Abstract
All the neurons and glial cells of the central nervous system are generated from the neuroepithelial cells in the walls of the embryonic neural tube, the ‘embryonic neural stem cells’. The stem cells seem to be equivalent to the so-called ‘radial glial cells’, which for many years had been regarded as a specialized type of glial cell. These radial cells generate different classes of neurons in a position-dependent manner. They then switch to producing glial cells (oligodendrocytes and astrocytes). It is not known what drives the neuron–glial switch, although downregulation of pro-neural basic helix–loop–helix transcription factors is one important step. This drives the stem cells from a neurogenic towards a gliogenic mode. The stem cells then choose between developing as oligodendrocytes or astrocytes, of which there might be intrinsically different subclasses. This review focuses on the different extracellular signals and intracellular responses that influence glial generation and the choice between oligodendrocyte and astrocyte fates.
Keywords: CNS, oligodendrocyte, astrocyte, development
1. Introduction
The nervous system is composed of neurons and glial cells. Glial cells in the central nervous system (CNS) include both the ‘macroglia’, which are derived from the neural tube, and the ‘microglia’, which are derived from haemopoietic precursors. Microglia are the resident macrophages of the CNS and play a key role in immune surveillance and defence. This review is concerned with the two major classes of macroglia: astrocytes and oligodendrocytes. These cells were first visualized over a century ago (Andriezen 1893) and were given names that described their morphology in Golgi tissue preparations: astrocytes (‘star-like’) and oligodendrocytes (‘sparsely branching’). The term ‘glial’ originally referred to the sticky nature of the substance we now know as white matter, the myelinated axon tracts. This seeded the idea that glial cells, which are abundant in white matter, have a purely passive support role—the ‘glue’ holding the neurons together (for a historical review see Pasik & Pasik 2004). Today, it is recognized that glial cells play active roles in nervous system function, even forming synaptic contacts with neurons and contributing in some way to information storage or processing. Oligodendrocytes have a well-defined function—formation of the insulating myelin sheaths around CNS axons—but they and their precursors also express voltage- and ligand-gated ion channels and have synaptic input from neurons. Astrocytes have many described functions: (i) they provide structural and trophic support for neurons, (ii) they interact intimately with blood vessels and induce formation of the blood–brain barrier, and (iii) they regulate CNS synaptogenesis and synaptic transmission. Despite the functional versatility of glial cells and the fact that they significantly outnumber neurons in most parts of the CNS, we know relatively little about their intrinsic diversity and how this arises during development.
The multitude of neuronal and glial cell types of the mature CNS are generated from an initially homogeneous pool of naive neural precursors (NPCs) in the embryonic neural tube, the so-called neuroepithelial precursor cells (NEPs). The NEPs first generate neurons, followed by glial cells. At early stages, NEPs are multipotent with the capacity to generate all types of CNS neurons and glia. With time, their potential is restricted by exposure to surrounding environmental signals that transmit positional information about both dorsal–ventral and anterior–posterior axes. These positional signals include morphogens such as Sonic hedgehog (Shh) and members of the bone morphogenetic protein (BMP) family. How this positional information is interpreted by precursor cells and how it leads them to adopt specific cell fates is one of the major questions in developmental biology today.
This review documents some of what we presently know about glial cell specification, where they are specified, and the key extracellular signals and the intracellular signal transduction pathways that are involved.
2. Oligodendrocytes
(a) The origins of oligodendrocytes
Unlike neurons, which become post-mitotic soon after they are specified, oligodendrocytes are initially specified as proliferative, migratory precursor cells. These were first identified in cultures of rat optic nerve cells (Raff et al. 1983), at which time they were named oligodendrocyte-type-2 astrocyte progenitor cells (O-2A progenitors) to denote the fact that they can differentiate into either oligodendrocytes or astrocytes (type-2 subtype), depending on the culture medium. However, it has so far not proved possible to identify cells with the antigenic phenotype of type-2 astrocytes in vivo, so the precursor cells are now more usually referred to simply as oligodendrocyte precursor or progenitor cells (OLPs or OPCs (OLP used herein)). These go through several identifiable maturation stages characterized by distinct morphological and antigenic changes, before finally associating with axons and elaborating myelin. Since mature myelinating oligodendrocytes are ubiquitous throughout the mature CNS, they were originally presumed to be derived more-or-less equally from all parts of the embryonic germinal zones—the so-called ventricular zone (VZ) that surrounds the ventricles of the brain and the central canal of the spinal cord. However, with the discovery of early markers of the oligodendrocyte lineage, such as the platelet-derived growth factor receptor-alpha (Pdgfr-α; Pringle & Richardson 1993), the myelin proteolipid protein isoform DM20 (Timsit et al. 1995) and the O4 antigen (Noll & Miller 1993), came the realization that OLPs might be generated from defined, spatially localized subsets of NEPs.
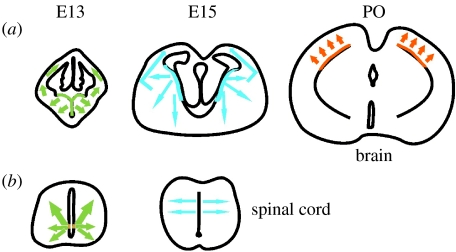
In the spinal cord, the first OLPs are generated from ventral NEPs (Warf et al. 1991; Noll & Miller 1993; Pringle & Richardson 1993; Yu et al. 1994; Timsit et al. 1995) from a specific domain of the VZ called pMN that also generates motor neurons (MNs; Richardson et al. 1997; Sun et al. 1998; Lu et al. 2002; Takebayashi et al. 2002; Zhou & Anderson 2002). The proliferating OLPs migrate away and rapidly populate the entire spinal cord before differentiating into myelin-forming oligodendrocytes. Recent studies using mouse mutants that lack this ventral source of OLPs showed that dorsal NEPs also generate some OLPs (Cai et al. 2005; Vallstedt et al. 2005). This was confirmed by Cre-lox fate mapping in wild-type mice (Fogarty et al. 2005). The dorsal half of the spinal cord neuroepithelium contributes approximately 20% of all oligodendrocytes in the cervical spinal cord and up to 50% of the oligodendrocytes in the dorsal funiculus (dorsal axon tracts; Fogarty (2005); for a review see Richardson et al. (2006); figure 1). In the mouse, the dorsally derived OLPs start to appear around embryonic day 15, approximately 2 days later than their ventrally derived counterparts. When they first appear, they have a distinctive radial morphology and co-label with the radial glial marker RC2 (Fogarty et al. 2005), verifying previous suggestions that oligodendrocytes as well as astrocytes might form by direct transformation of radial glia (Choi et al. 1983; Hirano & Goldman 1988; Voigt 1989; Culican et al. 1990; Barry & McDermott 2005).
Figure 1.
Developmental sites of origin of OLPs. The diagrams depict the sites of origin and migration pathways of OLPs during embryonic and perinatal development. (a) Drawings of coronal sections through the developing telencephalon at the indicated ages. Colour marks the sites of origin of OLPs in the VZ and arrows show their presumed migration pathways. (b) Transverse sections of the cervical spinal cord showing the early ventral and later dorsal sources along with their dispersal routes. Note: The brain sections are drawn to scale relative to each other, as are the spinal cord sections, but the spinal cord is drawn disproportionately large compared with the brain. The ventral sources of OLPs are induced by Shh from the floor plate of the spinal cord or the VZ of the ventral forebrain. The dorsal sources (including the cerebral cortex of the brain) are thought to be Shh independent, possibly relying on fibroblast growth factor. Dorsal OLP generation might also be helped by BMP downregulation in dorsal structures during later embryonic development. See text for a more complete description. E13, embryonic day 13; E15, embryonic day 15; PO, postnatal day 0.
A similar theme is observed at more anterior levels of the neuraxis. In the forebrain, the first Pdgfr-α-positive OLPs appear in the ventral neuroepithelium (anterior entopeduncular area, AEP; medial ganglionic eminence, MGE), then spread throughout the forebrain by migration and proliferation (Spassky et al. 1998; Olivier et al. 2001; Spassky et al. 2001; Tekki-Kessaris et al. 2001). Subsequent waves of OLP generation emanate from more dorsal regions (the lateral and the caudal ganglionic eminences (LGE/CGE)) and finally from within the cortex (Kessaris et al. 2006). Thus, there is a progressive wave of OLP production from ventral to dorsal regions (figure 1). The very first OLPs generated in the ventral forebrain (MGE/AEP) almost disappear during post-natal life and are replaced by other populations including the LGE/CGE-derived population. This might reflect some selective advantage of more recently generated OLPs that leads to competitive elimination of ventrally derived cells. Alternatively, continual turnover (loss and replacement) of oligodendrocytes during adult life might lead to gradual loss of the MGE/AEP-derived population because stem cells in the adult subventricular zone (SVZ)—the ultimate source of new oligodendrocytes in the adult (Levison & Goldman 1993, 1997; Luskin & McDermott 1994)—are derived from the embryonic LGE/CGE and cortex with little contribution from the MGE/AEP (Young et al. submitted).
(b) Oligodendrocyte specification: cell-extrinsic signals
(i) Induction of oligodendrogenesis
In the spinal cord, Shh is secreted from the notochord and subsequently from the floor plate at the ventral midline of the neural tube. Diffusion of Shh from these regions generates a morphogenetic gradient and, together with complementary signals from the roof plate, initiates a programme of NEP cell fate specification. This involves activation or repression of a set of homeobox, paired-box (Pax) and basic helix–loop–helix (bHLH) transcription factor genes, which have been classified as either class I or class II depending on whether they are activated (class II) or repressed (class I) by Shh (Briscoe et al. 2000). The combinatorial expression of these genes defines progenitor domains along the dorsal–ventral axis and influences the neuronal subtypes they generate. Shh is thus essential for the generation of different classes of ventral spinal cord neurons. Several studies have shown that Shh also promotes the generation of pMN-derived OLPs in the spinal cord (Poncet et al. 1996; Pringle et al. 1996; Orentas et al. 1999) and is a potent inducer of OLP formation in primary cultures of dissociated forebrain NEPs (Tekki-Kessaris et al. 2001; Kessaris et al. 2004). In the absence of Shh signalling, ventrally derived OLPs fail to form in the spinal cord or forebrain (Orentas et al. 1999; Nery et al. 2001; Tekki-Kessaris et al. 2001). Furthermore, transplanting Shh-expressing tissue adjacent to the dorsal neural tube in vivo (Orentas & Miller 1996; Poncet et al. 1996; Pringle et al. 1996), or adding Shh to spinal cord explants in vitro (Agius et al. 2004), induces the formation of ectopic OLPs, highlighting the importance of spatially restricted Shh signalling in directing OLP generation.
The mechanism by which Shh acts in vertebrates is still unclear (Ingham & McMahon 2001). Shh binds to its transmembrane receptor Patched (Ptc), causing disinhibition of its co-receptor Smoothened (Smo), a seven-pass transmembrane G-protein-coupled receptor (GPCR). In Drosophila, this eventually promotes nuclear translocation of the full-length form of the transcription factor Cubitus interruptus (Ci) which acts as a transcriptional activator. In the absence of Shh, Ci is proteolytically cleaved to a repressor form of the protein. The mammalian homologues of Ci are the zinc finger proteins of the Gli family (Gli1–Gli3). Owing to the existence of the three closely related members, it has been difficult to decipher their individual contributions to Shh signalling (Jacob & Briscoe 2003). Mouse Gli1 is not proteolytically cleaved and acts as a transcriptional activator with redundant functions during development. Although Gli2 possesses both activator and repressor activities, it is the activator properties that are required for ventral neural tube patterning. Gli3, on the other hand, is thought to act as an inhibitor of Shh signalling. Indeed, several aspects of Shh signalling are restored in mouse mutants lacking both Shh and Gli3 (Litingtung & Chiang 2000), indicative of Gli3 having a repressor function. The current model for Shh patterning in the neural tube is that Shh signalling leads to the formation of a ventral-to-dorsal (high–low) gradient of Gli activities with activator functions predominating ventrally and repressor functions dorsally (Jacob & Briscoe 2003). The requirements for Gli activity within the pMN domain are still unclear, as all three mouse Gli mutants lack defects in MN differentiation. However, mice lacking Gli2 function exhibit delayed and reduced oligodendrogenesis from the pMN domain (Qi et al. 2003). The proliferation of OLPs is unaffected in the Gli2 null mice and normal oligodendrocyte numbers are restored before birth, indicating a specific function for Gli2 at the early stages of oligodendrocyte generation (Qi et al. 2003).
NEPs from Shh or Smo null mice, the latter lacking all hedgehog (Hh) signalling, can still generate OLPs in culture (Chandran et al. 2003; Cai et al. 2005). This clearly demonstrates the existence of a Shh-independent pathway to oligodendrocyte specification. In vitro evidence suggests that FGF2 might be an alternative inducer of OLPs (Chandran et al. 2003; Gabay et al. 2003; Kessaris et al. 2004). NEPs from the cortex and the dorsal spinal cord can generate OLPs if cultured in the presence of fibroblast growth factor-2 (FGF2) and cyclopamine, a potent inhibitor of Hh signalling (Chandran et al. 2003; Gabay et al. 2003; Kessaris et al. 2004). It is therefore possible that FGF might be responsible for inducing oligodendrocytes in the dorsal spinal cord and/or forebrain, which are unlikely to be within range of Shh from the floor plate (spinal cord) or MGE (forebrain). However, conditional deletion of FGFR1 in the cortical neuroepithelium did not affect the capacity of these cells to generate oligodendrocytes in vivo (our own unpublished observations). Whether this indicates that FGF is not involved in oligodendrocyte generation from this region or else reflects redundancy among members of the FGF receptor family remains to be determined.
In addition to Shh and FGF2, insulin-like growth factor I (IGF-I) has also been suggested to influence oligodendrocyte specification (Hsieh et al. 2004). Previous evidence had shown that multipotent adult neural progenitors can be induced to generate oligodendrocytes in vitro when cultured in the presence of IGF-I, IGF-II or insulin. This effect is independent of the activity of IGFs on oligodendrocyte maturation or survival. The oligodendrogenic activity of IGF-I is thought to be mediated at least in part by inhibition of BMP signalling (see §2b(ii)). Whether insulin/IGF plays an important role in the oligodendrocyte specification in vivo is not yet known, although several studies have demonstrated the importance of this family of growth factors in oligodendrocyte maturation (Carson et al. 1993; Beck et al. 1995; Ye et al. 1995, 2002)
(ii) Inhibition of oligodendrogenesis
BMPs are members of an extended family of transforming growth factor-beta (TGF-β)-related proteins that also includes activins, dorsalin-1 (DSL1), growth/differentiation factors (GDFs) and glial cell-derived neurotrophic factor (GDNF). Several of these proteins (BMPs, GDFs and DSL1) are expressed in the dorsal part of the neural tube including the roof plate, where they are required for specification of subclasses of dorsal spinal interneurons (Liem et al. 1997; Lee et al. 1998). Members of the BMP family have been proposed to act as negative regulators of oligodendrocyte specification. Forebrain or spinal cord neural stem cells cultured in the presence of BMPs fail to develop oligodendrocytes, even in the presence of Shh or FGF2 (Gross et al. 1996; Mekki-Dauriac et al. 2002; Yung et al. 2002; Vallstedt et al. 2005). This raises the question of whether BMPs also regulate oligodendrocyte development in vivo. Ectopic expression of BMPs in chick ventral spinal cord inhibits OLP development (Mekki-Dauriac et al. 2002); conversely, inhibition of BMPs with the BMP antagonist Noggin causes an expansion of the oligodendrogenic domain. The same effect is also caused by surgical removal of the roof plate (Mekki-Dauriac et al. 2002). In all these experiments, effects on OLP and astrocyte production are reciprocal (see §3). Collectively, the data suggest that endogenous BMP signals spatially restrict neurogenesis and oligodendrogenesis to defined neuroepithelial regions (Hall & Miller 2004).
There is evidence that, in addition, BMPs also inhibit differentiation of OLPs by inducing expression of the inhibitors of differentiation (Id) genes (Samanta & Kessler 2004). The encoded Id proteins are bHLH proteins related to Olig1/2 (see §2c) but lacking a DNA-binding domain; they act as dominant-negative inhibitors of Olig1/2 and other bHLH proteins by binding to them and preventing their interaction with target genes. BMP induces cultured mouse NPCs to express all four Id proteins (Id1–4), but it is mainly Id4 (and to a lesser extent Id2) that inhibits OLP differentiation and expression of maturation markers of the oligodendrocyte lineage such as MBP and CNPase. This inhibition effect is mediated by binding and sequestering not only the Olig proteins, but also their required bHLH cofactors E12 and E47 (Samanta & Kessler 2004). In these experiments the Olig/Id complexes were retained in the cytoplasm and excluded from the cell nucleus.
Members of the Wnt family of proteins are also secreted from the roof plate and might influence gliogenesis in the dorsal neural tube. Unlike BMPs, Wnts are not involved in patterning the dorsoventral axis. They are known to be mitogenic for NPCs (Megason & McMahon 2002), although Wnt-3A was found not to promote proliferation of OLPs in vitro (Shimizu et al. 2005). Recent spinal cord explant studies indicate that Wnts do not influence OLP specification per se, but regulate the differentiation of OLPs to more mature oligodendrocytes (Shimizu et al. 2005).
(iii) Delta–Notch signalling in oligodendrocyte development
Activation of the Notch receptor through its ligands Delta and Serrate is one of the key mechanisms controlling cell fate decisions in the embryo. In the CNS, neurons are generated before glia and active Notch signalling is essential to maintain a population of proliferative NEPs into the gliogenic period. Accordingly, zebrafish embryos that have deficient Notch signalling have excess neurons in the spinal cord at the expense of OLPs (Appel et al. 2001; Park & Appel 2003). Conversely, expression of a constitutively active form of the Notch receptor resulted in excess OLPs at the expense of MNs (Park & Appel 2003). This led to the suggestion that Notch signalling is essential at the early stages of specification of the oligodendrocyte lineage. Notch plays another role later in the oligodendrocyte lineage; cell culture studies showed that constitutive activation of Notch signalling can inhibit oligodendrocyte maturation in vitro (Wang et al. 1998). Subsequently, it was found that loss of Notch signalling leads to premature differentiation of OLPs to oligodendrocytes in vivo (Genoud et al. 2002; Givorgi et al. 2002), while expression of an active form of the Notch receptor in transgenic zebrafish embryos prevented OLP differentiation (Park & Appel 2003). Collectively, these data indicate that Notch signalling might regulate both the specification of OLPs and their subsequent developmental maturation to oligodendrocytes. Remyelination studies in rodents, however, have shown that the expression of Notch1 in adult OLPs and its ligand Jagged in surrounding cells has no effect on the rate of remyelination, suggesting that distinct mechanisms of OLP maturation may operate during development and in the adult (Stidworthy et al. 2004).
(c) Oligodendrocyte specification: cell-intrinsic determinants
(i) bHLH genes
Olig1 and Olig2 were originally identified as two bHLH transcription factors that mark the oligodendrocyte lineage from the very early stages (Lu et al. 2000; Takebayashi et al. 2000; Zhou et al. 2000). In the spinal cord, Olig1 and Olig2 transcripts can be detected from embryonic day 9 onwards. Their expression is induced by Shh signalling. Transcripts are initially observed in the ventral third of the spinal cord but soon become restricted to the pMN domain. Olig2 expression persists in pMN throughout MN development; it is rapidly extinguished in post-mitotic MNs but remains on in OLPs. Expression of Olig1 is more dynamic at early times prior to the appearance of OLPs. Both the Olig genes are expressed throughout the oligodendrocyte lineage including in mature oligodendrocytes.
Functional studies in mice have demonstrated the significance of Olig2 in both MN and oligodendrocyte specification (Lu et al. 2002; Takebayashi et al. 2002; Zhou & Anderson 2002). In the absence of functional Olig2 protein, all spinal MNs and oligodendrocytes fail to develop. Embryos lacking Olig2 die at birth or shortly after due to the absence of motor function. At more anterior levels of the neuraxis, small pockets of oligodendrocytes develop, especially within the fore- and hindbrain, suggesting redundancy between Olig1 and Olig2 (Lu et al. 2002). Embryos lacking the function of both the Olig genes show complete absence of all oligodendrocytes throughout the CNS (Zhou & Anderson 2002).
Olig2 is thought to have multiple roles in the precursors of MNs. Initially, Olig2 promotes pMN identity by repression of alternative fates (Mizuguchi et al. 2001; Novitch et al. 2001). For example, Olig2 represses expression of Irx3, a transcription factor that promotes V2 interneuron identity in the ventral spinal cord. Ventral expansion of the p2 domain and the V2 interneuron population can thus be observed in embryos lacking Olig2 function. Olig2 also acts to maintain the MN precursor pool by repressing the expression of MN differentiation factors within the neuroepithelium. By binding directly to E-box elements within the gene encoding MN determinant HB9, Olig2 can repress its expression and inhibit premature differentiation of MNs (Lee et al. 2005). In addition, Olig2 is thought to promote the expression of Neurogenin-2 (Ngn2), a pro-neural gene that acts to promote neurogenesis (Lee et al. 2005). Olig2 and Ngn2 compete for binding to E-box elements as well as to other bHLH partners, creating a negative feedback loop that controls the differentiation of MNs and the maintenance of the pMN pool for subsequent generation of oligodendrocytes (Lee et al. 2005). Although Olig2 is expressed in MN precursors, its expression is downregulated in differentiating MNs. Unlike pro-neural proteins and other bHLH transcription factors, which mostly act as transcriptional activators, Olig2 appears to function as a transcriptional repressor (Bertrand et al. 2002). Direct targets of Olig2 presumably include MN determinants (Lee et al. 2005) and astrocyte specification factors (see §3), although such targets are as yet poorly characterized.
Unlike Olig2, which seems to have essential roles in oligodendrocyte development, initial evidence indicated a redundant, non-essential function for Olig1. Mice lacking Olig1 showed a slight delay in oligodendrocyte maturation but were otherwise normal. By contrast, Olig2 mutant mice lacked all oligodendrocytes and MNs in the spinal cord (Lu et al. 2002), although oligodendrogenesis in the fore- and hindbrain was apparently normal. However, more recent work with the Olig1 mutant mice questions the previous data and suggests that Olig1 is in fact a central regulator of oligodendrocyte maturation and myelinogenesis (Xin et al. 2005). These authors found that, in the absence of Olig1, the mice developed severe neurological deficits and died within the third post-natal week. While oligodendrocyte progenitors developed normally, maturation was severely affected and markers of myelinating oligodendrocytes were absent. Although the reason for the discrepancies in different loss-of-function studies is not clear, all data support a functional role for Olig1 in oligodendrocyte maturation. This is also consistent with the recent striking finding that remyelination does not occur in Olig1 null adult mice with experimentally induced focal demyelination (Arnett et al. 2004).
Another bHLH gene that has recently been found to play a role in oligodendrocyte development is the pro-neural gene Mash1 (Parras et al. 2004). In the early telencephalon, Mash1 is co-expressed with Olig2 at the time of oligodendrocyte specification in the MGE/AEP and its expression is briefly maintained in initial populations of migrating OLPs (Parras et al. 2005). At later stages, Mash1-negative OLPs populate the telencephalon. In newborn mice, expression of Mash1 can be observed in a large fraction of OLPs, and Mash1-deficient pups have severely reduced numbers of oligodendrocytes in the olfactory bulb (Parras et al. 2004). Although the data so far indicate that there are both Mash1-dependent and Mash1-independent OLPs in the telencephalon, the precise function of Mash1 in the oligodendrocyte lineage is still unknown.
(ii) Homeobox genes
Nkx6.1 and Nkx6.2 (also known as Gtx1) are two homeodomain transcription factors that are expressed in the ventral spinal cord under the inductive influence of Shh. Expression is restricted to the ventral neuroepithelium where they are thought to have several patterning roles. Within the pMN domains, Nkx6.1 and Nkx6.2 appear to regulate the expression of Olig2 in a dose-dependent manner (Vallstedt et al. 2001, 2005). Loss-of-function studies in mouse have suggested a redundant or non-essential function for Nkx6.2 in the spinal cord, whereas, in the absence of Nkx6.1, expression of Olig2 is delayed and greatly diminished and the generation of MNs (and more dorsally located interneurons) is severely impaired (Sander et al. 2000; Liu et al. 2003). In compound Nkx6.1/Nkx6.2 mutants, all the MNs and oligodendrocytes generated from the pMN domain are eliminated demonstrating an essential role for these two genes in oligodendrogenesis from this region (Cai et al. 2005; Vallstedt et al. 2005). Interestingly, oligodendrocyte development from dorsal precursors is unaffected, perhaps indicating a significant difference in oligodendrocyte specification from dorsal and ventral precursors (Cai et al. 2005; Vallstedt et al. 2005). Whether this difference simply reflects alternative patterning requirements within different neuroepithelial regions or a specific function for Nkx6 proteins in ventral oligodendrogenesis is not known. In addition to its patterning role, Nkx6.2 is expressed in myelinating oligodendrocytes, where it appears to be involved in regulating axon–glial interactions at myelin paranodes (Southwood et al. 2004).
Another homeobox transcription factor that is involved in oligodendrogenesis in the neural tube is Nkx2.2. The expression of Nkx2.2 is induced by Shh at the time of neural plate induction and is restricted to a small region of the VZ adjacent to the floor plate (the p3 domain). Until the end of neurogenesis, the expression domains of Olig2 and Nkx2.2 are non-overlapping. This mutually exclusive relationship is established by cross-repressive interactions between the two genes (Mizuguchi et al. 2001; Novitch et al. 2001; Sun et al. 2001, 2003; Zhou et al. 2001). With the onset of oligodendrogenesis, expression of Nkx2.2 expands dorsally into the pMN domain, suggesting a switch from a cross-repressive to a cooperative relationship between Nkx2.2 and Olig2. However, despite this suggestive expression pattern, it is now clear that Nkx2.2 does not play an important role in oligodendrocyte specification per se, although it is important during the maturation of oligodendrocytes from OLPs (Qi et al. 2001; for further discussion see Richardson et al. 2006).
(iii) Sox genes
The Sox family of transcription factors has a prominent role in oligodendrocyte development. There are at least 20 mammalian Sox proteins, three of which have so far been implicated in oligodendrocyte generation. Sox9, Sox8 and Sox10 are expressed at different stages of oligodendrocyte development. Sox9 is expressed in the entire neuroepithelium of the spinal cord at embryonic day 10.5 (Stolt et al. 2003). At later stages, it is expressed in proliferating oligodendrocyte progenitors as well as in astrocytes, where its expression persists throughout life (Stolt et al. 2003). The expression of Sox8 slightly precedes that of Sox10, and both are restricted to the oligodendrocyte lineage from the early stages of their development until maturation (Stolt et al. 2002, 2004, 2005). Mice lacking Sox9 have reduced numbers of oligodendrocytes (and astrocytes) indicating a requirement for this factor in glial specification (Stolt et al. 2003). Compound Sox8/Sox9 deficient mice exhibit complete loss of oligodendrocytes suggesting a redundancy between the two genes (Stolt et al. 2005). The loss of Sox10, on the other hand, results in normal numbers of oligodendrocyte progenitors but a failure to mature, indicating that Sox10 is required for terminal differentiation (Stolt et al. 2002).
(d) Oligodendrocyte diversity?
Oligodendrocyte heterogeneity has been reported on the basis of morphology and the observation that mature cells myelinate either a single large diameter axon or several small axons (Bunge 1968; Bjartmar et al. 1994; Butt et al. 1997). It is not clear whether this variation reflects intrinsic differences among oligodendrocyte subtypes or phenotypic plasticity. Heterogeneity has also been proposed on the basis of expression of markers such as Pdgfra or PLP/DM20 (Le Bras et al. 2005). The possibility that there might be different functional subclasses of oligodendrocyte becomes more intriguing owing to the discovery of dorsally derived as well as ventrally derived OLPs and the likelihood that these are specified by different signals (Chandran et al. 2003; Kessaris et al. 2004). Therefore, a major question in the field is whether different signalling pathways generate different oligodendrocyte subtypes. An attempt was made to address this question by ablating oligodendrocyte subpopulations in the forebrain, based on their region of origin in the VZ (Kessaris et al. 2006). This did not reveal any obvious phenotypic consequences that could point to different functional subclasses of oligodendrocytes. This tentatively suggests that the described morphological and/or molecular differences among oligodendrocytes might reflect phenotypic plasticity (Kessaris et al. 2006). Nevertheless, this question is still far from settled.
3. Astrocytes
(a) Astrocytes: one cell type or several?
Astrocytes were originally described more than 100 years ago as star-like cells in Golgi tissue preparations. Two morphological variants were later described: ‘fibrous’ astrocytes in white matter tracts and ‘protoplasmic’ astrocytes in grey matter. It is not clear whether the different morphology of fibrous versus protoplasmic astrocytes simply reflects their different physical environments—fibrous astrocytes in white matter being constrained to align with axons, for example—or whether they are intrinsically different cells that are specified independently, follow different developmental pathways and perform different functions in the mature CNS. Since morphology is an unreliable way of classifying cells, astrocytes later came to be defined by their high content of cytoplasmic intermediate filaments (Vaughn & Peters 1967; Choi & Lapham 1978), a major component of which is the glial fibrillary acidic protein (GFAP). GFAP immuno-reactivity subsequently became their defining mark (Bignami & Dahl 1974, 1977; Antanitus et al. 1975). However, defining a cell population on the basis of one protein is unsatisfactory as it could mislead us into lumping different types of cells together as one. Moreover, there are cells in grey matter with a protoplasmic astrocyte-like morphology that do not normally express GFAP, but can upregulate the protein after local CNS injury or disease (Gabbott & Stewart 1987; Stichel et al. 1991; Walz & Lang 1998; Scotti 2003). These cells have been called ‘GFAP-negative protoplasmic astrocytes’ (Walz 2000). To better characterize astrocyte-like glial cells, two approaches have been used: (i) there has been a search for alternative molecular markers that can either unify or subdivide the astrocyte population and (ii) there have been efforts to define glia on the basis of their electrophysiological properties in vitro and in situ.
Molecular markers that are useful for defining astrocyte-like glia are still few in number. They include the calcium-binding protein S100β (Ludwin et al. 1976), the enzyme glutamine synthetase (GS; Norenberg & Martinez-Hernandez 1979), the glutamate transporter GLAST (Schmitt et al. 1997), fibroblast growth factor receptor-3 (Fgfr3; Pringle et al. 2003), the water channel protein aquaporin-4 (Aqp4; Vitellaro-Zuccarello et al. 2005) and others. Not all these are completely specific (e.g. Cammer 1990; Hachem et al. 2005) and it is still far from clear whether they can define astrocytes as a single population of cells or several subgroups. However, there are indications that astrocytes might be heterogeneous. For example, in the post-natal rat spinal cord, there are regionally restricted populations of GFAP-positive white matter astrocytes that either do or do not co-express GLAST (Barry & McDermott 2005).
On top of this, there are regionally specialized types of GFAP-positive glial cells such as Bergmann glia in the cerebellum and Műller glia in the retina. Moreover, it has recently become clear that in some animal species, GFAP is also expressed on radial glial cells in the VZ of the embryonic neural tube and in ‘sub-ependymal astrocytes’ in the adult (Dahl et al. 1981; Levitt et al. 1981; Walz & Lang 1998; Malatesta et al. 2000; Andrae et al. 2001; Barry & McDermott 2005). Radial glia and sub-ependymal astrocytes are now thought to correspond to multipotent NPC (stem) cells in the embryo and adult, respectively; they can generate neurons and oligodendrocytes as well as differentiated astrocytes in vivo (Doetsch et al. 1999; Hartfuss et al. 2001; Götz et al. 2002; Kriegstein & Gotz 2003; Anthony et al. 2004; Merkle et al. 2004). Whether all these cell types should be regarded as specialized subtypes of astrocytes based purely on their expression of GFAP is debatable.
Ultimately what is needed are functional definitions of astrocytes, but we are still some way from that. For example, electrophysiological studies of astrocytes are confusing and inconclusive. Early work on adult optic nerve astrocytes led to the conclusion that all astrocytes are electrically ‘passive’, lacking voltage-gated currents and displaying a linear voltage–current relationship (Kuffler et al. 1966). Subsequent studies on astrocytes from younger animals revealed more complex electrophysiology, including inward and outward K+ currents and an inward Na+ current (for review see Barres 1991). This has led to the notion that there are two astrocyte subtypes: passive and complex. However, different laboratories disagree as to the relative frequencies of the passive and complex forms, whether and how their ratio changes with CNS maturation, whether the two phenotypes can be assigned to GFAP-positive and GFAP-negative molecular subgroups and so on. Some of the controversies might be attributable to different methodologies, regional variation, the criteria according to which cells were selected for recording (e.g. on the basis of morphology) and the bias that this might introduce (Steinhauser et al. 1992; Matthias et al. 2003; for review see Walz 2000). It is also possible that the passive and complex subtypes can interconvert rapidly and this might be influenced by the way cells or tissue slices are treated prior to and during recording. An additional complication results from the fact that astrocytes are extensively coupled via gap junctions in vivo, which tends to linearize current–voltage relationships (passive phenotype). A recent study concluded that the complex pattern is found only during early post-natal development and that most or all astrocytes in the mature hippocampus are of the passive variety (Zhou et al. 2006). This suggests that complex astrocytes might be glial precursors, and passive astrocytes their differentiated progeny.
It should be clear from the above discussion that, when seeking to define the mechanisms of astrocyte specification and subsequent development, we are on uncertain ground. As long as we remain unsure whether astrocytes comprise one group of phenotypically plastic cells or several intrinsically distinct subgroups, it will be difficult to know whether different laboratories are studying the same or different developmental pathways. In practice, most workers rely on induction of GFAP expression in vivo or in neural cell cultures as a surrogate for astrocyte specification. This raises additional questions about whether we are all studying normal developmental mechanisms (astrogenesis) or the acute activation of GFAP expression that accompanies CNS injury or disease (astrocytosis).
(b) The embryonic origins of astrocytes
Astrocytes, like oligodendrocytes and neurons, are ultimately formed from the NEPs that line the lumen of the embryonic neural tube. However, there is a persuasive body of evidence that astrocytes—at least fibrous astrocytes—are formed indirectly from NEPs via radial glial cells. The evidence supporting this idea was largely circumstantial. First, immunohistochemical studies have identified cells with antigenic and morphological phenotypes intermediate between radial glia and astrocytes (Choi et al. 1983; Hirano & Goldman 1988; Voigt 1989; Barry & McDermott 2005). Second, radial glial cells begin to disappear during development at about the same time as astrocytes appear (Choi & Lapham 1978; Schmechel & Rakic 1979; Misson et al. 1988) and the cells interpreted as intermediate forms are also observed during this period. Recently, more direct evidence has come from genetic fate mapping experiments. When introduced into transgenic mice, a short promoter fragment from the human GFAP gene drives reporter gene expression not only in differentiated astrocytes, but also earlier in radial glia. When these radial glia were purified from transgenic brains and cultured in vitro, they gave rise to astrocytes as well as neurons, providing the first indication that radial glia can be pluripotent NPCs (Malatesta et al. 2000). It has now been confirmed that radial glia are also pluripotent precursors in vivo (Noctor et al. 2002; Malatesta et al. 2003; Anthony et al. 2004; Fogarty et al. 2005).
It is not known whether all astrocytes are generated directly from radial glia. It is also not known whether all radial glia generate astrocytes, or whether they generate different subtypes of astrocytes from different parts of the VZ. In the embryonic spinal cord, oligodendrocytes and astrocytes are generated preferentially from different parts of the VZ, with most oligodendrocytes being derived from the pMN domain (Sun et al. 1998; Lu et al. 2002; Takebayashi et al. 2002; Zhou & Anderson 2002). Other progenitor domains generate predominantly astrocytes (Pringle et al. 1998; Lu et al. 2002; Pringle et al. 2003; Fogarty et al. 2005). By Cre-lox fate mapping in mice, we showed that the Dbx1-expressing domain (p1, p0, dP6 and dP5—four progenitor domains centred on the dorsoventral midline) generates significant numbers of protoplasmic astrocytes, the majority of which lie in grey matter and do not express detectable GFAP immuno-reactivity. The Dbx1 domain also generates fibrous astrocytes in the white matter and a small number of oligodendrocytes, in addition to neurons (Fogarty et al. 2005). Furthermore, the Nkx2.2-expressing domain (p3, the ventral-most domain abutting the floor plate) generates protoplasmic and fibrous astrocytes in addition to neurons and small numbers of oligodendrocytes (R.Taveira-Marques, N. Kessaris & W. D. Richardson 2006, unpublished observations). The protoplasmic astrocytes that are generated from Dbx1 and Nkx2.2 domains are morphologically similar, but occupy different territories in the adult spinal cord. It remains to be determined whether they are functionally or physiologically distinguishable.
(c) Astrocyte specification
As discussed previously, oligodendrocyte specification in the ventral spinal cord and telencephalon relies on the induction of Olig gene expression by Shh. A hedgehog-independent, FGF-dependent mechanism of Olig gene induction and oligodendrocyte specification has also been identified. What are the extracellular signals and cell-intrinsic factors that specify astrocytes? Two principal signalling systems have been identified: (i) the BMPs and (ii) cytokines such as ciliary neurotrophic factor (CNTF), leukaemia inhibitory factor (LIF) and interleukin-6 (IL-6). These two signalling pathways can act independently of one another but also synergize in some circumstances.
(i) Mutually antagonistic effects of BMPs and Shh on astrocyte and oligodendrocyte specification
Several BMPs (BMP2, 4, 5 and 7) have been shown to induce multipotent NPCs from the perinatal mouse forebrain SVZ to differentiate as GFAP-positive astrocytes in culture (Gross et al. 1996; Mehler et al. 2000). In these experiments, GFAP upregulation was accompanied by rapid and irreversible changes in cell shape and size as well as inhibition of cell division, suggesting that BMPs induced astrocyte specification rather than proliferation of pre-existing astrocyte precursors. Concomitantly, oligodendrocyte production was strongly inhibited (Gross et al. 1996).
The inducing activity of Shh on oligodendrocytes has been clearly demonstrated and the inhibitory activity of BMPs on oligodendrocyte specification has been confirmed both in explant cultures of embryonic chick spinal cords (Wada et al. 2000; Mekki-Dauriac et al. 2002) and in vivo (Mekki-Dauriac et al. 2002; see §2). In complementary experiments, Agius et al. (2004) showed that Shh not only induces OLP production, but also simultaneously inhibits astrocyte generation: when Shh is added to cultured ‘open book’ chick spinal cord explants, Olig2 expression and OLP production expand dorsally from pMN into p2 and beyond, and astrocyte production is shut down, judging by the loss of Fgfr3 and GLAST expression. Blocking Shh signalling with cyclopamine had the opposite effect: Olig2 expression was obliterated and astrocyte production expanded ventrally from p2 through pMN to the floor plate. These experiments clearly demonstrate that BMPs and Shh are mutually antagonistic as far as gliogenesis is concerned, and suggest that the normal pattern of astrocyte and OLP production along the dorsal–ventral axis is controlled by a fine balance between Shh, BMPs and BMP inhibitors from both dorsal and ventral sources (Mekki-Dauriac et al. 2002; Vallstedt et al. 2005). It has also been suggested that the later generation of OLPs from precursors in the dorsal spinal cord might be triggered by temporal downregulation of dorsally derived BMPs (Vallstedt et al. 2005). This interpretation implies that precursor cells at all dorsal–ventral levels might be inherently capable of generating either astrocytes or OLPs (or both) once neuron production is over, but can be biased towards one or the other cell type by locally prevailing signals. This would be compatible with a lineage model whereby all NEPs generate neurons followed by glial-restricted precursors (GRPs or glioblasts; Rao et al. 1998; Noble et al. 2004), which could then respond to positional signals to generate predominantly astrocytes or oligodendrocytes.
(ii) The Jak–Stat pathway
When OLPs, derived from perinatal rat optic nerve or other parts of the CNS, are cultured in defined medium, they differentiate into oligodendrocytes. In contrast, in the presence of 10% foetal calf serum (FCS), they start to express GFAP and adopt a process-bearing morphology; these cells were called ‘type-2 astrocytes’ to distinguish them from flat, fibroblast-like ‘type-1 astrocytes’ that are abundant in cultures established from many parts of the CNS (Raff et al. 1983). The type-2 astrocyte-inducing activity in FCS can be mimicked by the cytokine CNTF, in combination with extracellular matrix (ECM). CNTF by itself induces reversible GFAP expression, ECM being required for full morphological transformation and stable GFAP expression (Lillien et al. 1990). Since type-2 astrocytes have not yet been clearly identified in vivo (Fulton et al. 1992), it is not clear whether OLPs are exposed to CNTF during normal development. However, it has subsequently been shown that CNTF and the related cytokines IL-6 and LIF can also induce NEPs, growing as neurospheres or dissociated cell cultures, to express GFAP (Johe et al. 1996; Bonni et al. 1997; Rajan & McKay 1998). The response of primary NEPs to cytokines depends on their developmental stage, mirroring the normal developmental progression of NEPs from neurogenesis to gliogenesis in vivo. Thus, cells taken from the neurogenic embryonic day 11 mouse cerebral cortex do not activate GFAP during short-term (2-day) culture with cytokines. However, embryonic day 11 cortical NEPs acquire the ability to express GFAP in response to cytokines after 5 days in culture (the equivalent of embryonic day 16; He et al. 2005).
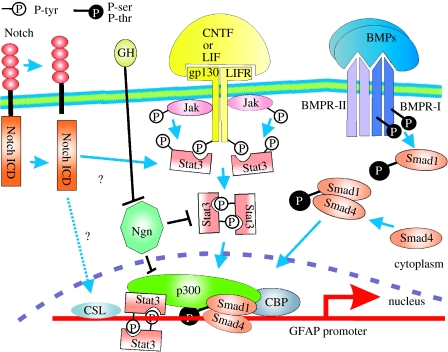
CNTF, IL-6 and LIF all bind to cell-surface receptors that share the same co-receptor gp130. Following ligand binding and dimerization, gp130 triggers activation of cytoplasmic tyrosine kinases of the Janus kinase (Jak) family. These in turn phosphorylate and activate Stat family transcription factors, Stat1 and Stat3 (for reviews see Darnell 1997; Levy & Darnell 2002). The activated Stat proteins dimerize, translocate to the nucleus and regulate transcription by binding to specific sites in the DNA. In addition, Stats can bind cofactors of the p300/CREB-binding protein (CBP) group (figure 2). These large (approx. 2500 residues) proteins bridge between the Stats (or other transcription factors) and RNA polymerase II, forming a complex that increases transcription, possibly by loosening chromatin structure in the vicinity of the gene promoter (Darnell 1997). This might occur because p300 has intrinsic histone acetylase activity and also can bind other acetylases.
Figure 2.
Signalling pathways involved in the induction of astrocyte specification/differentiation. Astrocyte differentiation is thought to be induced via cytokine and/or BMP signalling pathways. Cytokines (LIF or CNTF) induce dimerization of the LIF receptor (LIFR) with co-receptor gp130, leading to phosphorylation and activation of Janus kinases (Jaks). Receptor dimerization also creates docking sites for the transcription factor Stat3, which is tyrosine phosphorylated by Jaks. The phosphorylated Stats dimerize and move into the nucleus, recruiting p300 and binding to specific sequences in the GFAP promoter. BMP dimers induce the tetramerization of the BMPR-I and BMPR-II receptors, which are serine–threonine kinases. BMPR-II then phosphorylates and activates BMPR-I, which phosphorylates Smad1. Phosphorylated Smad1 associates with Smad4 (unknown stoichiometry) and the complex moves into the nucleus, recruiting p300 and CBP, which probably bind directly to the GFAP promoter and initiate transcription. Smads and Stats bind opposite ends of p300, which presumably allows synergistic integration of LIF/CNTF and BMP signalling pathways. These signalling pathways can be inhibited by neurogenins, which are thought to act by blocking Stat dimerization and also by sequestering p300. The activity of neurogenin can be inhibited, in an undefined manner, by growth hormone. Notch signalling (via the cleaved/released intracellular domain) might also interact with the Stat pathway. See text for a fuller description.
He et al. (2005) showed that the GFAP-inducing activity of LIF in NEP cultures was dependant on Stat1/3. The GFAP 1.9 kb promoter region contains a canonical-binding site for Stats and binding of phosphorylated Stat1 or Stat3 to this element was demonstrated in electrophoretic mobility shift assays (Bonni et al. 1997; He et al. 2005). A GFAP promoter-driven reporter gene was not LIF responsive in early (neurogenic) cortical progenitors, but became responsive to LIF after several days in culture, just like the endogenous GFAP gene. Stat1/3 activity was enriched in the micro-dissected cortical VZ/SVZ of late (P0), compared to early (embryonic day 12), embryos, supporting the idea that late activation of the Jak–Stat pathway contributes to gliogenesis in vivo. This is also supported by the fact that mice deficient in components of cytokine signalling (gp130, LIF and LIF receptor) have perturbed astrocyte development (Bugga et al. 1998; Koblar et al. 1998; Nakashima et al. 1999a). As endothelial cells are a demonstrated source of LIF in the CNS and can induce astrocyte differentiation in cultured rat optic nerve precursors (Mi et al. 2001), it is possible that blood vessels are also important for triggering astrocyte development.
Among the genes activated by Stat1/3 in late embryonic NEPs were several components of the Jak–Stat pathway itself, including gp130, Jak1, Stat1 and Stat3. Thus, Stat signalling triggers an autoregulatory loop that reinforces itself and presumably tends to consolidate and stabilize the astrocyte phenotype. This, taken together with previous work, suggests a model for how the neuron–astrocyte fate switch might be controlled. At early times, neurogenic bHLH transcription factors such as Ngn1 and Mash1 repress gliogenesis (Nieto et al. 2001; Sun et al. 2001) by inhibiting the activity of the Jak–Stat pathway. They achieve this by competing with Stat1/3 for binding to p300 and CBP that are present in limiting quantities in the cells and also by directly inhibiting Stat activation (Sun et al. 2001). In the absence of a robust positive feedback loop, the Jak–Stat pathway remains at a low basal level, insufficient for activating astrocyte-specific gene expression. As expression of Ngn1/2 diminishes during later embryonic development, repression of the Stat1/3 pathway is relieved and Stat-mediated transcription of GFAP and other astrocyte genes is activated. It remains to be determined what leads to Ngn downregulation. It has been shown that growth hormone (GH), which acts through the Jak–Stat pathway to activate the inhibitory Stat5, can downregulate Ngn1 expression in cultured neurospheres. GH and its receptor are expressed in the rodent brain from as early as embryonic day 10, peaking just before birth (Hojvat et al. 1982; Turnley et al. 2002); so it is conceivable that GH might play a role in the neuron–astrocyte switch (figure 2). However, this would not easily explain the fact that neuron–glial switching occurs approximately on schedule in primary NEPs in the absence of added GH (see above). Another alternative is that robust Jak–Stat signalling might itself inhibit Ngn expression.
(iii) The relationship between BMP and cytokine signalling
How can we reconcile the observations that both BMPs and LIF-like cytokines can induce astrocyte specification? Perhaps these two pathways induce different subtypes of astrocytes, or perhaps they operate independently of one another in different parts of the CNS, or perhaps they act together to reinforce one another.
The latter idea is supported by the recent demonstration that sub-effective doses of BMPs and cytokines can synergize with one another for astrocyte induction (Nakashima et al. 1999b). Once again, p300/CBP is central to the story. BMPs act through a hetero-tetrameric complex of type I and type II serine–threonine kinase cell-surface receptor subunits. Following BMP binding, the receptor phosphorylates and activates Smad proteins, which are transcriptional regulators. Nakashima et al. (1999b) showed that the GFAP promoter region contains separate binding sites for Stat3 and Smad1 and postulated that a direct or indirect physical interaction between the two factors might underlie their synergism. Knowing that Smads can associate with p300/CBP family members (e.g. Janknecht et al. 1998), they investigated the idea that p300 bridges Stat3 and Smad1. They discovered that indeed both factors bind non-competitively to p300, Stat3 mainly to the amino-terminal half and Smad1 to the carboxy-terminal region. This provides a simple and elegant explanation of how BMPs and cytokines might collaborate to induce astrocyte generation (figure 2).
(iv) The Notch signalling pathway
As mentioned previously, Notch signalling is integral to appropriate MN and oligodendrocyte generation. It has also been implicated in astrocyte induction (Tanigaki et al. 2001; Ge et al. 2002; Grandbarbe et al. 2003). In the canonical Notch signalling pathway, binding of a Notch ligand (e.g. Delta) to Notch at the cell surface leads to proteolytic cleavage and nuclear translocation of the released Notch intracellular domain, where it associates with the CSL DNA-binding protein and its partner SKIP. This converts CSL–SKIP from a transcriptional repressor to an activator of target genes, including the hes genes. A CSL-independent pathway also exists (for review see Louvi & Artavanis-Tsakonas 2006). A constitutively active Notch intracellular domain, transduced into cultured adult rat hippocampal progenitors, induced astrocyte production (GFAP expression) in a majority of the expressing cells (Tanigaki et al. 2001). In these experiments, the Notch-activated pathway was presumed to be independent of Jak–Stat signalling, because mutation of the Stat-binding element within the GFAP promoter did not impair its ability to be activated by Notch. However, it was later shown that in NEPs from mouse or rat embryonic cortex, a dominant-negative form of Stat3 blocked the ability of Notch to activate GFAP, implying that Notch signalling does interact with the Jak–Stat pathway (Ge et al. 2002). The form of the interaction remains unclear because the CSL-binding site in the GFAP promoter proved not to be necessary for Notch-mediated activation. Either a CSL-independent mechanism is responsible or a CSL-dependent mechanism might act indirectly on the GFAP promoter. In any case, Notch signalling seems not to be absolutely required for astrocyte development because astrocytes can still be generated by neural stem cells derived from the CSL null mutant embryo; however, their appearance is delayed relative to wild-type cells (Ge et al. 2002).
4. Conclusions
There has been a great deal of progress recently in identifying the molecular mechanisms of both oligodendrocyte and astrocyte specification. We know the major signalling pathways that specify the cells from their precursors: (i) Shh and possibly FGF for oligodendrocytes and (ii) BMPs and cytokines for astrocytes. We also have made significant headway in identifying the intracellular signal transduction pathways and transcription factors that elicit the earliest steps in astrocyte or oligodendrocyte differentiation. Inevitably, many questions remain. High on the list is the question of glial diversity: are there different ‘shades’ of oligodendrocytes and astrocytes and, if so, are these determined at the moment of specification, by subtle variations in the signalling environment of particular groups of precursor cells in the VZ, or later, by signals from the neurons and other glia with which they intermingle after leaving their birthplace? These are basic questions of anatomy and physiology that still have to be addressed. The discovery of oligodendrocyte lineage-specific transcription factors (Olig and Sox) has been a major boost not only for the insight they have given into the internal mechanism of cell specification, but also because they provide reliable histological markers for tracking the cells during development and regeneration, both in vitro and in vivo. Something similar for astrocytes is desperately needed and surely cannot be very far away. The discovery of the central importance of the Stat3 transcription factor for astrocyte gene transcription is exciting and will accelerate our understanding of general mechanisms of gene regulation and cell fate selection, revolving around the important role of the p300/CBP adaptor proteins in coordinating and integrating different signalling pathways including, and perhaps especially, epigenetic pathways.
Footnotes
One contribution of 14 to a Theme Issue ‘Stem cells and brain repair’.
References
- Agius E, Soukkarieh C, Danesin C, Kan P, Takebayashi H, Soula C, Cochard P. Converse control of oligodendrocyte and astrocyte lineage development by Sonic hedgehog in the chick spinal cord. Dev. Biol. 2004;270:308–321. doi: 10.1016/j.ydbio.2004.02.015. doi:10.1016/j.ydbio.2004.02.015 [DOI] [PubMed] [Google Scholar]
- Andrae J, Bongcam-Rudloff E, Hansson I, Lendahl U, Westermark B, Nistér M. A 1.8 kb GFAP-promoter fragment is active in specific regions of the embryonic CNS. Mech. Dev. 2001;107:181–185. doi: 10.1016/s0925-4773(01)00460-9. doi:10.1016/S0925-4773(01)00460-9 [DOI] [PubMed] [Google Scholar]
- Andriezen W.L. The neuroglia elements in the human brain. Brain J. Med. 1893;2:227–230. doi: 10.1136/bmj.2.1700.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antanitus D.S, Choi B.H, Lapham L.W. Immunofluorescence staining of astrocytes in vitro using antiserum to glial fibrillary acidic protein. Brain Res. 1975;89:363–367. doi: 10.1016/0006-8993(75)90729-5. doi:10.1016/0006-8993(75)90729-5 [DOI] [PubMed] [Google Scholar]
- Anthony T.E, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. doi:10.1016/S0896-6273(04)00140-0 [DOI] [PubMed] [Google Scholar]
- Appel B, Givan L.A, Eisen J.S. Delta-Notch signaling and lateral inhibition in zebrafish spinal cord development. BMC Dev. Biol. 2001;1:13. doi: 10.1186/1471-213X-1-13. doi:10.1186/1471-213X-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett H.A, et al. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. doi:10.1126/science.1103709 [DOI] [PubMed] [Google Scholar]
- Barres B.A. New roles for glia. J. Neurosci. 1991;11:3685–3694. doi: 10.1523/JNEUROSCI.11-12-03685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry D, McDermott K. Differentiation of radial glia from radial precursor cells and transformation into astrocytes in the developing rat spinal cord. Glia. 2005;50:187–197. doi: 10.1002/glia.20166. doi:10.1002/glia.20166 [DOI] [PubMed] [Google Scholar]
- Beck K.D, Powell-Braxton L, Widmer H.R, Valverde J, Hefti F. Igf1 gene disruption results in reduced brain size, CNS hypomyelination, and loss of hippocampal granule and striatal parvalbumin-containing neurons. Neuron. 1995;14:717–730. doi: 10.1016/0896-6273(95)90216-3. doi:10.1016/0896-6273(95)90216-3 [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro D.S, Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. doi:10.1038/nrn874 [DOI] [PubMed] [Google Scholar]
- Bignami A, Dahl D. Astrocyte-specific protein and neuroglial differentiation. An immunofluorescence study with antibodies to the glial fibrillary acidic protein. J. Comp. Neurol. 1974;153:27–38. doi: 10.1002/cne.901530104. doi:10.1002/cne.901530104 [DOI] [PubMed] [Google Scholar]
- Bignami A, Dahl D. Specificity of the glial fibrillary acidic protein for astroglia. J. Histochem. Cytochem. 1977;25:466–469. doi: 10.1177/25.6.69656. [DOI] [PubMed] [Google Scholar]
- Bjartmar C, Hildebrand C, Loinder K. Morphological heterogeneity of rat oligodendrocytes: electron microscopic studies on serial sections. Glia. 1994;11:235–244. doi: 10.1002/glia.440110304. doi:10.1002/glia.440110304 [DOI] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank D.A, Rozovsky I, Stahl N, Yancopoulos G.D, Greenberg M.E. Regulation of gliogenesis in the central nervous system by the JAK–STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. doi:10.1126/science.278.5337.477 [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell T.M, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. doi:10.1016/S0092-8674(00)80853-3 [DOI] [PubMed] [Google Scholar]
- Bugga L, Gadient R.A, Kwan K, Stewart C.L, Patterson P.H. Analysis of neuronal and glial phenotypes in brains of mice deficient in leukemia inhibitory factor. J. Neurobiol. 1998;36:509–524. doi: 10.1002/(sici)1097-4695(19980915)36:4<509::aid-neu5>3.0.co;2-#. doi:10.1002/(SICI)1097-4695(19980915)36:4<509::AID-NEU5>3.0.CO;2-# [DOI] [PubMed] [Google Scholar]
- Bunge R. Glial cells and the central myelin sheath. Physiol. Rev. 1968;48:197–251. doi: 10.1152/physrev.1968.48.1.197. [DOI] [PubMed] [Google Scholar]
- Butt A.M, Ibrahim M, Berry M. The relationship between developing oligodendrocyte units and maturing axons during myelinogenesis in the anterior medullary velum of neonatal rats. J. Neurocytol. 1997;27:327–338. doi: 10.1023/a:1018556702353. doi:10.1023/A:1018556702353 [DOI] [PubMed] [Google Scholar]
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45:41–53. doi: 10.1016/j.neuron.2004.12.028. doi:10.1016/j.neuron.2004.12.028 [DOI] [PubMed] [Google Scholar]
- Cammer W. Glutamine synthetase in the central nervous system is not confined to astrocytes. J. Neuroimmunol. 1990;26:173–178. doi: 10.1016/0165-5728(90)90088-5. doi:10.1016/0165-5728(90)90088-5 [DOI] [PubMed] [Google Scholar]
- Carson M.J, Behringer R.R, Brinster R.L, McMorris F.A. Insulin-like growth factor I increases brain growth and central nervous system myelination in transgenic mice. Neuron. 1993;10:729–740. doi: 10.1016/0896-6273(93)90173-o. doi:10.1016/0896-6273(93)90173-O [DOI] [PubMed] [Google Scholar]
- Chandran S, Kato H, Gerreli D, Compston A, Svendsen C.N, Allen N.D. FGF-dependent generation of oligodendrocytes by a hedgehog-independent pathway. Development. 2003;130:6599–6609. doi: 10.1242/dev.00871. doi:10.1242/dev.00871 [DOI] [PubMed] [Google Scholar]
- Choi B.H, Lapham L.W. Radial glia in the human fetal cerebrum: a combined Golgi, immunofluorescent and electron microscopic study. Brain Res. 1978;148:295–311. doi: 10.1016/0006-8993(78)90721-7. doi:10.1016/0006-8993(78)90721-7 [DOI] [PubMed] [Google Scholar]
- Choi B.H, Kim R.C, Lapham L.W. Do radial glia give rise to both astroglial and oligodendroglial cells? Dev. Brain Res. 1983;8:119–130. doi:10.1016/0165-3806(83)90163-3 [Google Scholar]
- Culican S.M, Baumrind N.L, Yamamoto M, Pearlman A.L. Cortical radial glia: identification in tissue culture and evidence for their transformation into astrocytes. J. Neurosci. 1990;10:684–692. doi: 10.1523/JNEUROSCI.10-02-00684.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl D, Rueger D.C, Bignami A, Weber K, Osborn M. Vimentin, the 57 000 molecular weight protein of fibroblast filaments, is the major cytoskeletal component in immature glia. Eur. J. Cell Biol. 1981;24:191–196. [PubMed] [Google Scholar]
- Darnell J.E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. doi:10.1126/science.277.5332.1630 [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim D.A, Garcia-Verdugo J.M, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. doi:10.1016/S0092-8674(00)80783-7 [DOI] [PubMed] [Google Scholar]
- Fogarty, M. H. (2005). Fate-mapping the neural tube by Cre-loxP transgenesis. Ph.D. thesis, University of London.
- Fogarty M, Richardson W.D, Kessaris N. A subset of oligodendrocytes generated from radial glia in the dorsal spinal cord. Development. 2005;132:1951–1959. doi: 10.1242/dev.01777. doi:10.1242/dev.01777 [DOI] [PubMed] [Google Scholar]
- Fulton B.P, Burne J.F, Raff M.C. Visualization of O-2A progenitor cells in developing and adult rat optic nerve by quisqualate-stimulated cobalt uptake. J. Neurosci. 1992;12:4816–4833. doi: 10.1523/JNEUROSCI.12-12-04816.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay L, Lowell S, Rubin L.L, Anderson D.J. Deregulation of dorsoventral patterning by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron. 2003;40:485–499. doi: 10.1016/s0896-6273(03)00637-8. doi:10.1016/S0896-6273(03)00637-8 [DOI] [PubMed] [Google Scholar]
- Gabbott P.L, Stewart M.G. Distribution of neurons and glia in the visual cortex (area 17) of the adult albino rat: a quantitative description. Neuroscience. 1987;21:833–845. doi: 10.1016/0306-4522(87)90040-6. doi:10.1016/0306-4522(87)90040-6 [DOI] [PubMed] [Google Scholar]
- Ge W, Martinowich K, Wu X, He F, Miyamoto A, Fan G, Weinmaster G, Sun Y.E. Notch signaling promotes astrogliogenesis via direct CSL-mediated glial gene activation. J. Neurosci. Res. 2002;69:848–860. doi: 10.1002/jnr.10364. doi:10.1002/jnr.10364 [DOI] [PubMed] [Google Scholar]
- Genoud S, Lappe-Siefke C, Goebbels S, Radtke F, Aguet M, Scherer S.S, Suter U, Nave K.A, Mantei N. Notch1 control of oligodendrocyte differentiation in the spinal cord. J. Cell Biol. 2002;158:709–718. doi: 10.1083/jcb.200202002. doi:10.1083/jcb.200202002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givogri M.I, Costa R.M, Schonmann V, Silva A.J, Campagnoni A.T, Bongarzone E.R. Central nervous system myelination in mice with deficient expression of Notch1 receptor. J. Neurosci. Res. 2002;67:309–320. doi: 10.1002/jnr.10128. doi:10.1002/jnr.10128 [DOI] [PubMed] [Google Scholar]
- Götz M, Hartfuss E, Malatesta P. Radial glial cells as neuronal precursors: a new perspective on the correlation of morphology and lineage restriction in the developing cerebral cortex of mice. Brain Res. Bull. 2002;57:777–788. doi: 10.1016/s0361-9230(01)00777-8. [DOI] [PubMed] [Google Scholar]
- Grandbarbe L, Bouissac J, Rand M, de Hrabe A.M, Artavanis-Tsakonas S, Mohier E. Delta-Notch signaling controls the generation of neurons/glia from neural stem cells in a stepwise process. Development. 2003;130:1391–1402. doi: 10.1242/dev.00374. doi:10.1242/dev.00374 [DOI] [PubMed] [Google Scholar]
- Gross R.E, Mehler M.F, Mabie P.C, Zang Z, Santschi L, Kessler J.A. Bone morphogenetic proteins promote astroglial lineage commitment by mammalian subventricular zone progenitor cells. Neuron. 1996;17:595–606. doi: 10.1016/s0896-6273(00)80193-2. doi:10.1016/S0896-6273(00)80193-2 [DOI] [PubMed] [Google Scholar]
- Hachem S, Aguirre A, Vives V, Marks A, Gallo V, Legraverend C. Spatial and temporal expression of S100B in cells of oligodendrocyte lineage. Glia. 2005;51:81–97. doi: 10.1002/glia.20184. doi:10.1002/glia.20184 [DOI] [PubMed] [Google Scholar]
- Hall A.K, Miller R.H. Emerging roles for bone morphogenetic proteins in central nervous system glial biology. J. Neurosci. Res. 2004;76:1–8. doi: 10.1002/jnr.20019. doi:10.1002/jnr.20019 [DOI] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Dev. Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. doi:10.1006/dbio.2000.9962 [DOI] [PubMed] [Google Scholar]
- He F, et al. A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat. Neurosci. 2005;8:616–625. doi: 10.1038/nn1440. doi:10.1038/nn1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Goldman J.E. Gliogenesis in the rat spinal cord: Evidence for origin of astrocytes and oligodendrocytes from radial precursors. J. Neurosci. Res. 1988;21:155–167. doi: 10.1002/jnr.490210208. doi:10.1002/jnr.490210208 [DOI] [PubMed] [Google Scholar]
- Hojvat S, Emanuele N, Baker G, Connick E, Kirsteins L, Lawrence A.M. Growth hormone (GH), thyroid-stimulating hormone (TSH), and luteinizing hormone (LH)-like peptides in the rodent brain: non-parallel ontogenetic development with pituitary counterparts. Brain Res. 1982;256:427–434. doi: 10.1016/0165-3806(82)90186-9. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Aimone J.B, Kaspar B.K, Kuwabara T, Nakashima K, Gage F.H. IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J. Cell Biol. 2004;164:111–122. doi: 10.1083/jcb.200308101. doi:10.1083/jcb.200308101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P.W, McMahon A.P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. doi:10.1101/gad.938601 [DOI] [PubMed] [Google Scholar]
- Jacob J, Briscoe J. Gli proteins and the control of spinal-cord patterning. EMBO Rep. 2003;4:761–765. doi: 10.1038/sj.embor.embor896. doi:10.1038/sj.embor.embor896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R, Wells N.J, Hunter T. TGF-beta-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johe K.K, Hazel T.G, Muller T, Dugich-Djordjevic M.M, McKay R.D. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 1996;10:3129–3140. doi: 10.1101/gad.10.24.3129. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Jamen F, Rubin L, Richardson W.D. Cooperation between sonic hedgehog and fibroblast growth factor/MAPK signalling pathways in neocortical precursors. Development. 2004;131:1289–1298. doi: 10.1242/dev.01027. doi:10.1242/dev.01027 [DOI] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson W.D. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat. Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. doi:10.1038/nn1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblar S.A, Turnley A.M, Classon B.J, Reid K.L, Ware C.B, Cheema S.S, Murphy M, Bartlett P.F. Neural precursor differentiation into astrocytes requires signaling through the leukemia inhibitory factor receptor. Proc. Natl Acad. Sci. USA. 1998;95:3178–3181. doi: 10.1073/pnas.95.6.3178. doi:10.1073/pnas.95.6.3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A.R, Gotz M. Radial glia diversity: a matter of cell fate. Glia. 2003;43:37–43. doi: 10.1002/glia.10250. doi:10.1002/glia.10250 [DOI] [PubMed] [Google Scholar]
- Kuffler S.W, Nicholls J.G, Orkland R.K. Physiological properties of glial cells in the central nervous system of amphibia. J. Neurophysiol. 1966;29:768–787. doi: 10.1152/jn.1966.29.4.768. [DOI] [PubMed] [Google Scholar]
- Le Bras B, Chatzopoulou E, Heydon K, Martinez S, Ikenaka K, Prestoz L, Spassky N, Zalc B, Thomas J.L. Oligodendrocyte development in the embryonic brain: the contribution of the plp lineage. Int. J. Dev. Biol. 2005;49:209–220. doi: 10.1387/ijdb.041963bl. doi:10.1387/ijdb.041963bl [DOI] [PubMed] [Google Scholar]
- Lee K.J, Mendelsohn M, Jessell T.M. Neuronal patterning by BMPs: a requirement for GDF7 in the generation of a discrete class of commissural interneurons in the mouse spinal cord. Genes Dev. 1998;12:3394–3407. doi: 10.1101/gad.12.21.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K, Lee B, Ruiz E.C, Pfaff S.L. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. doi:10.1101/gad.1257105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levison S.W, Goldman J.E. Both oligodendrocytes and astrocytes develop from progenitors in the subventricular zone of postnatal rat forebrain. Neuron. 1993;10:201–212. doi: 10.1016/0896-6273(93)90311-e. doi:10.1016/0896-6273(93)90311-E [DOI] [PubMed] [Google Scholar]
- Levison S.W, Goldman J.E. Multipotential and lineage restricted precursors coexist in the mammalian perinatal subventricular zone. J. Neurosci. Res. 1997;48:83–94. doi:10.1002/(SICI)1097-4547(19970415)48:2<83::AID-JNR1>3.0.CO;2-8 [PubMed] [Google Scholar]
- Levitt P, Cooper M.L, Rakic P. Coexistence of neuronal and glial precursor cells in the cerebral ventricular zone of the fetal monkey: an ultrastructural immunoperoxidase analysis. J. Neurosci. 1981;1:27–39. doi: 10.1523/JNEUROSCI.01-01-00027.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.E, Darnell J.E., Jr Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. doi:10.1038/nrm909 [DOI] [PubMed] [Google Scholar]
- Liem K.F, Jr, Tremml G, Jessell T.M. A role for the roof plate and its resident TGFbeta-related proteins in neuronal patterning in the dorsal spinal cord. Cell. 1997;91:127–138. doi: 10.1016/s0092-8674(01)80015-5. doi:10.1016/S0092-8674(01)80015-5 [DOI] [PubMed] [Google Scholar]
- Lillien L.E, Sendtner M, Raff M.C. Extracellular matrix-associated molecules collaborate with ciliary neurotrophic factor to induce type-2 astrocyte development. J. Cell Biol. 1990;111:635–644. doi: 10.1083/jcb.111.2.635. doi:10.1083/jcb.111.2.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y, Chiang C. Specification of ventral neuron types is mediated by an antagonistic interaction between Shh and Gli3. Nat. Neurosci. 2000;3:979–985. doi: 10.1038/79916. doi:10.1038/79916 [DOI] [PubMed] [Google Scholar]
- Liu R, Cai J, Hu X, Tan M, Qi Y, German M, Rubenstein J, Sander M, Qiu M. Region-specific and stage-dependent regulation of Olig gene expression and oligodendrogenesis by Nkx6.1 homeodomain transcription factor. Development. 2003;130:6221–6231. doi: 10.1242/dev.00868. doi:10.1242/dev.00868 [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. doi:10.1038/nrn1847 [DOI] [PubMed] [Google Scholar]
- Lu Q.R, Yuk D, Alberta J.A, Zhu Z, Pawlitzky I, Chan J, McMahon A.P, Stiles C.D, Rowitch D.H. Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. doi:10.1016/S0896-6273(00)80897-1 [DOI] [PubMed] [Google Scholar]
- Lu Q.R, Sun T, Zhu Z, Ma N, Garcia M, Stiles C.D, Rowitch D.H. Common developmental requirement for oligodendrocyte lineage gene (Olig) function indicates a motor neuron/oligodendrocyte lineage connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. doi:10.1016/S0092-8674(02)00678-5 [DOI] [PubMed] [Google Scholar]
- Ludwin S.K, Kosek J.C, Eng L.F. The topographical distribution of S-100 and GFA proteins in the adult rat brain: an immunohistochemical study using horseradish peroxidase-labelled antibodies. J. Comp. Neurol. 1976;165:197–207. doi: 10.1002/cne.901650206. doi:10.1002/cne.901650206 [DOI] [PubMed] [Google Scholar]
- Luskin M.B, McDermott K. Divergent lineages for oligodendrocytes and astrocytes originating in the neonatal forebrain subventricular zone. Glia. 1994;11:211–226. doi: 10.1002/glia.440110302. doi:10.1002/glia.440110302 [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hack M.A, Hartfuss E, Kettenmann H, Klinkert W, Kirchhoff F, Gotz M. Neuronal or glial progeny: regional differences in radial glia fate. Neuron. 2003;37:751–764. doi: 10.1016/s0896-6273(03)00116-8. doi:10.1016/S0896-6273(03)00116-8 [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- Matthias K, Kirchhoff F, Seifert G, Huttmann K, Matyash M, Kettenmann H, Steinhauser C. Segregated expression of AMPA-type glutamate receptors and glutamate transporters defines distinct astrocyte populations in the mouse hippocampus. J. Neurosci. 2003;23:1750–1758. doi: 10.1523/JNEUROSCI.23-05-01750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason S.G, McMahon A.P. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- Mehler M.F, Mabie P.C, Zhu G, Gokhan S, Kessler J.A. Developmental changes in progenitor cell responsiveness to bone morphogenetic proteins differentially modulate progressive CNS lineage fate. Dev. Neurosci. 2000;22:74–85. doi: 10.1159/000017429. doi:10.1159/000017429 [DOI] [PubMed] [Google Scholar]
- Mekki-Dauriac S, Agius E, Kan P, Cochard P. Bone morphogenetic proteins negatively control oligodendrocyte precursor specification in the chick spinal cord. Development. 2002;129:5117–5130. doi: 10.1242/dev.129.22.5117. [DOI] [PubMed] [Google Scholar]
- Merkle F.T, Tramontin A.D, Garcia-Verdugo J.M, Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc. Natl Acad. Sci. USA. 2004;101:17 528–17 532. doi: 10.1073/pnas.0407893101. doi:10.1073/pnas.0407893101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Haeberle H, Barres B.A. Induction of astrocyte differentiation by endothelial cells. J. Neurosci. 2001;21:1538–1547. doi: 10.1523/JNEUROSCI.21-05-01538.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J.P, Edwards M.A, Yamamoto M, Caviness V.S., Jr Identification of radial glial cells within the developing murine central nervous system: studies based upon a new immunohistochemical marker. Brain Res. Dev. Brain Res. 1988;44:95–108. doi: 10.1016/0165-3806(88)90121-6. doi:10.1016/0165-3806(88)90121-6 [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, Nabeshima Y, Shimamura K, Nakafuku M. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757–771. doi: 10.1016/s0896-6273(01)00413-5. doi:10.1016/S0896-6273(01)00413-5 [DOI] [PubMed] [Google Scholar]
- Nakashima K, et al. Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J. Neurosci. 1999a;19:5429–5434. doi: 10.1523/JNEUROSCI.19-13-05429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by Stat3-Smad1 complex bridged by p300. Science. 1999b;284:479–482. doi: 10.1126/science.284.5413.479. doi:10.1126/science.284.5413.479 [DOI] [PubMed] [Google Scholar]
- Nery S, Wichterle H, Fishell G. Sonic hedgehog contributes to oligodendrocyte specification in the mammalian forebrain. Development. 2001;128:527–540. doi: 10.1242/dev.128.4.527. [DOI] [PubMed] [Google Scholar]
- Nieto M, Schuurmans C, Britz O, Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. doi:10.1016/S0896-6273(01)00214-8 [DOI] [PubMed] [Google Scholar]
- Noble M, Proschel C, Mayer-Proschel M. Getting a GR(i)P on oligodendrocyte development. Dev. Biol. 2004;265:33–52. doi: 10.1016/j.ydbio.2003.06.002. doi:10.1016/j.ydbio.2003.06.002 [DOI] [PubMed] [Google Scholar]
- Noctor S.C, Flint A.C, Weissman T.A, Wong W.S, Clinton B.K, Kriegstein A.R. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J. Neurosci. 2002;22:3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll E, Miller R.H. Oligodendrocyte precursors originate at the ventral ventricular zone dorsal to the ventral midline region in the embryonic rat spinal cord. Development. 1993;118:563–573. doi: 10.1242/dev.118.2.563. [DOI] [PubMed] [Google Scholar]
- Norenberg M.D, Martinez-Hernandez A. Fine structural localization of glutamine synthetase in astrocytes of rat brain. Brain Res. 1979;161:303–310. doi: 10.1016/0006-8993(79)90071-4. doi:10.1016/0006-8993(79)90071-4 [DOI] [PubMed] [Google Scholar]
- Novitch B.G, Chen A.I, Jessell T.M. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. doi:10.1016/S0896-6273(01)00407-X [DOI] [PubMed] [Google Scholar]
- Olivier C, Cobos Silleros I, Perez Villegas E.M, Spassky N, Zalc B, Martinez S, Thomas J.-L. Monofocal origin of telencephalic oligodendrocytes in the chick embryo: the entopeduncular area. Development. 2001;128:1757–1769. doi: 10.1242/dev.128.10.1757. [DOI] [PubMed] [Google Scholar]
- Orentas D.M, Miller R.H. The origin of spinal cord oligodendrocytes is dependent on local influences from the notochord. Dev. Biol. 1996;177:43–53. doi: 10.1006/dbio.1996.0143. doi:10.1006/dbio.1996.0143 [DOI] [PubMed] [Google Scholar]
- Orentas D.M, Hayes J.E, Dyer K.L, Miller R.H. Sonic hedgehog signaling is required during the appearance of spinal cord oligodendrocyte precursors. Development. 1999;126:2419–2429. doi: 10.1242/dev.126.11.2419. [DOI] [PubMed] [Google Scholar]
- Park H.C, Appel B. Delta–Notch signaling regulates oligodendrocyte specification. Development. 2003;130:3747–3755. doi: 10.1242/dev.00576. doi:10.1242/dev.00576 [DOI] [PubMed] [Google Scholar]
- Parras C.M, et al. Mash1 specifies neurons and oligodendrocytes in the postnatal brain. EMBO J. 2004;23:4495–4505. doi: 10.1038/sj.emboj.7600447. doi:10.1038/sj.emboj.7600447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras, C. M., Hunt, C. & Guillemot, F. 2005 Mash1 function in generation of oligodendrocyte precursors in the developing forebrain. Program No. 551.10 2005. Abstract viewer/itinerary planner 2005. Washington, DC: Society for Neuroscience. See http://sfn.scholarone.com/itin2005/
- Pasik P, Pasik T. Cajal, Achúcarro, Río Hortego, and the early exploration of neuroglia. In: Lazzarini R.A, Griffin J.W, Miller R.H, Lassman H, Nave K, Trapp B.D, editors. Myelin biology and disorders. Elsevier Academic Press; San Diego, CA; London, UK: 2004. pp. xxiii–xLi. [Google Scholar]
- Poncet C, Soula C, Trousse F, Kan P, Hirsinger E, Pourquié O, Duprat A.-M, Cochard P. Induction of oligodendrocyte precursors in the trunk neural tube by ventralizing signals: effects of notochord and floor plate grafts, and of sonic hedgehog. Mech. Dev. 1996;60:13–32. doi: 10.1016/s0925-4773(96)00595-3. doi:10.1016/S0925-4773(96)00595-3 [DOI] [PubMed] [Google Scholar]
- Pringle N.P, Richardson W.D. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development. 1993;117:525–533. doi: 10.1242/dev.117.2.525. [DOI] [PubMed] [Google Scholar]
- Pringle N.P, Yu W.-P, Guthrie S, Roelink H, Lumsden A, Peterson A.C, Richardson W.D. Determination of neuroepithelial cell fate: induction of the oligodendrocyte lineage by ventral midline cells and Sonic hedgehog. Dev. Biol. 1996;177:30–42. doi: 10.1006/dbio.1996.0142. doi:10.1006/dbio.1996.0142 [DOI] [PubMed] [Google Scholar]
- Pringle N.P, Guthrie S, Lumsden A, Richardson W.D. Dorsal spinal cord neuroepithelium generates astrocytes but not oligodendrocytes. Neuron. 1998;20:883–893. doi: 10.1016/s0896-6273(00)80470-5. doi:10.1016/S0896-6273(00)80470-5 [DOI] [PubMed] [Google Scholar]
- Pringle N.P, Yu W.P, Howell M, Colvin J.S, Ornitz D.M, Richardson W.D. Fgfr3 expression by astrocytes and their precursors: evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development. 2003;130:93–102. doi: 10.1242/dev.00184. doi:10.1242/dev.00184 [DOI] [PubMed] [Google Scholar]
- Qi Y, et al. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128:2723–2733. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- Qi Y, Tan M, Hui C.C, Qiu M. Gli2 is required for normal Shh signaling and oligodendrocyte development in the spinal cord. Mol. Cell. Neurosci. 2003;23:440–450. doi: 10.1016/s1044-7431(03)00067-8. doi:10.1016/S1044-7431(03)00067-8 [DOI] [PubMed] [Google Scholar]
- Raff M.C, Miller R.H, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature. 1983;303:390–396. doi: 10.1038/303390a0. doi:10.1038/303390a0 [DOI] [PubMed] [Google Scholar]
- Rajan P, McKay R.D. Multiple routes to astrocytic differentiation in the CNS. J. Neurosci. 1998;18:3620–3629. doi: 10.1523/JNEUROSCI.18-10-03620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M.S, Noble M, Mayer-Proschel M. A tripotential glial precursor cell is present in the developing spinal cord. Proc. Natl Acad. Sci. USA. 1998;95:3996–4001. doi: 10.1073/pnas.95.7.3996. doi:10.1073/pnas.95.7.3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson W.D, Pringle N.P, Yu W.-P, Hall A.C. Origins of spinal cord oligodendrocytes: possible developmental and evolutionary relationships with motor neurons. Dev. Neurosci. 1997;19:54–64. doi: 10.1159/000111186. [DOI] [PubMed] [Google Scholar]
- Richardson W.D, Kessaris N, Pringle N. Oligodendrocyte wars. Nat. Rev. Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. doi:10.1038/nrn1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Kessler J.A. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. doi:10.1242/dev.01273 [DOI] [PubMed] [Google Scholar]
- Sander M, Paydar S, Ericson J, Briscoe J, Berber E, German M, Jessell T.M, Rubenstein J.L. Ventral neural patterning by Nkx homeobox genes: Nkx6.1 controls somatic motor neuron and ventral interneuron fates. Genes Dev. 2000;14:2134–2139. doi: 10.1101/gad.820400. doi:10.1101/gad.820400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmechel D.E, Rakic P. A golgi study of radial glial cells in developing monkey telencephalon: morphogenesis and transformation into astrocytes. Anat. Embryol. (Berl) 1979;156:115–152. doi: 10.1007/BF00300010. doi:10.1007/BF00300010 [DOI] [PubMed] [Google Scholar]
- Schmitt A, Asan E, Puschel B, Kugler P. Cellular and regional distribution of the glutamate transporter GLAST in the CNS of rats: nonradioactive in situ hybridization and comparative immunocytochemistry. J. Neurosci. 1997;17:1–10. doi: 10.1523/JNEUROSCI.17-01-00001.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti C.L. Evidence for astrocyte heterogeneity: a distinct subpopulation of protoplasmic-like glial cells is detected in transgenic mice expressing Lmo1-lacZ. Glia. 2003;43:195–207. doi: 10.1002/glia.10254. doi:10.1002/glia.10254 [DOI] [PubMed] [Google Scholar]
- Shimizu T, Kagawa T, Wada T, Muroyama Y, Takada S, Ikenaka K. Wnt signaling controls the timing of oligodendrocyte development in the spinal cord. Dev. Biol. 2005;282:397–410. doi: 10.1016/j.ydbio.2005.03.020. doi:10.1016/j.ydbio.2005.03.020 [DOI] [PubMed] [Google Scholar]
- Southwood C, He C, Garbern J, Kamholz J, Arroyo E, Gow A. CNS myelin paranodes require Nkx6-2 homeoprotein transcriptional activity for normal structure. J. Neurosci. 2004;24:11 215–11 225. doi: 10.1523/JNEUROSCI.3479-04.2004. doi:10.1523/JNEUROSCI.3479-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, et al. Multiple restricted origin of oligodendrocytes. J. Neurosci. 1998;18:8331–8343. doi: 10.1523/JNEUROSCI.18-20-08331.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, Heydon K, Mangatal A, Jankovski A, Olivier C, Queraud-Lesaux F, Goujet-Zalc C, Thomas J.L, Zalc B. Sonic hedgehog-dependent emergence of oligodendrocytes in the telencephalon: evidence for a source of oligodendrocytes in the olfactory bulb that is independent of PDGFRalpha signaling. Development. 2001;128:4993–5004. doi: 10.1242/dev.128.24.4993. [DOI] [PubMed] [Google Scholar]
- Steinhauser C, Berger T, Frotscher M, Kettenmann H. Heterogeneity in the membrane current pattern of identified glial cells in the hippocampal slice. Eur. J. Neurosci. 1992;4:472–484. doi: 10.1111/j.1460-9568.1992.tb00897.x. doi:10.1111/j.1460-9568.1992.tb00897.x [DOI] [PubMed] [Google Scholar]
- Stichel C.C, Muller C.M, Zilles K. Distribution of glial fibrillary acidic protein and vimentin immunoreactivity during rat visual cortex development. J. Neurocytol. 1991;20:97–108. doi: 10.1007/BF01279614. doi:10.1007/BF01279614 [DOI] [PubMed] [Google Scholar]
- Stidworthy M.F, Genoud S, Li W.W, Leone D.P, Mantei N, Suter U, Franklin R.J. Notch1 and Jagged1 are expressed after CNS demyelination, but are not a major rate-determining factor during remyelination. Brain. 2004;127:1928–1941. doi: 10.1093/brain/awh217. doi:10.1093/brain/awh217 [DOI] [PubMed] [Google Scholar]
- Stolt C.C, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. doi:10.1101/gad.215802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt C.C, Lommes P, Sock E, Chaboissier M.C, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. doi:10.1101/gad.259003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt C.C, Lommes P, Friedrich R.P, Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131:2349–2358. doi: 10.1242/dev.01114. doi:10.1242/dev.01114 [DOI] [PubMed] [Google Scholar]
- Stolt C.C, Schmitt S, Lommes P, Sock E, Wegner M. Impact of transcription factor Sox8 on oligodendrocyte specification in the mouse embryonic spinal cord. Dev. Biol. 2005;281:309–317. doi: 10.1016/j.ydbio.2005.03.010. doi:10.1016/j.ydbio.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Sun T, Pringle N.P, Hardy A.P, Richardson W.D, Smith H.K. Pax6 inluences the time and site of origin of glial precursors in the ventral neural tube. Mol. Cell. Neurosci. 1998;12:228–239. doi: 10.1006/mcne.1998.0711. doi:10.1006/mcne.1998.0711 [DOI] [PubMed] [Google Scholar]
- Sun T, Echelard Y, Lu R, Yuk D.I, Kaing S, Stiles C.D, Rowitch D.H. Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Curr. Biol. 2001;11:1413–1420. doi: 10.1016/s0960-9822(01)00441-9. doi:10.1016/S0960-9822(01)00441-9 [DOI] [PubMed] [Google Scholar]
- Sun T, Dong H, Wu L, Kane M, Rowitch D.H, Stiles C.D. Cross-repressive interaction of the Olig2 and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex. J. Neurosci. 2003;23:9547–9556. doi: 10.1523/JNEUROSCI.23-29-09547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, Nabeshima Y. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech. Dev. 2000;99:143–148. doi: 10.1016/s0925-4773(00)00466-4. doi:10.1016/S0925-4773(00)00466-4 [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor Olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr. Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. doi:10.1016/S0960-9822(02)00926-0 [DOI] [PubMed] [Google Scholar]
- Tanigaki K, Nogaki F, Takahashi J, Tashiro K, Kurooka H, Honjo T. Notch1 and Notch3 instructively restrict bFGF-responsive multipotent neural progenitor cells to an astroglial fate. Neuron. 2001;29:45–55. doi: 10.1016/s0896-6273(01)00179-9. doi:10.1016/S0896-6273(01)00179-9 [DOI] [PubMed] [Google Scholar]
- Tekki-Kessaris N, Woodruff R, Hall A.C, Gaffield W, Kimura S, Stiles C.D, Rowitch D.H, Richardson W.D. Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development. 2001;128:2545–2554. doi: 10.1242/dev.128.13.2545. [DOI] [PubMed] [Google Scholar]
- Timsit S, Martinez S, Allinquant B, Peyron F, Puelles L, Zalc B. Oligodendrocytes originate in a restricted zone of the embryonic ventral neural tube defined by DM-20 mRNA expression. J. Neurosci. 1995;15:1012–1024. doi: 10.1523/JNEUROSCI.15-02-01012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnley A.M, Faux C.H, Rietze R.L, Coonan J.R, Bartlett P.F. Suppressor of cytokine signaling 2 regulates neuronal differentiation by inhibiting growth hormone signaling. Nat. Neurosci. 2002;5:1155–1162. doi: 10.1038/nn954. doi:10.1038/nn954 [DOI] [PubMed] [Google Scholar]
- Vallstedt A, Muhr J, Pattyn A, Pierani A, Mendelsohn M, Sander M, Jessell T.M, Ericson J. Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron. 2001;31:743–755. doi: 10.1016/s0896-6273(01)00412-3. doi:10.1016/S0896-6273(01)00412-3 [DOI] [PubMed] [Google Scholar]
- Vallstedt A, Klos J.M, Ericson J. Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron. 2005;45:55–67. doi: 10.1016/j.neuron.2004.12.026. doi:10.1016/j.neuron.2004.12.026 [DOI] [PubMed] [Google Scholar]
- Vaughn J.E, Peters A. Electron microscopy of the early postnatal development of fibrous astrocytes. Am. J. Anat. 1967;121:131–152. doi: 10.1002/aja.1001210109. doi:10.1002/aja.1001210109 [DOI] [PubMed] [Google Scholar]
- Vitellaro-Zuccarello L, Mazzetti S, Bosisio P, Monti C, De Biasi S. Distribution of Aquaporin 4 in rodent spinal cord: relationship with astrocyte markers and chondroitin sulfate proteoglycans. Glia. 2005;51:148–159. doi: 10.1002/glia.20196. doi:10.1002/glia.20196 [DOI] [PubMed] [Google Scholar]
- Voigt T. Development of glial cells in the cerebral wall of ferrets: direct tracing of their transformation from radial glia into astrocytes. J. Comp. Neurol. 1989;289:74–88. doi: 10.1002/cne.902890106. doi:10.1002/cne.902890106 [DOI] [PubMed] [Google Scholar]
- Wada T, Kagawa T, Ivanova A, Zalc B, Shirasaki R, Murakami F, Iemura S, Ueno N, Ikenaka K. Dorsal spinal cord inhibits oligodendrocyte development. Dev. Biol. 2000;227:42–55. doi: 10.1006/dbio.2000.9869. doi:10.1006/dbio.2000.9869 [DOI] [PubMed] [Google Scholar]
- Walz W. Controversy surrounding the existence of discrete functional classes of astrocytes in adult gray matter. Glia. 2000;31:95–103. doi: 10.1002/1098-1136(200008)31:2<95::aid-glia10>3.0.co;2-6. doi:10.1002/1098-1136(200008)31:2<95::AID-GLIA10>3.0.CO;2-6 [DOI] [PubMed] [Google Scholar]
- Walz W, Lang M.K. Immunocytochemical evidence for a distinct GFAP-negative subpopulation of astrocytes in the adult rat hippocampus. Neurosci. Lett. 1998;257:127–130. doi: 10.1016/s0304-3940(98)00813-1. doi:10.1016/S0304-3940(98)00813-1 [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla A.D, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres B. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- Warf B.C, Fok-Seang J, Miller R.H. Evidence for the ventral origin of oligodendrocyte precursors in the rat spinal cord. J. Neurosci. 1991;11:2477–2488. doi: 10.1523/JNEUROSCI.11-08-02477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu F.F, Gow A, Lu Q.R. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J. Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. doi:10.1523/JNEUROSCI.3034-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Carson J, D’ Ercole A.J. In vivo actions of insulin-like growth factor-I (IGF-I) on brain myelination: studies of IGF-I and IGF binding protein-1 (IGFBP-1) transgenic mice. J. Neurosci. 1995;15:7344–7356. doi: 10.1523/JNEUROSCI.15-11-07344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Li L, Richards R.G, DiAugustine R.P, D'Ercole A.J. Myelination is altered in insulin-like growth factor-I null mutant mice. J. Neurosci. 2002;22:6041–6051. doi: 10.1523/JNEUROSCI.22-14-06041.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, K., Fogarty, M., Kessaris, N. & Richardson, W. D. Submitted. Adult SVZ stem cells are phenotypically heterogeneous wih origins in both the embryonic striatum and cerebral cortex.
- Yu W.-P, Collarini E.J, Pringle N.P, Richardson W.D. Embryonic expression of myelin genes: evidence for a focal source of oligodendrocyte precursors in the ventricular zone of the neural tube. Neuron. 1994;12:1353–1362. doi: 10.1016/0896-6273(94)90450-2. doi:10.1016/0896-6273(94)90450-2 [DOI] [PubMed] [Google Scholar]
- Yung S.Y, Gokhan S, Jurcsak J, Molero A.E, Abrajano J.J, Mehler M.F. Differential modulation of BMP signaling promotes the elaboration of cerebral cortical GABAergic neurons or oligodendrocytes from a common sonic hedgehog-responsive ventral forebrain progenitor species. Proc. Natl. Acad. Sci. USA. 2002;99:16 273–16 278. doi: 10.1073/pnas.232586699. doi:10.1073/pnas.232586699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson D.J. The bHLH transcription factors Olig2 and Olig1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. doi:10.1016/S0092-8674(02)00677-3 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson D.J. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. doi:10.1016/S0896-6273(00)80898-3 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Choi G, Anderson D.J. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. doi:10.1016/S0896-6273(01)00414-7 [DOI] [PubMed] [Google Scholar]
- Zhou M, Schools G.P, Kimelberg H.K. Development of GLAST(+) astrocytes and NG2(+) glia in rat hippocampus CA1: mature astrocytes are electrophysiologically passive. J. Neurophysiol. 2006;95:134–143. doi: 10.1152/jn.00570.2005. doi:10.1152/jn.00570.2005 [DOI] [PubMed] [Google Scholar]