Abstract
The attachment and interactions of analyte receptor biomolecules at solid–liquid interfaces are critical to development of hybrid biological–synthetic sensor devices across all size regimes. We use protein engineering approaches to engineer the sensing interface of biochemically modified field effect transistor sensors (BioFET). To date, we have deposited analyte receptor proteins on FET sensing channels by direct adsorption, used self-assembled monolayers to tether receptor proteins to planar FET SiO2 sensing gates and demonstrated interface biochemical function and electrical function of the corresponding sensors. We have also used phage display to identify short peptides that recognize thermally grown SiO2. Our interest in these peptides is as affinity domains that can be inserted as translational fusions into receptor proteins (antibody fragments or other molecules) to drive oriented interaction with FET sensing surfaces. We have also identified single-chain fragment variables (scFvs, antibody fragments) that recognize an analyte of interest as potential sensor receptors. In addition, we have developed a protein engineering technology (scanning circular permutagenesis) that allows us to alter protein topography to manipulate the position of functional domains of the protein relative to the BioFET sensing surface.
Keywords: biosensors, interface engineering, surface modifications, protein engineering
1. Introduction
For many sensing architectures based on distance-dependent bio(chemical) transduction methods (e.g. field effect transistor (FET)-, fluorescence resonance energy transfer (FRET)- or bioluminescence resonance energy transfer (BRET)-based sensors), signal magnitude is directly related to the efficiency of charge or energy transfer from a donor (analyte) to an acceptor (a sensing transducer). Efficiency of electronic or optical signal transduction is directly related to the nanometre-scale distance of between 2 and 6 nm (e.g. Debye length or Förster distance) between analyte and transducer (Bergveld 1996; Schoning & Poghossian 2002, 2006; Fan et al. 2005). We are pursuing a biosensor development programme aimed at improving the efficiency of charge transfer due to analyte binding to receptor proteins, the receptor itself bound to a sensing channel surface, in a biochemically modified field effect transistor (BioFET) sensor. In this sensor, the gate metal of a metal oxide semiconductor field effect transistor (MOSFET) is replaced with a protein (e.g. antibody molecule acting as receptor interface) whose binding cognate (antigen or analyte) is detected as the result of the changes in FET electrical properties induced by charges on the bound analyte molecule (Bhushan et al. 2005; Lee et al. 2005). Detection depends on analyte charge: a receptor immobilized on the FET surface binds a charged analyte resulting in accumulation or depletion of carriers (electrons or holes) in the FET that is measured as current. FET sensors potentially combine the specificity of biological affinity reagents with micro/nanoelectronics for label-free protein detection (i.e. direct electronic readout of charged biomolecular interactions; Gabig-Ciminska 2006).
Unfortunately, BioFETs have proven problematic for direct sensing of protein binding. For BioFETs and ImmunoFETs (BioFETs using antibody as receptor), the distance between the FET sensing surface and charges on analyte bound to the receptor has been considered to be a fundamental limitation. This is attributed predominantly to the size of intact antibody receptors (10–15 nm) relative to the Debye length (the distance over which a shielding electrical double layer of buffer ions would form between bound analyte and the sensing surface, 1–3 nm in most biological buffers, see Bergveld 1996; Schoning & Poghossian 2002, 2006; Fan et al. 2005). Efficiency of charge transfer can also be negatively affected if analyte binding to receptor on the sensing surface experiences steric interference. Both Debye and steric limitations might be addressed by protein engineering manipulations that modulate the relative orientation of protein analytes and their charges to the interface, though the approach has never been systematically addressed.
In the simplest approach, we have chosen to functionalize sensing surfaces with the well-studied model protein streptavidin by: (i) direct adsorption to the SiO2 surface and (ii) linking the protein and surface through a polymeric linker. We characterized these surfaces biochemically, electrically and by atomic force microscopy (AFM). Direct adsorption has the virtue of depositing receptor protein without distance intervening between the sensing channel and protein, and should offer the minimal distance between the analyte-binding site and channel that is possible for native protein (Lee et al. 2005). Adsorption depends on a variety of non-covalent interactions with the surface (Van Tassel et al. 1998), but these interactions may not be satisfactorily robust for all applications. Keeping this in mind, we have also used a combination of covalent linkage and non-covalent (but strong, Moy et al. 1994) streptavidin–biotin interaction to tether receptor streptavidin to SiO2 surfaces. Comparison of the electrical properties of FETs with these two types of interface reveals the impact of interfacial design on device properties, and reveals each approach to be unsatisfactory from either a device robustness or sensor sensitivity standpoint. We therefore are exploring two protein engineering approaches to systematically modify receptors of sensing interfaces to minimize the distance between FET sensing channels and receptor-bound analytes to maximize the efficiency of charge transduction to the FET sensing surfaces.
2. Experimental details
2.1 Microfabrication
FET devices and thermally grown oxide surfaces were prepared as for the gate of an n-channel metal oxide semiconductor FET (MOSFET), as previously described (Bhushan et al. 2005; Eteshola et al. 2005; Lee et al. 2005).
2.2 Surface modification/functionalization and characterization of grown silicon dioxide films
SiO2 surfaces were functionalized by direct adsorption of streptavidin or by covalent attachment of APTES and sulpho-NHS biotin as a SAM (self-assembled monolayer, described in Bhushan et al. (2005) and Lee et al. (2005)) to which streptavidin was subsequently bound. Surfaces were biochemically characterized using enzyme-linked immunosorbent assay to demonstrate the specific affinity properties. Morphology of deposited protein and polymer was studied using AFM phase angle imaging in tapping mode (Bhushan et al. 2005; Lee et al. 2005).
2.3 Selection of peptides and scFv fragments that bind thermally grown silicon dioxide films and a chemokine analyte, respectively
A combinatorial library of linear random peptides containing 12 amino acids and libraries of single-chain fragment variable (scFv, Tomlinson I and J) fused to the pIII minor coat protein of M13 bacteriophage were obtained, respectively, from New England Biolabs (Beverly, MA), and the Medical Research Council, Cambridge, England. The peptide and naive scFv libraries were subjected to affinity selection on the thermally oxidized silicon wafers or a biotinylated chemokine (protein) analyte, respectively. Affinity selection methods were a combination of those recommended by the supplier and methods developed in our laboratory (Eteshola et al. 2005, 2006).
3. Results and discussion
We have tried two different approaches to establish an interface for sensing charged analyte binding by a BioFET sensor: direct adsorption of streptavidin to the channel and binding of streptavidin to a biotinylated SAM on the channel. Direct adsorption is simple and affords the closest possible contact between streptavidin and the sensing surface.
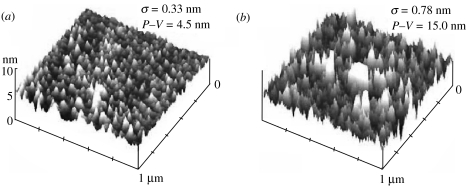
AFM images derived in PBS buffer (figure 1a) show that the peak-to-valley (P–V) values for flat SiO2 substrates with directly adsorbed streptavidin are a modest 4–5 nm. This value roughly corresponds to the expected diameter of streptavidin tetramers (Hendrickson et al. 1989) and suggests that streptavidin deposited on flat SiO2 surface may have been homogenously one molecule thick. However, streptavidin bound to the biotinylated surface (seen in figure 1b) is rougher than streptavidin directly deposited on the SiO2 surface (roughness of 0.78 versus 0.33 nm), as might be expected from a multilayer structure. In addition, the height of the streptavidin bound to biotinylated polymer (P–V=15.0 nm) is higher than on flat surface (P–V=4.5 nm).
Figure 1.
AFM images of streptavidin-coated SiO2 surfaces. (a) Streptavidin directly deposited on the surface, and (b) streptavidin deposited through interaction with a biotinylated SAM (accepted JRSI-2007-1033).
Direct adsorption allows the deposition of the streptavidin (and hence biotinylated analytes) at the closest distance to the sensing surface possible for the native protein, but we found those interfaces to be very fragile. The adhesion of streptavidin to the SiO2 surface is minimal and can be disrupted by solution turbulence (Bhushan et al. 2005; Lee et al. 2005). Therefore, directly deposited streptavidin interfaces on SiO2 are unlikely to be sufficiently robust for many FET sensor applications of interest to us.
Based on the strength of the biotin–streptavidin bond (Bhushan et al. 2005; Lee et al. 2005), the biotinylated SAM scheme we used provides the strongest possible non-covalent attachment of streptavidin to the SiO2 surface. The draw backs are: (i) the need for surface derivatization of the substrate and (ii) that the protein is recessed from the SiO2 sensing surface by the length of the polymeric constituents of APTES and sulpho-NHS biotin used. This is evident in P–V values for streptavidin on biotinylated surfaces (15 nm) as opposed to 4–5 nm for streptavidin directly deposited on SiO2. In some applications, a few nanometre distance between deposited protein and the substrate is not critical, but for the receptor interface for an ImmunoFET sensor, proximity between bound analyte charges and sensing surfaces is a key determinant of sensitivity. In most biological buffers, a signal-attenuating shielding layer of ions forms between bound charged analyte and FET sensing surfaces over distances (Debye lengths) of only a few nanometres (Bergveld 1996; Schoning & Poghossian 2002).
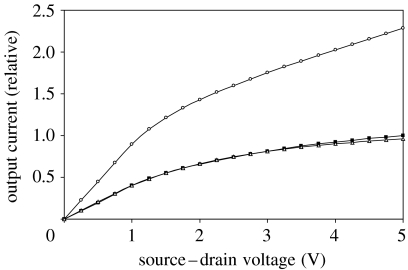
Thus, while the interface for chemically conjugated streptavidin is substantially more robust than those comprising directly deposited streptavidin, the increased distance between analyte and sensing channel diminished the sensitivity of the BioFET (see measured FET I–V characteristics resulting from direct streptavidin deposition at the interface versus streptavidin bound through a biotinylated SAM; figure 2). This technical challenge motivated exploration of several molecular biology approaches to engineer receptors for the sensing channel interface. If, as theory suggests, sensitivity of intact protein-based FET sensors can be improved by greater proximity between the sensing surface and the analyte, sensor sensitivity might be enhanced by judicious engineering of the receptor protein.
Figure 2.
Electrical response of insulated BioFET devices in phosphate-buffered saline (PBS, a biological buffer). Devices' responses to streptavidin are shown. In one device, receptor protein streptavidin was directly adsorbed to the sensing channel (open circles), and in another, streptavidin was attached to the channel by interaction with a biotinylated SAM on the surface (open triangles). The electrical properties of these devices are changed substantially by streptavidin directly bound to the sensing channel. Compare that device (open circles) with the device with no SAM (filled squares). Note that the electrical characteristics of a device with no SAM (and receiving no streptavidin, filled squares) are virtually identical to the characteristics of a device binding streptavidin via a biotinylated SAM (open triangles), indicative of the low sensitivity of the device when receptor and analyte are bound using this biotinylated SAM (accepted JRSI-2007-1033).
Multiple protein engineering approaches might be used to minimize the distance between charges of receptor-bound analytes and sensing surfaces. We illustrate two here. Firstly, we isolated affinity peptides recognizing thermally grown silica (Eteshola et al. 2005) that can be inserted at various positions within receptor sequences as protein fusions to derive receptors that bind silica in specific orientation determined by the position of peptide insertion. Secondly, we developed a high-throughput method to modify the topography of receptor proteins (scanning circular permutagenesis; Eteshola et al. 2006). When used in conjunction with a chemoselective conjugation method to deploy proteins on surfaces, different permuted variants of a single receptor hold analyte at different distances from the sensing surface.
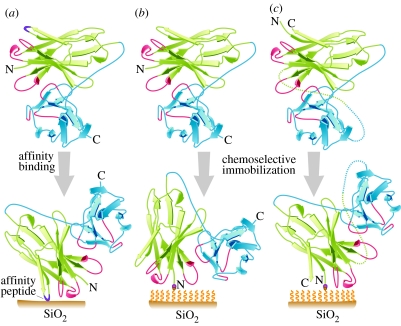
Dodecapeptides which specifically recognize thermally grown SiO2 layers were isolated from a peptide phage display library (Eteshola et al. 2005) and can be used to drive oriented interaction of receptor with the sensing channel. The peptide consensus we identified (e.g. HXXHXH) is structurally and functionally distinct from those previously reported to catalyse the precipitation of silica from silicilic acid (Naik et al. 2002). We have also affinity selected a series of scFv clones that bind a chemokine protein (an analyte of interest) from naive antibody libraries (results not shown). Consider the scFv (receptor) analyte-binding domain (antigen-combining site) as a site on the scFv surface, and further that interaction with the sensing channel SiO2 occurs between the inserted affinity peptides of the scFv and the sensing surface. Different positions of the inserted peptide relative to the antigen-combining site would ultimately result in differing proximities between bound analyte and sensing surface (figure 3a).
Figure 3.
Ribbon diagrams of scFvs (VH–VL configuration) to be deposited on a SiO2 surface. VH is in green, VL is in blue. Complementarity determining regions (CDRs) of VH and VL are in red. SiO2 surface is represented by a brown bar. A polymeric SAM on a SiO2 surface is represented by wavy yellow lines. N- and C-ends of scFvs are indicated. Chemoselective ligation between N-ends of scFvs and SAM is indicated. (a) Affinity peptide-scFv: SiO2 affinity peptide selected from a display library (Eteshola et al. 2005) is inserted into the scFv antibody fragment (purple line). The affinity peptide binds the SiO2 surface, effectively orienting the scFv and determining the proximity of the CDRs to the SiO2 surface. (b) Parent scFv: chemoselective conjugation of a modified scFv (with an N-terminal aldehyde; Lee et al. 2004) to oxyamine residues on the SAM. Note position of VH CDRs. (c) CP scFv: chemoselective conjugation of a circularly permuted (CP; Eteshola et al. 2006), but otherwise comparable, scFv. In (b) and (c), note that chemoselective conjugation produces a consistent orientation of scFvs, and that, relative to the parent scFv, CP alters the proximity of the CDRs to the SiO2 surface (accepted JRSI-2007-1033).
In a second approach, we apply a technology we developed (scanning circular permutation or SCP of proteins; Eteshola et al. 2006) in combination with chemoselective conjugation. The method allows alteration of protein topography so as to manipulate the position of the ends of the protein and any point on the protein surface (such as the antigen-combining site). In brief, circularly permuted (CP) proteins are made by changing the order of primary sequence amino acids of a parent protein by recombinant DNA methods to create topological variants of proteins. CP proteins have the same amino acid content as the parent proteins, but the protein primary sequence is reordered. The amino and carboxyl ends of the parent protein are joined covalently by a peptide linker and new N- and C-ends are introduced in alternate sites within the protein sequence. The result is alteration of protein primary structures, while leaving the secondary and tertiary structures intact (Thornton & Sibanda 1983; Paavola et al. 2006). CP variants often have folded structures and biological activities comparable with that of their parental proteins (Schwartz et al. 2004).
A hypothetical scFv fragment and a CP derivative of the scFv are shown in figure 3b,c. The spatial relationship between the antigen-combining site (red) and the N-end of the scFv is altered by CP. This is exploited when the receptor (scFv) is linked to the surface by chemoselective conjugation, which involves reaction between a mutually exclusively reactive electrophile–nucleophile pair and results in a consistent orientation of receptor molecules. The nucleophile is provided by oxyamine residues applied to the sensing channel as part of a SAM, and the electrophile is provided in the receptor molecules. This electrophile is produced by incorporation of an N-terminal serine in the scFv (and CP scFv) by genetic engineering. The N-terminal serine (but not serines elsewhere in the sequence) is oxidized with periodate to produce an aldehyde functionality (Lee et al. 2004). Conjugation produces oxime linkage between protein N-terminus and the SAM on the sensing channel.
As described above for insertion of SiO2 affinity peptides at various points on the receptor surface, the combination of CP with chemoselective conjugation allows functional domains of CP proteins (sites of analyte binding) to be moved into and out of proximity with a BioFET sensing channel, depending on which CP variant is used. CP can also relieve steric interferences (figure 3). Both of these effects, CP and chemoselective conjugation, effectively adjust the distance between analyte charges and sensing surface, and therefore should be expected to influence sensor sensitivity (Bergveld 1996; Schoning & Poghossian 2002, 2006; Fan et al. 2005).
4. Summary
We have demonstrated that streptavidin binding by direct adsorption to cleaned thermal SiO2 surfaces is so weak as to make the approach unsuitable for most BioFET applications of interest to us. Adhesion of streptavidin to surfaces via interaction with a biotinylated SAM is substantially stronger and such interfaces are more resistant to environmental insult than their directly adsorbed counterparts. However, the most critical limitation of the method is the distance of the streptavidin from the sensing channel. The 15 nm height of the features we observed would be expected to extend well above the shielding counter-ion layer that forms at physiological salt concentrations. To mitigate this critical limitation, we pursue protein engineering approaches to minimize the distance between analyte charges and FET sensing surfaces for more efficient charge transduction. We hypothesize that protein engineering strategies may lead to more sensitive FET protein detection than has yet been described. In addition to those proposed here, still other protein engineering approaches, some of which we may have not yet considered, might also achieve improved sensor sensitivity. In light of the long-standing dismissal of the feasibility of ImmunoFET and related technologies (Bergveld 1996; Schoning & Poghossian 2002), we believe this perspective warrants discussion and consideration by the sensor community.
Acknowledgments
This work was supported by the NIH/NIBIB Career Development grant number EB004960 and Program for International and Homeland Security (programme no. 14525).
References
- Bergveld P. The future of biosensors. Sensors Actuators A. 1996;56:65–73. doi: 10.1016/0924-4247(96)01275-7. [DOI] [Google Scholar]
- Bhushan B, Tokachichu D.R, Keener M.T, Lee S.C. Morphology and adhesion of biomolecules on silicon based surfaces. Acta Biomaterialia. 2005;1:327–341. doi: 10.1016/j.actbio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Eteshola E, Brillson L.J, Lee S.C. Selection and characteristics of peptides that bind thermally grown silicon dioxide films. Biomol. Eng. 2005;22:201–204. doi: 10.1016/j.bioeng.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Eteshola E, et al. Screening libraries of circularly permuted proteins by phage display to manipulate protein topographies. Proc. IMechE, Part N: J. Nanoeng. Nanosyst. 2006;219:45–55. doi: 10.1243/17403499JNN38. [DOI] [Google Scholar]
- Fan C, Plaxo K.W, Heeger A.J. Biosensors based on binding-modulated donor–acceptor distances. Trends Biotechnol. 2005;23:186–192. doi: 10.1016/j.tibtech.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Gabig-Ciminska M. Developing nucleic acid-based electrical detection systems. Microb. Cell Fact. 2006;5:9. doi: 10.1186/1475-2859-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W.A, Pahler A, Smith J.L, Satow Y, Merritt E.A, Phizackerley R.P. Crystal structure of core streptavidin determined from multiwavelength anomalous diffraction of synchrotron radiation. Proc. Natl Acad. Sci. USA. 1989;86:2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C, et al. Biochemical and immunological properties of cytokines conjugated to dendritic polymers. Biomed. Microdevices Biomems Biomed. Nanotechnol. 2004;6:191–201. doi: 10.1023/B:BMMD.0000042048.18186.ff. [DOI] [PubMed] [Google Scholar]
- Lee S.C, Keener M.T, Tokachichu D.R, Bhushan B, Barnes P.D, Cipriany B.R, Gao M, Brillson L.J. Protein binding on thermally grown silicon dioxide. J. Vac. Sci. Technol. B. 2005;23:1856–1865. doi: 10.1116/1.2006127. [DOI] [Google Scholar]
- Moy V.T, Florin E.L, Gaub H.E. Adhesion forces between individual ligand–receptor pairs. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- Naik R.R, Brott L.L, Clarson S.J, Stone M.O. Silica-precipitating peptides isolated from a combinatorial phage display peptide library. J. Nanosci. Nanotechnol. 2002;2:95–100. doi: 10.1166/jnn.2002.074. [DOI] [PubMed] [Google Scholar]
- Paavola C.D, Chan S.L, Li Y, Mazazarella K.M, McMillan R.A, Trent J.D. A versatile platform for nanotechnology based on circular permutation of a chaperonin protein. Nanotechnology. 2006;17:1171–1176. doi: 10.1088/0957-4484/17/5/001. [DOI] [Google Scholar]
- Schoning M.J, Poghossian A. Recent advances in biologically sensitive field-effect transistors (BioFETs) Analyst. 2002;127:1137–1151. doi: 10.1039/b204444g. [DOI] [PubMed] [Google Scholar]
- Schoning M.J, Poghossian A. Bio FEDs (field-effect devices): state-of-the-art and new directions. Electroanalysis. 2006;18:1893–1900. doi: 10.1002/elan.200603609. [DOI] [Google Scholar]
- Schwartz T.U, Walczak R, Blobel G. Circular permutation as a tool to reduce surface entropy triggers crystallization of the signal recognition particle receptor beta subunit. Protein Sci. 2004;13:2814–2818. doi: 10.1110/ps.04917504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton J.M, Sibanda B.L. Amino and carboxy-terminal regions in globular proteins. J. Mol. Biol. 1983;167:443–460. doi: 10.1016/S0022-2836(83)80344-1. [DOI] [PubMed] [Google Scholar]
- Van Tassel P.R, Guemouri L, Ramsden J.J, Tarjus G, Viot P, Talbot J. A particle-level model of irreversible protein adsorption with a postadsorption transition. J. Colloid Interface Sci. 1998;207:317–323. doi: 10.1006/jcis.1998.5781. [DOI] [PubMed] [Google Scholar]