD1, and once initiated, a positive autoregulatory loop acts, whereby STAT1/3 directly upregulates further components of the JAK-STAT pathway to strengthen STAT signalling and trigger astrogliogenesis (He et al. 2005). In early-stage NPCs, the association between STAT1/3 and CBP/p300 is suppressed by high levels of the neurogenic basic helix-loop-helix transcription factor neurogenin1, which also associates with CBP/p300 and SMAD1 and thereby sequesters CBP–SMAD1 away from glial-specific genes and inhibits STAT1/3-mediated gene transcription (figure 4a; Sun et al. 2001).
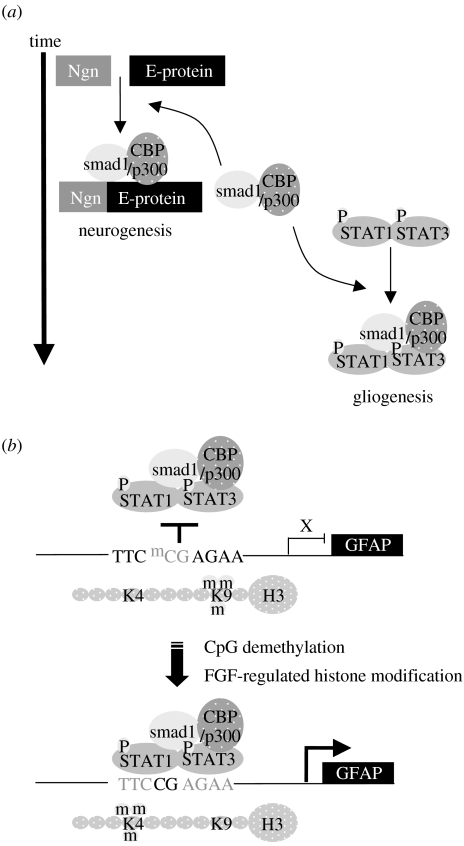
Figure 4.
Activation of the glial marker gene Gfap is regulated at multiple levels. (a) Gfap expression involves activation of STAT transcription factors and recruitment of SMAD and CBP/p300 coactivator complexes; at early time points, the availability of the SMAD and CBP/p300 coactivators are limiting due to competitive binding by neurogenic transcription factors. (b) The binding of activated STAT and coactivator complexes to the Gfap promoter is regulated by its epigenetic status. In neurogenic cells, the Gfap promoter is repressed by histone H3-lysine-9 methylation and CpG methylation within the STAT-binding site. In later-stage gliogenic cultures, there is an FGF-dependent chromatin remodelling that results in CpG demethylation, histone H3-lysine-9 demethylation and histone K4 methylation.
Significant insight into mechanisms of gliogenesis has come from studying regulation of glial fibrillar-associated protein (Gfap) gene expression as a marker of the glial lineage (figure 4b). Consistent with the role of STAT signalling in gliogenesis, Gfap promoter activity is dependent on upstream STAT-binding sites, with one site at −1518 to −1510 being critical (Bonni et al. 1997; Nakashima et al. 1999). To address the questions of astroglial competence and cell-type-specific and temporal regulation of Gfap expression, Taga and colleagues have performed extensive analyses of the DNA methylation status of the Gfap promoter (Takizawa et al. 2001). In early-stage NPCs and neuronal and non-neural lineage cells, the Gfap promoter region is hypermethylated compared with the pattern seen in astrocytes and late-stage astroglial competent NPCs. Although hypermethylation is indicative of a more closed or repressed chromatin state, Taga and colleagues recognized that the STAT-binding site at position −1518 to −1510 (TTCCGAGAA) contained a CpG dinucleotide and that methylation of this specific site was sufficient to block STAT-dependent expression of Gfap. In a second more recent study, Fan and colleagues have further examined the effect of DNA methylation on gliogenesis by studying mice with DNMT1 conditionally deleted from the neural lineage, and found that neural development in the absence of DNMT1 leads to precocious astroglial differentiation (Fan et al. 2005). The importance of this work has been to show a direct role for epigenetic programming in neural lineage fate restriction and, in particular, to provide a cell intrinsic mechanism that accounts for the competence of cells to respond to a specific extrinsic signal, in this case CNTF signalling and STAT activation.
Although it is clear that the DNA methylation status determines competence for Gfap expression, it is less clear what first determines the methylation status. The Gfap promoter is hypermethylated at all developmental stages and in all lineages, besides the late-stage NPCs and their astrocyte progeny. Therefore, one of the primary events in determining competence for Gfap expression (and by extrapolation gliogenesis) must be temporal and cell-type-specific demethylation. At present, mechanisms for demethylation are poorly characterized; since no demethylase enzyme has been unambiguously identified, the most accepted method for demethylation is the passive loss of CpG methylation that occurs following successive rounds of DNA replication in the absence of maintenance methylase activity, catalysed by DNMT1 (Bestor 2000). Alternatively, gliogenic cues could signal an active demethylation of target loci, for example by mechanisms that promote 5-methylcytosine deamination to create a T-G mismatch, which when converted back to a matched C-G base pair by DNA repair enzymes effectively results in demethylation (Morgan et al. 2004). Whether demethylation of the Gfap promoter is passive or active is unknown. In either case, chromatin-remodelling events in the glial lineage that control access of DNMT1 (or other enzymes) to glial-specific genes such as Gfap would lead to demethylation and allow their activation by extrinsic signals.
Extrinsic factors that affect glial competence of progenitor cells, particularly in in vitro studies, are fibroblast growth factor (FGF) and epidermal growth factor (EGF) signalling. Developmental and culture studies have shown that early foetal neural progenitors are initially responsive to FGF2 and that this signal primes cells to later become responsive to EGF (Tropepe et al. 1999; Lillien & Raphael 2000; Qian et al. 2000; Ciccolini 2001). For routine culture, neural progenitors (or ‘neurospheres’) are usually maintained using a combination of FGF2 and EGF. A recent study by Song & Ghosh (2004) has shown that FGF2 signalling is also directly responsible for the acquired competence of neural progenitors to respond to CNTF to promote astrocyte differentiation by regulating the histone methylation status of glia-specific promoters. Chromatin immunoprecipitation analyses showed that glial competence of cortical progenitor cultures was associated with increased H3-K4 methylation and suppression of H3-K9 methylation of chromatin at the STAT-binding site of the Gfap and S100b promoters. The inference is that FGF2 directly or indirectly regulates histone methyltransferase activities.
Taken together, regulation of the gliogenic switch involves the interaction or coupling of both extrinsic and intrinsic mechanisms. Extrinsic signals can be divided into two types: the first controls the progenitor cell state to establish competence for differentiation (e.g. Notch/FGF/EGF) and the second activates differentiation pathways themselves (e.g. CT/CNTF/BMP).
6. Developmental plasticity of neural precursor cell cultures
Landmark studies by Reynolds and Weiss, and Bartlett and colleagues in the early 1990s identified culture conditions to grow neural progenitor cells from foetal and adult germinal regions of the brain (Murphy et al. 1990; Reynolds & Weiss 1992, 1996; Reynolds et al. 1992). Cultured neural stem cells were shown to be multipotent, with the ability to clonally generate neurons, astrocytes and oligodendrocytes, and today the clonal generation of neurospheres (the unit of neural stem cell colony formation) remains the predominant in vitro assay for studying neural stem cells and neurogenesis (Reynolds & Rietze 2005). Although neurosphere cultures are multipotent, neurons that differentiate become restricted in their developmental potential and the efficiency of neuronal differentiation tends to decline with passage. A consistent finding is that neurons derived from passaged neurosphere cultures, derived from different brain regions and species, tend to differentiate towards a GABAergic neurotransmitter phenotype (Jain et al. 2003). A major concern, and area of some controversy, is the degree to which neurosphere cultures retain their starting positional identity. Some reports claim that regional specification of progenitors is maintained in culture and that intrinsic differences persist even in late passages (Hitoshi et al. 2002; Seaberg et al. 2005), while other studies suggest that in vitro culture leads to a significant relaxation of lineage restriction with major changes in gene expression. For example, neurospheres derived from foetal dorsal and ventral telencephalon have been shown to rapidly lose their original identity as examined by the expression of dorsal and ventral markers, respectively (Santa-Olalla et al. 2003; Hack et al. 2004; Machon et al. 2005). These studies suggest that maintenance of positional identity requires ongoing interaction with environmental cues that otherwise become disrupted in culture. Machon and colleagues found that within three passages, gene expression regulated by canonical wingless (WNT) signalling, in dorsal cultures, is downregulated and that dorsal character could be partially rescued by dominant activation of the WNT pathway (Machon et al. 2005), albeit only up to about passage 8.
The neurodevelopmental plasticity of neurosphere cultures is a significant issue for their potential downstream uses, for example for transplantation (Jain et al. 2003; Zietlow et al. 2005). To date, no studies have reported successful manipulation of neurosphere cultures to specify and differentiate progenitor cells with a desired neuronal phenotype. The failure to systematically manipulate the fate of neurosphere cultures (e.g. by instruction using patterning cues) suggests that the cells in culture are fate-restricted and refractory to patterning. In addition to changes in neurosphere properties over the initial few passages, cell potential continues to change upon prolonged culture. When assayed by intrastriatal transplantation, four-week expanded human and mouse cells show good graft survival at 12 weeks following transplantation; however, 12-week expanded cells fail to survive after transplantation (Zietlow et al. 2005).
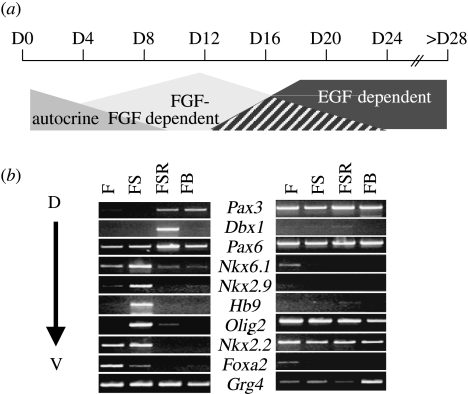
Problems associated with attempts to manipulate foetal and adult neural stem cell fates contrast significantly with studies using embryonic stem cell-derived neural progenitor cells (ES-NPCs). Neural differentiation of both mouse and human ES cells is very robust, particularly in serum-free medium, in which ES cells rapidly downregulate pluripotency-associated genes and upregulate proneural and neurogenic genes (Zhang et al. 2001; Aubert et al. 2003; Aiba et al. 2005; Bouhon et al. 2005; Itsykson et al. 2005; Watanabe et al. 2005; Joannides et al. 2006). Significantly, the growth and differentiation properties of ES-NPCs change with time in a strictly regulated manner (figure 5a). It is striking that the developmental plasticity of ES-NPCs closely follows that of endogenous neural development. Neural differentiation in serum-free conditions is dependent on autocrine FGF signalling and generates a population of SOX1-positive NPCs (Ying et al. 2003; Bouhon et al. 2005). By limiting potential extrinsic cues, ES-NPCs default to express markers characteristic of the anterior neural plate (e.g. OTX1/2, PAX6; Watanabe et al. 2005); however, at early stages, these cells are fully competent to respond to extrinsic developmental cues, and they can be manipulated accordingly to derive cells that express gene profiles which reflect distinct rostrocaudal and dorsoventral neural identities. Such a strategy of directed differentiation, based on an applied developmental biology of neural patterning, has been used to derive a number of specific neural lineages, including motor neurons, midbrain-like dopaminergic neurons, hypothalamic dopamine neurons and retinal ganglion neurons (Kim et al. 2002; Wichterle et al. 2002; Ikeda et al. 2005; Ohyama et al. 2005). However, like the situation discussed above (§3) for patterning neural progenitors in vivo, the responsiveness of ES-NPCs is also temporally restricted. Mouse ES-NPCs can be systematically patterned in their first 4–8 days of culture, as exemplified in figure 5b; however, later cultures, given the same treatments between days 16 and 20, are non-responsive and instead adopt a more constitutive pattern of gene expression (Bouhon et al. 2006). In human cultures, the responsiveness and potential of ES-NPCs for directed differentiation is similarly temporally regulated (Li et al. 2005).
Figure 5.
Temporal development of ES cell-derived neural progenitors in serum-free conditions. (a) Neural differentiation initially occurs under autocrine FGF control. Passage and proliferation of early-born neural progenitors is dependent on exogenous FGF2, while later-stage cultures acquire responsiveness to EGF and eventually become dependent on exogenous EGF for long-term propagation. (b) Only early-stage cultures (D8) are responsive to instructive patterning cues; the figure shows a semi-quantitative RT-PCR analysis of a panel of genes expressed in progressively dorsal to ventral progenitor domains of the neural tube being regulated in response to FGF2 alone (F), FGF2+SHH (FS), FGF2+SHH+retinoic acid (FSR) or FGF2+BMP4 (FB). Later-stage cultures (D20) become non-responsive and instead exhibit constitutive patterns of gene expression.
The mechanisms that limit the temporal responsiveness of cells to patterning cues are unknown. Given the apparent recapitulation of developmental mechanisms in ES cultures, one possibility is that early cell fates are maintained by similar mechanisms, for example involving programming by TLE co-repressor activity. In this regard, it is interesting to note that TLE expression is temporally regulated during ES-NPC development (N. D. Allen 2004, unpublished observations). Since extrinsic cues conferred by the spatial organization and interaction of cells that are otherwise present in vivo are not present in vitro, the tight temporal control over patterning seen in ES cell cultures suggests that programming is very much an intrinsic property of cells. Nevertheless, some extrinsic signals are present and may be important.
Developmental progression of mouse ES-NPCs also recapitulates the temporal regulation of gliogenesis that is seen in vivo and in foetal primary cell cultures. Early ES-NPCs are highly neurogenic, generating more than 90% differentiated β-tubulinIII-positive neurons, and only later-passaged cultures generate astrocytes or oligodendrocytes. Taga and colleagues have analysed the competence of ES-NPCs for gliogenesis, and showed that cells acquire responsiveness to LIF and STAT activated gliogenesis only in later-stage cultures (more than 14–18 days after initiating neural differentiation). Furthermore, as with primary foetal cultures, the onset of gliogenesis coincided with CpG demethylation of STAT-binding sites in the GFAP promoter. In ES cells and early ES-NPCs, and also non-neural cell types, the GFAP promoter was hypermethylated (Shimozaki et al. 2005). Human cultures show a similar pattern of temporal change in gliogenic competence, although compared with mouse cultures, the time-course of events is much more protracted (Itsykson et al. 2005).
7. Roles for FGF2 and EGF in shaping developmental competence
A recurring theme for temporal regulation of NPC fate, from both in vivo and in vitro studies, is the role played by FGF and EGF signalling (Ciccolini & Svendsen 1998; Tropepe et al. 1999; Lillien & Raphael 2000; Qian et al. 2000; Ciccolini 2001; Sun et al. 2005). Culture of early mouse ES-NPCs, like their embryonic counterparts, is dependent on FGF2; however, FGF2 alone is unable to sustain cultures for much more than 20 days, at which point proliferation starts to decline and cell death ensues. EGF receptor (EGFR) expression and EGF responsiveness are only acquired in later-stage cultures and are dependent on the prior FGF signalling. Interestingly, the temporal upregulation of EGFRs coincides with the time at which responsiveness to patterning cues is lost and with the transition to glial competence. FGF2 and EGF both signal through receptor tyrosine kinases (RTKs) and activate similar intracellular signalling pathways; however, their roles in NPC proliferation and fate determination are clearly distinct. The outcome of RTK signalling is influenced by many parameters (Yarden 2001; Jorissen et al. 2003). Phosphorylated tyrosine of activated receptors serves as docking sites for Src homology 2 (SH2) and phosphotyrosine-binding domain effector and adaptor proteins. A recent proteomic study of EGFR and related ErbB family members has revealed an extensive network of potential protein interactions, with varying affinity thresholds, and suggests a greater complexity in cell responses than hitherto suspected (Jones et al. 2005). Although there is currently little information about the downstream targets of FGF and EGF signalling during neural development, especially at different times during lineage progression, both signals activate pathways that can regulate gene expression directly and indirectly via chromatin modification. For EGFR, activation leads to the sequential phosphorylation of mitogen activated protein (MAP) kinases and effector kinases, such as RSK2. Following translocation to the nucleus, effector kinases regulate transcription by phosphorylating transcription factors and, in addition, have been shown to modify chromatin by phosphorylation of H3 histone tails, which then enhances their efficiency of acetylation by HAT complexes (Cheung et al. 2000).
Unlike FGF2, EGF is essential for the long-term propagation of ES-derived NPCs. Indeed, EGF promotes the selection of a late-stage EGF-responsive cell type, which is distinct in character and potential from the earlier neurogenic and patterning-responsive NPCs. Conti and co-workers have shown that EGF is sufficient to maintain long-term culture via continued symmetric self-renewal of cells initially derived from FGF2-propagated ES-NPCs and also from primary foetal and human neurosphere cultures (Conti et al. 2005). The phenotypic properties of these cells are distinct from those present in early-stage cultures. The so-called ‘NS’ cells share many properties with proliferative radial glia that have been identified in vivo as neural stem cells (Merkle et al. 2004; Gotz & Barde 2005). NS cells are a homogenous population of cells that express a panel of radial glial markers, including RC2, GLAST and BLBP; however, they also co-express a number of genes, such as Pax6, Emx2, Olig2 and Ascl1, that are not normally co-expressed in vivo, and this suggests a certain level of gene dysregulation (Conti et al. 2005).
The differential actions of FGF2 and EGF on NPC proliferation or NS cell self-renewal have not been defined. Part of the change in dependence of cells from FGF to EGF may be related to control of cell cycle regulators. p16INK4a and p19ARF are two tumour suppressors encoded by the Ink/Arf locus and function as negative regulators of cell proliferation by promoting activation of the Rb and p53 pathways (Lowe & Sherr 2003). In contrast, loss or repression of p16INK4a/p19ARF is associated with enhanced proliferation, including stem cell self-renewal. Furthermore, mutations in p16INK4a and p19ARF, together with EGFR activation, are hallmarks of developing gliomas (Bachoo et al. 2002). Repression of the Ink/Arf locus and consequent promotion of stem cell self-renewal are critically dependent on repression mediated by the Polycomb group (PcG) protein BMI1. In Bmi1 null mice, there is a gradual depletion of the stem cell pool that leads to defects in adult neurogenesis (Molofsky et al. 2003), and these defects can be partially rescued in mice that are doubly mutant for Bmi1 and Ink/Arf (Bruggeman et al. 2005; Molofsky et al. 2005).
8. Potential for epigenetic reprogramming and enhancement of neural potential
From the viewpoint of regenerative medicine and stem cell therapeutics, there is considerable interest in strategies that might promote reprogramming to enhance or expand the developmental potential of neural progenitor populations, especially endogenous populations present in vivo. Studies on reprogramming are in their infancy; nevertheless, some progress has been made. Different approaches to reprogramming can be considered and include physiological, pharmacological and genetic strategies.
Currently, the best-characterized example of reprogramming by a physiological means is the reversion of OPCs back to ‘neural stem-like cells’ (NSLCs) by treatment with FGF2 (Kondo & Raff 2000). Following foetal neurogenesis, neural progenitor cells proliferate as glial-restricted precursors, generating astrocytes and oligodendrocytes. OPCs harvested and cultured from the rat postnatal optic nerve (Barres & Raff 1994) have been established as a well-characterized lineage-restricted cell model to study mechanisms of fate determination and differentiation. OPCs proliferate in response to PDGF and can be induced to differentiate into either oligodendrocytes or type-2 astrocytes (2As) by treatment with thyroid hormone or bone morphogenetic proteins (BMPs), respectively (Mabie et al. 1997). Although the natural fate of OPCs is to become glia, Kondo & Raff (2000) found that treatment of OPC-derived type-2 astrocytes with FGF2 led to cell reprogramming, to derive cells with neural stem cell-like properties (NSCLs), that could self-renew and, most importantly, could be differentiated to derive neurons and type-1 astrocytes, in addition to oligodendrocytes and 2As. The reprogramming from OPC to NSCL is a multistep process, with BMP treatment and transition through the 2A being a necessary intermediate for responsiveness to the FGF2 signal. A key event is the BMP-dependent re-expression of the SoxB1 gene Sox2 in the type-2 astrocyte, and it is relevant, as there is increasing evidence for an essential role for SoxB1 genes for the maintenance of neural stem cells (Zappone et al. 2000; Bylund et al. 2003; Graham et al. 2003). Sox2 expression is repressed in OPCs, with H3 histones at the enhancer region being in an inactive form, H3K9 being methylated and unacetylated and H3K4 being unmethylated, while in the 2As, the histone H3 modification pattern is switched to an ‘active’ pattern. The mechanism by which BMP2 initiates this reprogramming is not known; however, it does involve chromatin remodelling by recruitment to the Sox2 promoter/enhancer region of the BRCA1- and BRM-containing SWI/SNF chromatin-remodelling complexes to reactivate the locus (Kondo & Raff 2004).
A second example of dedifferentiation within the neural lineage has been recognized by examination of glioblastomas. A hallmark of glioblastoma is the uncontrolled proliferation of cells with an immature astrocytic phenotype: genetically tumours are most frequently associated with EGFR activation and null mutations in the p16INK4a and p19ARF genes. Analysis of neural stem cell cultures from p16INK4a/p19ARF−/− showed that EGF responsiveness and proliferation were no different from wild-type cultures; however, when the growth of primary astrocyte cultures was examined, a significant phenotype was seen. Wild-type astrocyte cultures have limited growth potential, as cells begin to senesce after five to seven population doublings. However, p19/p16 null cells exhibit immortal cell growth, and furthermore, in response to EGF, astrocytes show a rapid dedifferentiation. Cells contract their cytoplasms, adopt a bipolar morphology and then detach from the culture dish and grow as substrate-independent aggregates, similarly to neurospheres. Dedifferentiation is suggested by the fact that cells lose their GFAP expression and re-express neural progenitor markers nestin and A2B5. Furthermore, some of these cells could be induced to undergo neural differentiation upon plating (Bachoo et al. 2002).
A third potential example of reprogramming, although less well characterized, is the neural differentiation of NS cells. NS cells self-renew in the presence of EGF and, upon growth factor withdrawal, they differentiate into astrocytes (Conti et al. 2005). However, if EGF is first withdrawn and differentiation is initiated in the continued presence of FGF2, then astrocyte differentiation is prevented and approximately 35% of cells undergo neurogenesis, yet these cultures differentiate into astrocytes if initiated in the presence of serum or BMP4. Although the mechanisms of NS cell neurogenesis need to be determined, there is a clear requirement for FGF signalling in the absence of EGF to ‘prime’ or reprogramme cells for neural competence, suggesting possible parallels to the role of FGF in OPC reprogramming discussed previously.
Other examples of enhanced potency by culture in FGF or EGF include induced ventral character and multipotency of dorsal spinal cord cultures when exposed to FGF2 in vitro (Chandran et al. 2003; Gabay et al. 2003), and include the conversion of restricted neuronal progenitors (type C cells) from the adult subventricular zone to multipotent stem cells as a response to EGF (Doetsch et al. 2002).
9. Strategies for directed reprogramming of neural cells
The aforementioned examples demonstrate dedifferentiation or reprogramming of cells towards earlier more potent progenitor states, which occurs in response to specific extrinsic cues or, in the case of glioblastoma, genetic change. It would seem reasonable that understanding, and then systematic reversal, of the pathways that lead to lineage restriction in normal development would enable more directed reprogramming of neural cells.
Reagents that affect chromatin structure either directly or indirectly are of particular interest to cell reprogramming. It is widely recognized that dysregulation of genes involved in tumorigenesis and tumour progression is associated with epigenetic changes. Consequently, there is currently a significant interest and effort in the identification of drugs that target enzymes involved in epigenetic programming, such as DNMTs, HDACs and HMTs (Kristeleit et al. 2004; Villar-Garea & Esteller 2004; Lyko & Brown 2005). Once these tools become available, testing their use in the context of cell reprogramming within the neural lineage will be of particular interest.
At present, only a few systematic studies on cell fate using currently available drugs have been reported, and have focused on the effects of HDAC inhibitors. Hseih and colleagues showed that valproic acid (VPA), an HDAC inhibitor, promoted the neuronal differentiation of adult hippocampal neural progenitors and inhibited astrocyte and oligodendrocyte differentiation, even in conditions that favoured gliogenesis. Although VPA might alter the expression of many genes, one target gene identified with a major neurogenic influence was the transcription factor, NeuroD (Hsieh et al. 2004). In separate studies, Casaccia-Bonnefil and colleagues had shown that oligodendrocyte lineage progression was dependent on HDAC activity (Marin-Husstege et al. 2002) and determined that VPA affected the timing of progenitor cell differentiation into myelin-forming oligodendrocytes in vivo (Shen et al. 2005). Postnatal administration of VPA resulted in significant hypomyelination in the developing corpus callosum with delayed expression of late differentiation markers and retained expression of progenitor markers. Interestingly, HDAC inhibition by VPA after the onset of myelination had no effect on myelin gene expression, and was consistent with further changes to nucleosomal histones from a state of reversible deacetylation to a more stable repressed state characterized by histone methylation and chromatin compaction.
The shortcoming of drugs such as HDAC, HMT or DNMT inhibitors is their general lack of specificity. Drugs such as VPA would have widespread effects on histone deacetylation across the genome, and are likely to have global effects of shifting inactive or poised chromatin towards more active states. This may be sufficient in some instances to trigger or promote reprogramming events; however, more directed reprogramming is likely to require further manipulation of specific targets of extrinsic cues. First, in contrast to drug action, specificity for the majority of potential targets could be achieved using RNA interference (RNAi) to knockdown gene expression. Importantly, advances in RNAi technology have established libraries of vectors with extensive genome coverage, thereby establishing the ability to conduct screens for gene knockdowns that might promote reprogramming or enhance developmental plasticity (Dykxhoorn & Lieberman 2005; Silva et al. 2005). Second, a better understanding of signalling pathways that regulate chromatin structure might facilitate reprogramming; for example, Paro and colleagues have found that cell reprogramming during the process of transdetermination in Drosophila involves downregulation of PcG protein function, and this is directly controlled by the activation of the Jun amino-terminal kinase (JNK) signalling pathway in the cells undergoing regeneration (Lee et al. 2005). Moreover, this interaction is conserved in mammalian cells, suggesting that a role for JNK signalling in remodelling cellular fates might be conserved. Systematic manipulation of signalling pathways that modulate chromatin structure may provide more physiological routes to reprogramming and enhancing progenitor cell plasticity.
Acknowledgments
I am particularly grateful to colleagues Hidemasa Kato, Siddharthan Chandran, Alexis Joannides and Alysia Battersby for stimulating discussion, and for help with the manuscript. N.D.A. is supported by grants from the BBSRC and MRC.
Footnotes
One contribution of 14 to a Theme Issue ‘Stem cells and brain repair’.
References
- Agarwala S, Sanders T.A, Ragsdale C.W. Sonic hedgehog control of size and shape in midbrain pattern formation. Science. 2001;291:2147–2150. doi: 10.1126/science.1058624. [DOI] [PubMed] [Google Scholar]
- Aiba K, Sharov A.A, Carter M.G, Foroni C, Vescovi A.L, Ko M.S. Defining a developmental path to neural fate by global expression profiling of mouse embryonic stem cells and adult neural stem/progenitor cells. Stem Cells. 2005;4:889–895. doi: 10.1634/stemcells.2005-0332. [DOI] [PubMed] [Google Scholar]
- Ajamian F, Suuronen T, Salminen A, Reeben M. Upregulation of class II histone deacetylases mRNA during neural differentiation of cultured rat hippocampal progenitor cells. Neurosci. Lett. 2003;346:57–60. doi: 10.1016/s0304-3940(03)00545-7. [DOI] [PubMed] [Google Scholar]
- Arney K.L, Fisher A.G. Epigenetic aspects of differentiation. J. Cell Sci. 2004;117:4355–4363. doi: 10.1242/jcs.01390. [DOI] [PubMed] [Google Scholar]
- Aubert J, et al. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-gfp knock-in mice. Proc. Natl Acad. Sci. USA. 2003;100(Suppl. 1):11 836–11 841. doi: 10.1073/pnas.1734197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachoo R.M, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr. Opin. Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu D.D, Speh J.C, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Ballestar E, Wolffe A.P. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur. J. Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- Bannister A.J, Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- Barnabe-Heider F, Wasylnka J.A, Fernandes K.J, Porsche C, Sendtner M, Kaplan D.R, Miller F.D. Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron. 2005;48:253–265. doi: 10.1016/j.neuron.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Barres B.A, Raff M.C. Control of oligodendrocyte number in the developing rat optic nerve. Neuron. 1994;12:935–942. doi: 10.1016/0896-6273(94)90305-0. [DOI] [PubMed] [Google Scholar]
- Bestor T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Bjorklund A, Lindvall O. Cell replacement therapies for central nervous system disorders. Nat. Neurosci. 2000;3:537–544. doi: 10.1038/75705. [DOI] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank D.A, Rozovsky I, Stahl N, Yancopoulos G.D, Greenberg M.E. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Bouhon I.A, Kato H, Chandran S, Allen N.D. Neural differentiation of mouse embryonic stem cells in chemically defined medium. Brain Res. Bull. 2005;68:62–75. doi: 10.1016/j.brainresbull.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Bouhon I.A, Joannides A, Kato H, Chandran S, Allen N.D. Embryonic stem cell derived neural progenitors display temporal restriction to neural patterning. Stem Cells. 2006;24:1908–1913. doi: 10.1634/stemcells.2006-0031. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Novitch B.G. Regulatory pathways linking progenitor patterning, cell fates, and neurogenesis in the ventral neural tube. Phil. Trans. R. Soc. B. 2007;363:57–70. doi: 10.1098/rstb.2006.2012. doi:10.1098/rstb.2006.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell T.M, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Bruggeman S.W, et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers W.A, Fuks F, Kouzarides T. DNA methyltransferases get connected to chromatin. Trends Genet. 2002;18:275–277. doi: 10.1016/S0168-9525(02)02667-7. [DOI] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch B.G, Muhr J. Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat. Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Caballero I.M, Hendrich B. MeCP2 in neurons: closing in on the causes of Rett syndrome. Hum. Mol. Genet. 2005;14:R19–R26. doi: 10.1093/hmg/ddi102. Spec No. 1. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J, Utley R.T, Workman J.L, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco G, Sousa K.M, Bryja V, Pinto L, Wagner J, Arenas E. Ventral midbrain glia express region-specific transcription factors and regulate dopaminergic neurogenesis through Wnt-5a secretion. Mol. Cell. Neurosci. 2005;31:251–262. doi: 10.1016/j.mcn.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Casarosa S, Kageyama R, Guillemot F. Hes genes regulate sequential stages of neurogenesis in the olfactory epithelium. Development. 2000;127:2323–2332. doi: 10.1242/dev.127.11.2323. [DOI] [PubMed] [Google Scholar]
- Chandran S, Compston A. Neural stem cells as a potential source of oligodendrocytes for myelin repair. J. Neurol. Sci. 2005;233:179–181. doi: 10.1016/j.jns.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Chandran S, Kato H, Gerreli D, Compston A, Svendsen C.N, Allen N.D. FGF-dependent generation of oligodendrocytes by a hedgehog-independent pathway. Development. 2003;130:6599–6609. doi: 10.1242/dev.00871. [DOI] [PubMed] [Google Scholar]
- Chang Q, Khare G, Dani V, Nelson S, Jaenisch R. The disease progression of mecp2 mutant mice is affected by the level of BDNF expression. Neuron. 2006;49:341–348. doi: 10.1016/j.neuron.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Chen G, Courey A.J. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- Chen W.G, Chang Q, Lin Y, Meissner A, West A.E, Griffith E.C, Jaenisch R, Greenberg M.E. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner K.G, Cheung W.L, Sassone-Corsi P, Denu J.M, Allis C.D. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Ciccolini F. Identification of two distinct types of multipotent neural precursors that appear sequentially during CNS development. Mol. Cell Neurosci. 2001;17:895–907. doi: 10.1006/mcne.2001.0980. [DOI] [PubMed] [Google Scholar]
- Ciccolini F, Svendsen C.N. Fibroblast growth factor 2 (FGF-2) promotes acquisition of epidermal growth factor (EGF) responsiveness in mouse striatal precursor cells: identification of neural precursors responding to both EGF and FGF-2. J. Neurosci. 1998;18:7869–7880. doi: 10.1523/JNEUROSCI.18-19-07869.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin J.G, Gaiano N, Machold R.P, Langston A, Fishell G. The Gsh2 homeodomain gene controls multiple aspects of telencephalic development. Development. 2000;127:5007–5020. doi: 10.1242/dev.127.23.5007. [DOI] [PubMed] [Google Scholar]
- Daniel J.M, Spring C.M, Crawford H.C, Reynolds A.B, Baig A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30:2911–2919. doi: 10.1093/nar/gkf398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Petreanu L, Caille I, Garcia-Verdugo J.M, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002;36:1021–1034. doi: 10.1016/s0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn D.M, Lieberman J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu. Rev. Med. 2005;56:401–423. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- Edlund T, Jessell T.M. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- Fan G, Hutnick L. Methyl-CpG binding proteins in the nervous system. Cell Res. 2005;15:255–261. doi: 10.1038/sj.cr.7290294. [DOI] [PubMed] [Google Scholar]
- Fan G, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J. Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–832. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis C.D. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Fisher A.G. Cellular identity and lineage choice. Nat. Rev. Immunol. 2002;2:977–982. doi: 10.1038/nri958. [DOI] [PubMed] [Google Scholar]
- Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd P.J, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003a;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd P.J, Wolf D, Nan X, Bird A.P, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003b;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Gabay L, Lowell S, Rubin L.L, Anderson D.J. Deregulation of dorsoventral patterning by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron. 2003;40:485–499. doi: 10.1016/s0896-6273(03)00637-8. [DOI] [PubMed] [Google Scholar]
- Gasperowicz M, Otto F. Mammalian Groucho homologs: redundancy or specificity? J. Cell Biochem. 2005;95:670–687. doi: 10.1002/jcb.20476. [DOI] [PubMed] [Google Scholar]
- Goll M.G, Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2004;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gotz M, Barde Y.A. Radial glial cells defined and major intermediates between embryonic stem cells and CNS neurons. Neuron. 2005;46:369–372. doi: 10.1016/j.neuron.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Hack M.A, Sugimori M, Lundberg C, Nakafuku M, Gotz M. Regionalization and fate specification in neurospheres: the role of Olig2 and Pax6. Mol. Cell Neurosci. 2004;25:664–678. doi: 10.1016/j.mcn.2003.12.012. [DOI] [PubMed] [Google Scholar]
- He F, et al. A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat. Neurosci. 2005;8:616–625. doi: 10.1038/nn1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–277. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Guy J, Ramsahoye B, Wilson V.A, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson O, Jepsen K, Rosenfeld M.G. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- Hitoshi S, Tropepe V, Ekker M, van der Kooy D. Neural stem cell lineages are regionally specified, but not committed, within distinct compartments of the developing brain. Development. 2002;129:233–244. doi: 10.1242/dev.129.1.233. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage F.H. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc. Natl Acad. Sci. USA. 2004;101:16 659–16 664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H, et al. Generation of Rx+/Pax6+ neural retinal precursors from embryonic stem cells. Proc. Natl Acad. Sci. USA. 2005;102:11 331–11 336. doi: 10.1073/pnas.0500010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsykson P, Ilouz N, Turetsky T, Goldstein R.S, Pera M.F, Fishbein I, Segal M, Reubinoff B.E. Derivation of neural precursors from human embryonic stem cells in the presence of noggin. Mol. Cell Neurosci. 2005;30:24–36. doi: 10.1016/j.mcn.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jain M, Armstrong R.J, Tyers P, Barker R.A, Rosser A.E. GABAergic immunoreactivity is predominant in neurons derived from expanded human neural precursor cells in vitro. Exp. Neurol. 2003;182:113–123. doi: 10.1016/s0014-4886(03)00055-4. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Rosenfeld M.G. Biological roles and mechanistic actions of co-repressor complexes. J. Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Jessell T.M. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including hairy and engrailed. Genes Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joannides, A. J. et al 2006 A scaleable and defined system for generating neural stem cells from human embryonic stem cells. Stem Cells 9 November 2006 [Epub ahead of print]. [DOI] [PubMed]
- Jones P.L, Veenstra G.J, Wade P.A, Vermaak D, Kass S.U, Landsberger N, Strouboulis J, Wolffe A.P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Jones R.B, Gordus A, Krall J.A, Macbeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2005;439:168–174. doi: 10.1038/nature04177. doi:10.1038/nature04177 [DOI] [PubMed] [Google Scholar]
- Jorissen R.N, Walker F, Pouliot N, Garrett T.P, Ward C.W, Burgess A.W. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp. Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- Ju B.G, Solum D, Song E.J, Lee K.J, Rose D.W, Glass C.K, Rosenfeld M.G. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp. Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Kaludov N.K, Wolffe A.P. MeCP2 driven transcriptional repression in vitro: selectivity for methylated DNA, action at a distance and contacts with the basal transcription machinery. Nucleic Acids Res. 2000;28:1921–1928. doi: 10.1093/nar/28.9.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao H.Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C.R, Evans R.M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H, et al. Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson's disease. Nature. 2002;418:50–56. doi: 10.1038/nature00900. [DOI] [PubMed] [Google Scholar]
- Klose R.J, Sarraf S.A, Schmiedeberg L, McDermott S.M, Stancheva I, Bird A.P. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Nishikawa K, Suzuki T, Yamamoto M. The homeobox protein Six3 interacts with the Groucho corepressor and acts as a transcriptional repressor in eye and forebrain formation. Dev. Biol. 2001;232:315–326. doi: 10.1006/dbio.2001.0185. [DOI] [PubMed] [Google Scholar]
- Kobayashi D, Kobayashi M, Matsumoto K, Ogura T, Nakafuku M, Shimamura K. Early subdivisions in the neural plate define distinct competence for inductive signals. Development. 2002;129:83–93. doi: 10.1242/dev.129.1.83. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science. 2000;289:1754–1757. doi: 10.1126/science.289.5485.1754. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes Dev. 2004;18:2963–2972. doi: 10.1101/gad.309404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristeleit R, Stimson L, Workman P, Aherne W. Histone modification enzymes: novel targets for cancer drugs. Expert Opin. Emerg. Drugs. 2004;9:135–154. doi: 10.1517/eoed.9.1.135.32947. [DOI] [PubMed] [Google Scholar]
- Lai K, Kaspar B.K, Gage F.H, Schaffer D.V. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat. Neurosci. 2003;6:21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- Lee N, Maurange C, Ringrose L, Paro R. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature. 2005;438:234–237. doi: 10.1038/nature04120. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor T.H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li X.J, Du Z.W, Zarnowska E.D, Pankratz M, Hansen L.O, Pearce R.A, Zhang S.C. Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 2005;23:215–221. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- Lillien L, Raphael H. BMP and FGF regulate the development of EGF-responsive neural progenitor cells. Development. 2000;127:4993–5005. doi: 10.1242/dev.127.22.4993. [DOI] [PubMed] [Google Scholar]
- Lindvall O, Kokaia Z, Martinez-Serrano A. Stem cell therapy for human neurodegenerative disorders—how to make it work. Nat. Med. 2004;10:S42–S50. doi: 10.1038/nm1064. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lowe S.W, Sherr C.J. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J. Natl Cancer Inst. 2005;97:1498–1506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- Mabie P.C, Mehler M.F, Marmur R, Papavasiliou A, Song Q, Kessler J.A. Bone morphogenetic proteins induce astroglial differentiation of oligodendroglial–astroglial progenitor cells. J. Neurosci. 1997;17:4112–4120. doi: 10.1523/JNEUROSCI.17-11-04112.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machon O, Backman M, Krauss S, Kozmik Z. The cellular fate of cortical progenitors is not maintained in neurosphere cultures. Mol. Cell Neurosci. 2005;30:388–397. doi: 10.1016/j.mcn.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J. Neurosci. 2002;22:10 333–10 345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- Merkle F.T, Tramontin A.D, Garcia-Verdugo J.M, Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc. Natl Acad. Sci. USA. 2004;101:17 528–17 532. doi: 10.1073/pnas.0407893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller J.M, Schneider R, Peters A.H, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Molofsky A.V, Pardal R, Iwashita T, Park I.K, Clarke M.F, Morrison S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A.V, He S, Bydon M, Morrison S.J, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H.D, Dean W, Coker H.A, Reik W, Petersen-Mahrt S.K. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 2004;279:52 353–52 360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Muhr J, Andersson E, Persson M, Jessell T.M, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Muroyama Y, Fujiwara Y, Orkin S.H, Rowitch D.H. Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature. 2005;438:360–363. doi: 10.1038/nature04139. [DOI] [PubMed] [Google Scholar]
- Murphy M, Drago J, Bartlett P.F. Fibroblast growth factor stimulates the proliferation and differentiation of neural precursor cells in vitro. J. Neurosci. Res. 1990;25:463–475. doi: 10.1002/jnr.490250404. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by STAT3–Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng H.H, Johnson C.A, Laherty C.D, Turner B.M, Eisenman R.N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nuthall H.N, Husain J, McLarren K.W, Stifani S. Role for Hes1-induced phosphorylation in Groucho-mediated transcriptional repression. Mol. Cell Biol. 2002;22:389–399. doi: 10.1128/MCB.22.2.389-399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthall H.N, Joachim K, Stifani S. Phosphorylation of serine 239 of Groucho/TLE1 by protein kinase CK2 is important for inhibition of neuronal differentiation. Mol. Cell Biol. 2004;24:8395–8407. doi: 10.1128/MCB.24.19.8395-8407.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma S, Harris W.A. Neurogenesis and the cell cycle. Neuron. 2003;40:199–208. doi: 10.1016/s0896-6273(03)00632-9. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Ellis P, Kimura S, Placzek M. Directed differentiation of neural cells to hypothalamic dopaminergic neurons. Development. 2005;132:5185–5197. doi: 10.1242/dev.02094. [DOI] [PubMed] [Google Scholar]
- Prokhortchouk A, et al. Kaiso-deficient mice show resistance to intestinal cancer. Mol. Cell Biol. 2006;26:199–208. doi: 10.1128/MCB.26.1.199-208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie S.K, He W, Capela A, Davis A.A, Temple S. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Rajan P, McKay R.D. Multiple routes to astrocytic differentiation in the CNS. J. Neurosci. 1998;18:3620–3629. doi: 10.1523/JNEUROSCI.18-10-03620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallu M, Corbin J.G, Fishell G. Parsing the prosencephalon. Nat. Rev. Neurosci. 2002a;3:943–951. doi: 10.1038/nrn989. [DOI] [PubMed] [Google Scholar]
- Rallu M, Machold R, Gaiano N, Corbin J.G, McMahon A.P, Fishell G. Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and Hedgehog signaling. Development. 2002b;129:4963–4974. doi: 10.1242/dev.129.21.4963. [DOI] [PubMed] [Google Scholar]
- Reynolds B.A, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Reynolds B.A, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev. Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- Reynolds B.A, Rietze R.L. Neural stem cells and neurospheres—re-evaluating the relationship. Nat. Methods. 2005;2:333–336. doi: 10.1038/nmeth758. [DOI] [PubMed] [Google Scholar]
- Reynolds B.A, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J. Neurosci. 1992;12:4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K.D. DNA methylation and chromatin—unraveling the tangled web. Oncogene. 2002;21:5361–5379. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- Robertson K.D, Wolffe A.P. DNA methylation in health and disease. Nat. Rev. Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- Roth S.Y, Denu J.M, Allis C.D. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Rowitch D.H. Glial specification in the vertebrate neural tube. Nat. Rev. Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Sandberg M, Kallstrom M, Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat. Neurosci. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- Santa-Olalla J, Baizabal J.M, Fregoso M, del Carmen Cardenas M, Covarrubias L. The in vivo positional identity gene expression code is not preserved in neural stem cells grown in culture. Eur. J. Neurosci. 2003;18:1073–1084. doi: 10.1046/j.1460-9568.2003.02824.x. [DOI] [PubMed] [Google Scholar]
- Sarraf S.A, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Seaberg R.M, Smukler S.R, van der Kooy D. Intrinsic differences distinguish transiently neurogenic progenitors from neural stem cells in the early postnatal brain. Dev. Biol. 2005;278:71–85. doi: 10.1016/j.ydbio.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J. Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine J.R, Cole P.A, Casero R.A, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y.J, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Shimozaki K, Namihira M, Nakashima K, Taga T. Stage- and site-specific DNA demethylation during neural cell development from embryonic stem cells. J. Neurochem. 2005;93:432–439. doi: 10.1111/j.1471-4159.2005.03031.x. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff S.L. Transcriptional codes and the control of neuronal identity. Annu. Rev. Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Silva J.M, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Song M.R, Ghosh A. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat. Neurosci. 2004;7:229–235. doi: 10.1038/nn1192. [DOI] [PubMed] [Google Scholar]
- Song H, Hasson P, Paroush Z, Courey A.J. Groucho oligomerization is required for repression in vivo. Mol. Cell Biol. 2004;24:4341–4350. doi: 10.1128/MCB.24.10.4341-4350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl N, Yancopoulos G.D. The tripartite CNTF receptor complex: activation and signaling involves components shared with other cytokines. J. Neurobiol. 1994;25:1454–1466. doi: 10.1002/neu.480251111. [DOI] [PubMed] [Google Scholar]
- Stillman B, Stewart D.J. Cold Spring Harbor Symposia on Quantitative Biology. vol. 69. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 2004. Epigenetics. [Google Scholar]
- Strahl B.D, Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin M.Z, Zubiaga A, Hua X, Fan G, Greenberg M.E. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Sun Y, Goderie S.K, Temple S. Asymmetric distribution of EGFR receptor during mitosis generates diverse CNS progenitor cells. Neuron. 2005;45:873–886. doi: 10.1016/j.neuron.2005.01.045. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Tannahill D, Harris L.W, Keynes R. Role of morphogens in brain growth. J. Neurobiol. 2005;64:367–375. doi: 10.1002/neu.20163. [DOI] [PubMed] [Google Scholar]
- Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S, Cheng K.H, Lee H.J, Mineva N, Thiagalingam A, Ponte J.F. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann. N. Y. Acad. Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- Tropepe V, Sibilia M, Ciruna B.G, Rossant J, Wagner E.F, van der Kooy D. Distinct neural stem cells proliferate in response to EGF and FGF in the developing mouse telencephalon. Dev. Biol. 1999;208:166–188. doi: 10.1006/dbio.1998.9192. [DOI] [PubMed] [Google Scholar]
- Tsukada Y.I, Fang J, Erdjument-Bromage H, Warren M.E, Borchers C.H, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2005;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Tudor M, Akbarian S, Chen R.Z, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc. Natl Acad. Sci. USA. 2002;99:15 536–15 541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Villar-Garea A, Esteller M. Histone deacetylase inhibitors: understanding a new wave of anticancer agents. Int. J. Cancer. 2004;112:171–178. doi: 10.1002/ijc.20372. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat. Neurosci. 2005;8:288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya R.J, Scott M.P. Control of neuronal precursor proliferation in the cerebellum by sonic hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter J.A, Jessell T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Wilson S.I, Edlund T. Neural induction: toward a unifying mechanism. Nat. Neurosci. 2001;4:1161–1168. doi: 10.1038/nn747. [DOI] [PubMed] [Google Scholar]
- Wu S, Wu Y, Capecchi M.R. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development. 2006;433:581–590. doi: 10.1242/dev.02236. [DOI] [PubMed] [Google Scholar]
- Yang X.J, Gregoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol. Cell Biol. 2005;25:2873–2884. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y. The EGFR family and its ligands in human cancer. Signalling mechanisms and therapeutic opportunities. Eur. J. Cancer. 2001;37(Suppl. 4):S3–S8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- Ying Q.L, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat. Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- Zaki P.A, Quinn J.C, Price D.J. Mouse models of telencephalic development. Curr. Opin. Genet. Dev. 2003;13:423–437. doi: 10.1016/s0959-437x(03)00084-4. [DOI] [PubMed] [Google Scholar]
- Zappone M.V, et al. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000;127:2367–2382. doi: 10.1242/dev.127.11.2367. [DOI] [PubMed] [Google Scholar]
- Zhang S.C. Embryonic stem cells for neural replacement therapy: prospects and challenges. J. Hematother. Stem Cell Res. 2003;12:625–634. doi: 10.1089/15258160360732650. [DOI] [PubMed] [Google Scholar]
- Zhang S.C, Wernig M, Duncan I.D, Brustle O, Thomson J.A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- Zhao X, et al. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl Acad. Sci. USA. 2003;100:6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C.C, Dyer M.A, Uchikawa M, Kondoh H, Lagutin O.V, Oliver G. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development. 2002;129:2835–2849. doi: 10.1242/dev.129.12.2835. [DOI] [PubMed] [Google Scholar]
- Zietlow R, Pekarik V, Armstrong R.J.E, Tyers P, Dunnett S.B, Rosser A.E. The survival of neural precursor cell grafts is influenced by in vitro expansion. J. Anat. 2005;207:227–240. doi: 10.1111/j.1469-7580.2005.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]